Abstract
Enterococcus faecium strain 10/96A (VanD4) was the first vancomycin-resistant enterococcus (VRE) isolated in Brazil. Subsequent Brazilian VRE strains have all had the VanA phenotype. Multilocus sequence typing showed that strain 10/96A was isolated sporadically, has a unique sequence type (ST 281), and was not the progenitor of the VRE strains isolated from hospital outbreaks in Brazil.
Vancomycin-resistant enterococci (VRE) have been isolated since the late 1980s in Europe (17) and in the United States (15). In Brazil, the first VRE strain was isolated in 1996, when a vancomycin-resistant strain of Enterococcus faecium was isolated from the blood of a 9-year-old girl with aplastic anemia (7). The VanD-like phenotype of this isolate was later classified as VanD4 (6). Subsequent to this initial case, there have been several reports of VRE strains in Brazil, including reports of intra- and interhospital spread of strains (3, 5, 13, 18). However, most of these subsequent strains have had the VanA phenotype; no other VanD strains have been reported in Brazil, although they have been identified, albeit rarely, elsewhere (1, 2, 14).
Acquired VanD-type vancomycin resistance is due to the constitutive production of peptidoglycan precursors ending in D-alanine-D-lactate, except for E. faecium A902 (VanD2), which is reported to have inducible resistance (9, 11). The organization of the vanD operon, which is located on the chromosome of the strains thus far examined, is similar to that of vanA and vanB, except for the vanX and vanW genes (9). In vitro transfer of vanD operons by conjugation to other enterococci strains has not been demonstrated. VanD strains have negligible D,D-dipeptidase (VanX) activity. This is essential in other vancomycin-resistant phenotypes to eliminate the intracellular pool of peptidoglycan precursors terminating in D-alanyl-D-alanine (8). However, D,D-dipeptidase activity appears to be unnecessary in most VanD strains; their native Ddl ligases—required to generate the D-alanyl-D-alanine precursor pool—are inactive, owing to mutations in the chromosomal ddl gene (9). Most VanD enterococci strains have mutations in the VanSD sensor or VanRD regulator proteins, and their vanD clusters are expressed constitutively (9), which eliminates the need for vancomycin induction for synthesis of critical cell wall precursors and circumvents vancomycin dependence.
The vanD gene clusters and ddl genes have been examined in several VanD phenotype strains, and several mutations have been described (5). However, it remains unclear how or when the vanD gene cluster was acquired. We have used multilocus sequence typing (MLST) to further analyze the original vancomycin-resistant VanD4 isolate from Brazil, E. faecium strain 10/96A, and to compare it with VanA strains isolated in Brazil (4) and elsewhere (http://efaecium.mlst.net/). The value of this MLST approach is that the predicted founder and descent of clusters of related isolates can be identified (10). Hence, we sought to determine the relatedness of strain 10/96A to subsequent VanA strains of E. faecium from Brazil and to determine whether it represents the progenitor of the strains responsible for outbreaks in Brazilian hospitals.
DNA was isolated from E. faecium strain 10/96A (VanD4) by detergent cell lysis as described previously (16). For MLST, internal fragments of seven housekeeping genes (adk, atpA, ddl, gyd, gdh, purK, and pstS) were amplified with specific primers, using reaction conditions as previously described (12). PCR-amplified fragments were sequenced on both strands, using the ABI PRISM Big Dye Cycle Sequencing Ready Reaction kit (Perkin-Elmer) and an ABI 3100 sequencer (Perkin-Elmer). The nucleotide sequences were compared with alleles deposited in the E. faecium MLST database (http://efaecium.mlst.net/). A sequence type (ST) was assigned, and a diagram of the most-related STs was constructed, using tools available at http://efaecium.mlst.net/. The sequences of E. faecium ddl genes were also compared, and the relative position of the strain 10/96A allele on the diagram was determined using PhyloDraw version 0.8 (http://pearl.cs.pusan.ac.kr/phylodraw/#test).
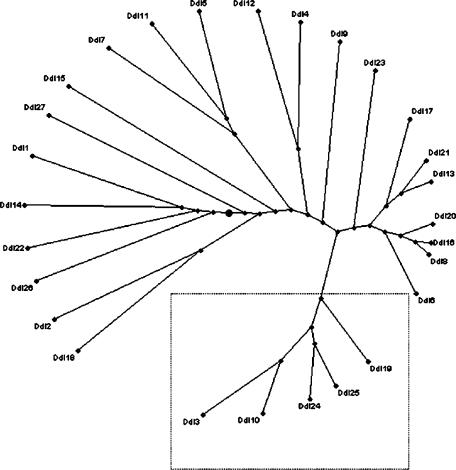
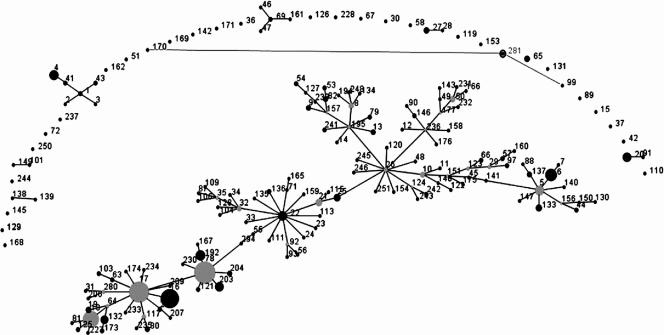
To our knowledge, E. faecium strain 10/96A is the first VanD strain subjected to MLST and deposited in the database. The ddl gene of strain 10/96A represented a novel allele, owing to a unique G→A transversion (9), and has been designated allele Ddl24. This allele differed from 25 other ddl alleles in the database but clustered most closely with alleles 25, 3, 10, and 19 (Fig. 1). Based on analysis of all seven MLST loci, strain 10/96A has the novel allelic profile AtpA9, Ddl24, Gdh1, PurK18, Gyd1, PstS1, and Adk3 (or 9, 24, 1, 18, 1, 1, 3) and represents a new ST, which has been designated ST 281. Only two of 298 STs in the E. faecium MLST database were types closely related to ST 281. These types were ST 99 (linked to one strain from Poland) and ST 170 (linked to one strain from Tanzania), both of which share five of seven alleles with ST 281 and are double-locus variants of it. ST 99 and ST 170 differ from ST 281 by having alleles Ddl5 and PstS21 and alleles Ddl3 and PurK5, respectively. An eBURST analysis of all STs in the database was constructed (Fig. 2); the algorithms judged ST 99 to be evolutionarily more closely related to ST 281 than ST 170.
FIG. 1.
Phylogenetic tree showing the relationships between 26 ddl alleles available at the E. faecium MLST database. The radial tree was constructed by the neighbor-joining method with PhyloDraw, version 0.8, after clustering using ClustalW 1.8. The box highlights Ddl24 and the most-related alleles.
FIG. 2.
eBURST analysis of known E. faecium STs showing the ST 281 singleton and its two DLVs, ST 99 and ST 170.
E. faecium 10/96A (VanD4) was isolated in 1996; no other VanD4-type strains have been isolated in Brazil. Consistent with this, strain 10/96A represented a unique ST, ST 281. Most subsequent Brazilian VRE strains have a VanA phenotype, and most are from hospitals in the south or southeast of the country (5). Camargo et al. recently typed by MLST 56 vancomycin-sensitive and -resistant E. faecium strains from Brazil, isolated from infected and colonized hospitalized patients, colonized volunteers in the community, and animals (4). Twenty-six STs, which represented three groups of related strains and 19 singletons (eBURST analysis), were identified. Most vancomycin-resistant E. faecium were ST 114, including those from the first outbreak of VRE in Brazil (4). We have shown here that strain 10/96A is not related to the VanA E. faecium strains from Brazilian hospitals.
Indeed, we identified only two E. faecium strains in the MLST database with STs closely related to ST 281, and although the strains corresponding to ST 170 and ST 99 are clinical isolates, neither is described as glycopeptide resistant. Both of these closest relatives were double-locus variants of ST 281. Moreover, neither possessed the Ddl24 allele of strain 10/96A. This finding suggests that either (i) the ddl gene was exchanged horizontally with more-distantly related strains, such as those that exhibited greater sequence identity within this locus (Fig. 1), (ii) or that, following acquisition of the vancomycin-resistance gene cluster, under subsequent glycopeptide selection, spontaneous mutations in ddl were favored (because of the lack of a homolog to the vanX and vanW D-D dipeptidase), leading independently to the outgrowth of the ddl mutation observed in the VanD strain 10/96A. Further studies are being directed toward determining the rate and nature of mutation in ddl in VanD strains, as well as the mechanism of acquisition or exchange of the VanD gene cluster.
Acknowledgments
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, 02/11518-6), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, BEX1684/03-5), and Conselho Nacional de Desenvolvimento Científico e Technológico (CNPq).
REFERENCES
- 1.Boyd, D. A., J. Conly, H. Dedier, G. Peters, L. Robertson, E. Slater, and M. R. Mulvey. 2000. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant enterococcus isolated in Canada. J. Clin. Microbiol. 38:2392-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D. A., P. Kibsey, D. Roscoe, and M. R. Mulvey. 2004. Enterococcus faecium N03-0072 carries a new VanD-type vancomycin resistance determinant: characterization of the VanD5 operon. J. Antimicrob. Chemother. 54:680-683. [DOI] [PubMed] [Google Scholar]
- 3.Camargo, I. L., P. F. Del Peloso, C. F. Da Costa Leite, G. H. Goldman, and A. L. Darini. 2004. Identification of an unusual VanA element in glycopeptide-resistant Enterococcus faecium in Brazil following international transfer of a bone marrow transplant patient. Can. J. Microbiol. 50:767-770. [DOI] [PubMed] [Google Scholar]
- 4.Camargo, I. L., M. S. Gilmore, and A. L. Darini. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium strains isolated in Brazil. Clin. Microbiol. Infect., in press. [DOI] [PubMed]
- 5.Cordeiro, J. C., S. Silbert, A. O. Reis, and H. S. Sader. 2004. Inter-hospital dissemination of glycopeptide-resistant Enterococcus faecalis in Brazil. Clin. Microbiol. Infect. 10:260-262. [DOI] [PubMed] [Google Scholar]
- 6.Dalla Costa, L. M., P. E. Reynolds, H. A. Souza, D. C. Souza, M. F. Palepou, and N. Woodford. 2000. Characterization of a divergent vanD-type resistance element from the first glycopeptide-resistant strain of Enterococcus faecium isolated in Brazil. Antimicrob. Agents Chemother. 44:3444-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalla Costa, L. M., D. C. Souza, L. T. Martins, R. C. Zanella, M. C. Brandilone, S. Bokermann, H. S. Sader, and H. A. Souza. 1998. Vancomycin-resistant Enterococcus faecium: first case in Brazil. Braz. J. Infect. Dis. 2:160-163. [PubMed] [Google Scholar]
- 8.Depardieu, F., M. Kolbert, H. Pruul, J. Bell, and P. Courvalin. 2004. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 48:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depardieu, F., P. E. Reynolds, and P. Courvalin. 2003. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob. Agents Chemother. 47:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gholizadeh, Y., M. Prevost, F. Van Bambeke, B. Casadewall, P. M. Tulkens, and P. Courvalin. 2001. Sequencing of the ddl gene and modeling of the mutated D-alanine:D-alanine ligase in glycopeptide-dependent strains of Enterococcus faecium. Protein Sci. 10:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretti, M. L., O. J. Bratfich, R. B. Stucchi, C. Levi, A. S. Levin, G. M. Duboc, E. Vormittag, and D. Blum-Menezes. 2004. Clonal dissemination of VanA-type glycopeptide-resistant Enterococcus faecalis between hospitals of two cities located 100 km apart. Braz. J. Med. Biol. Res. 37:1339-1343. [DOI] [PubMed] [Google Scholar]
- 14.Perichon, B., P. Reynolds, and P. Courvalin. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet 1:57-58. [DOI] [PubMed] [Google Scholar]
- 18.Zanella, R. C., M. C. Brandileone, S. Bokermann, S. C. Almeida, F. Valdetaro, F. Vitorio, F. Moreira Mde, M. Villins, R. Salomao, and A. C. Pignatari. 2003. Phenotypic and genotypic characterization of VanA Enterococcus isolated during the first nosocomial outbreak in Brazil. Microb. Drug Resist. 9:283-291. [DOI] [PubMed] [Google Scholar]