Abstract
The arbuscular mycorrhizal (AM) symbiosis is responsible for huge fluxes of photosynthetically fixed carbon from plants to the soil. Carbon is transferred from the plant to the fungus as hexose, but the main form of carbon stored by the mycobiont at all stages of its life cycle is triacylglycerol. Previous isotopic labeling experiments showed that the fungus exports this storage lipid from the intraradical mycelium (IRM) to the extraradical mycelium (ERM). Here, in vivo multiphoton microscopy was used to observe the movement of lipid bodies through the fungal colony and to determine their sizes, distribution, and velocities. The distribution of lipid bodies along fungal hyphae suggests that they are progressively consumed as they move toward growing tips. We report the isolation and measurements of expression of an AM fungal expressed sequence tag that encodes a putative acyl-coenzyme A dehydrogenase; its deduced amino acid sequence suggests that it may function in the anabolic flux of carbon from lipid to carbohydrate. Time-lapse image sequences show lipid bodies moving in both directions along hyphae and nuclear magnetic resonance analysis of labeling patterns after supplying 13C-labeled glycerol to either extraradical hyphae or colonized roots shows that there is indeed significant bidirectional translocation between IRM and ERM. We conclude that large amounts of lipid are translocated within the AM fungal colony and that, whereas net movement is from the IRM to the ERM, there is also substantial recirculation throughout the fungus.
Arbuscular mycorrhizal (AM) fungi colonize the roots of more than 80% of land plants and take up a significant fraction of all photosynthetically fixed carbon (for recent reviews, see Douds et al., 2000; Graham, 2000). Photosynthate sustains the growth and development of the extraradical mycelium (ERM) and the intraradical fungal mycelium (Ho and Trappe, 1973), so there must be large fluxes of carbon along long narrow coenocytic fungal hyphae. Despite their importance in the global carbon economy, we know very little about these fluxes.
In this mutualistic symbiosis, the fungus acquires carbon as hexose within the root (Shachar-Hill et al., 1995; Solaiman and Saito, 1997), but at all stages of the life cycle carbon is stored predominantly as triacylglycerol (TAG; Cox et al., 1975; Beilby and Kidby, 1980; Beilby, 1983; Jabaji-Hare, 1988; Gaspar et al., 1994). Lipid bodies have been observed in arbuscular trunks, intercellular hyphae, extraradical spores, and germ-tubes by electron microscopy (Sward, 1981; Bonfante-Fasolo, 1984; Bonfante et al., 1994), but this does not tell us directly about transport. Labeling experiments indicate that TAG is synthesized within the intraradical mycelium (IRM) and is exported to the ERM (Pfeffer et al., 1999) where it is stored and used to sustain anabolism both in the ERM (Pfeffer et al., 1999; Lammers et al., 2001) and during spore germination (Bago et al., 1999a). Accordingly, recently proposed models of carbon movement in the AM symbiosis include translocation of fungal lipids as a central route of carbon flow during both symbiotic and spore germination (asymbiotic) phases of the fungal life cycle (Bago et al., 2000).
Multiphoton microscopy (Denk et al., 1990, 1995; Williams et al., 1994; Xu et al., 1996; Xu and Webb, 1996) is well suited to testing the idea that lipid movement is important to carbon flow in the AM symbiosis, because it allows cytology and transport to be observed in vivo. This approach has been used to describe nuclear dynamics in AM fungi (Bago et al., 1998c, 1999b), and its capacity for rapidly and non-destructively obtaining thin optical slices allows volume integrations and time-lapse photographic “movies” of transport along the cytoplasm to be made. These allow the rate of lipid translocation within the fungus to be directly estimated.
Given the substantial movement of carbon from the roots of one plant through AM fungi to the roots of other plants (Francis and Read, 1984; Graves et al., 1997) and the ongoing debate about the significance of such transfer (Robinson and Fitter, 1999), it is also important to establish whether lipid movement between IRM and ERM is unidirectional or bidirectional. Our earlier labeling experiments indicate that lipid is translocated from the IRM where it is made, to the ERM where it is not (Pfeffer et al., 1999), but those experiments did not determine whether some of this lipid might return from the ERM to the IRM of the same or another root. Experiments designed to label lipids in the ERM and to follow their subsequent fates are needed to address this ecologically important question.
Once TAG is moved out of the spore to the germ tube, or from the IRM to the ERM, it may undergo one of four divergent fates. It may be stored, it may continue to circulate around the fungal mycelium, it may be catabolized via the tricarboxylic acid cycle, or else it may serve as an anabolic substrate by entering the glyoxylate cycle (Lammers et al., 2001). Understanding the regulation of this crucial branch point requires the identification and characterization of the enzymes involved, particularly those of storage lipid utilization. The discovery and characterization of expressed fungal sequences that are homologous to the sequences of known lipid breakdown enzymes would be consistent with a flux of carbon from lipid into both anabolism and catabolism.
RESULTS
Lipid Distribution along Germ Tubes
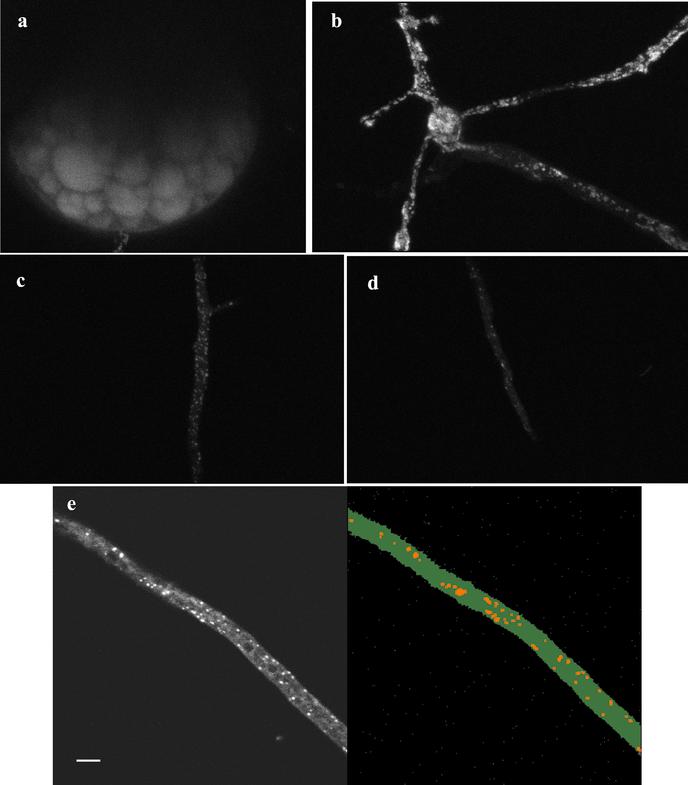
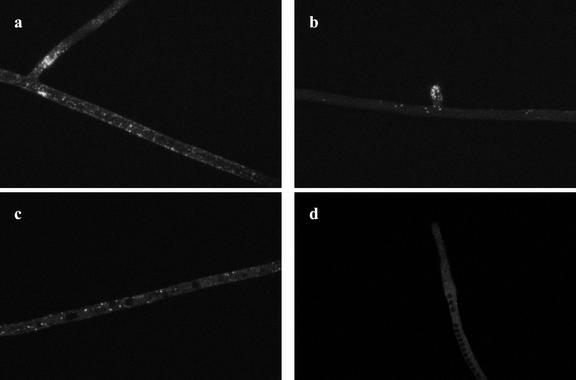
In vivo microscopy of Glomus intraradices and Gigaspora rosea germ tubes reveals brightly stained lipid bodies (Figs. 1 and 2). The large lipid deposits within germinating spores of G. intraradices were also visible (Fig. 1a). In the germ tubes of G. intraradices the numbers of lipid bodies were always higher near the fungal spore (Fig. 1b) and progressively lower further from the spore (Fig. 1c) with almost no lipid globules at the germ-tube tip (Fig. 1d). Three-dimensional digital reconstruction of optical slices (see “Materials and Methods”; Fig. 1e) allowed us to estimate the percentage of the hyphal volume occupied by lipid droplets. For G. intraradices germ tubes, values obtained range from 15.6% ± 1.9 in zones closer to the spore, down to 0.3% ± 0.1 at the hyphal tip. Fewer lipid droplets were visible along the germ tubes of Gi. rosea (Fig. 2) than in G. intraradices (compare Figs. 1 and 2). A gradient of storage lipid distribution along germ tubes of Gi. rosea was also detected, ranging from 4.5% ± 0.6 next to the spore (Fig. 2a) down to no detected lipid bodies at the apex (Fig. 2d). In this fungus, lipid bodies accumulate at the base of existing branches (Fig. 2a) and at the apex of developing germ-tube branches (Fig. 2b). Closer to the hyphal tip, stain-excluding vacuoles were observed (Fig. 2, c and d), and in some cases the lipid droplets seemed to be in close association with these (Fig. 2c).
Figure 1.
Two-photon microscopy of storage lipids in the AM fungus G. intraradices growing asymbiotically. a, Storage lipid deposits within a mature spore; b, lipid bodies in germ tubes are most abundant in the close proximity of the spore; c, further along the germ-tube apex less lipid bodies are visualized; d, close to the hyphal tip almost no lipid bodies are observed; e, projection of a z-series of a G. intraradices germ-tube (left) and visualization of the same image after applying the software for lipid globules volume measurements (right).
Figure 2.
Two-photon microscopy of storage lipids in the AM fungus Gi. rosea growing in the absence of a host root. Fewer lipid globules are observed as compared with G. intraradices germ-tubes. Lipid globules preferentially accumulate at hyphal branching zones (a and b) and are almost absent at germ-tube zones close to the apex (d). Note the presence of black vacuoles in zones closer to the apex (c and d).
Lipid Movement within Germ Tubes
Time-lapse series of micrographs of G. intraradices and Gi. rosea germ-tubes show that lipid bodies move along the hyphae. These movies may be seen at the www.plantphysiol.org or at http://darwin.nmsu.edu/∼plammers/glomus/AM%20Movies.html. Movement is more rapid close to the germinating spore (movie 1). Not all lipid bodies are translocated; some remain near the edges of the hypha and barely move, whereas others in the middle of the germ-tube move more rapidly. Not all movement along the germ-tube is smooth or in one direction (movie 2); sometimes it occurs in pulses, and frequently it does not appear to follow the cytoplasmic streaming. Irregular motion was more frequent in zones closer to hyphal tips (movies 3 and 4).
Lipid Distribution along Extraradical Hyphae
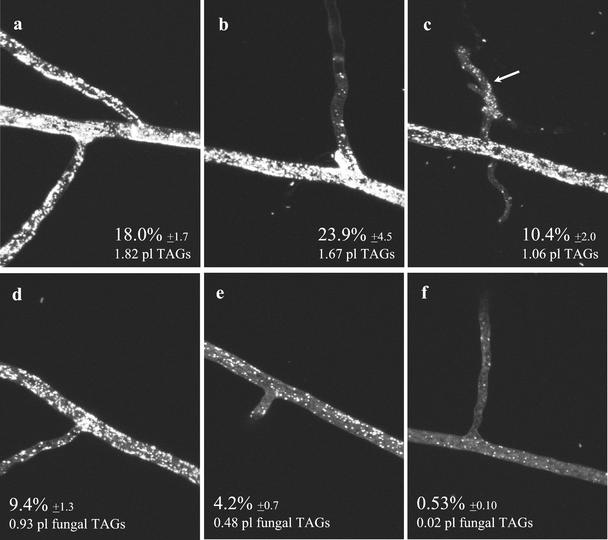
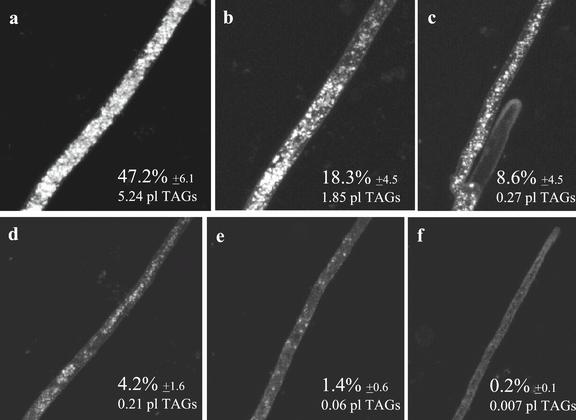
Figure 3 shows a series of micrographs taken along extraradical runner hyphae of G. intraradices, from close to the root (Fig. 3a) toward the growing tip (Fig. 3f). In each micrograph, the left side of the image is closer to the root, whereas the right side is closer to the apex. The fraction of the hyphal volume occupied by lipid in the ERM of this AM fungus is higher than in germ tubes (contrast Figs. 1 and 3). In the ERM the percentage of hyphal volume occupied by storage lipids ranged from approximately 24% close to the root (Fig. 3b) to 0.5% closer to the growing front (Fig. 3f). In this fungus, branched absorbing structures (BAS) contained the fewest lipid bodies in the fungal colony (Figs. 3c, arrow, and 4a), whereas the accumulation of lipid bodies in newly developing spores is evident (Fig. 4b).
Figure 3.
Two-photon microscopy of storage lipids in the ERM of G. intraradices during symbiosis. Lipid globules are most abundant in hyphal zones closer to the root (a–c), then its number progressively decrease as we move to zones closer to the hyphal growing front (d–f). Storage lipid globules are least abundant in zones of high C consumption, as BAS (c). Percentages represent the fraction of total hyphal volume occupied by lipid globules ± se. An estimate of total TAGs volume present in each image is also provided.
Figure 4.
Storage lipid bodies in a BAS (a) and a developing group of spores (b) of symbiotic G. intraradices.
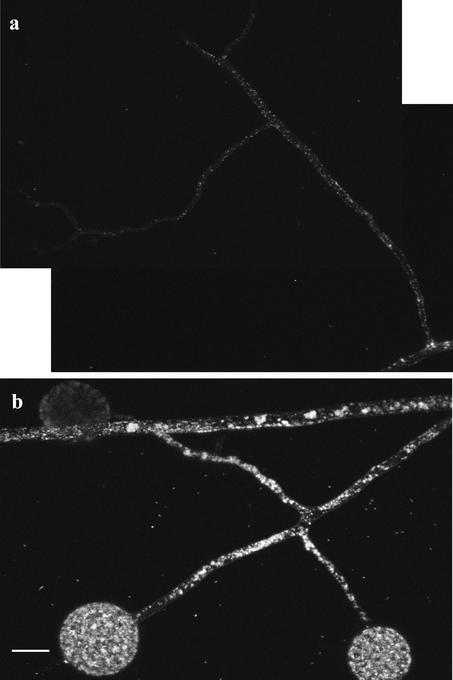
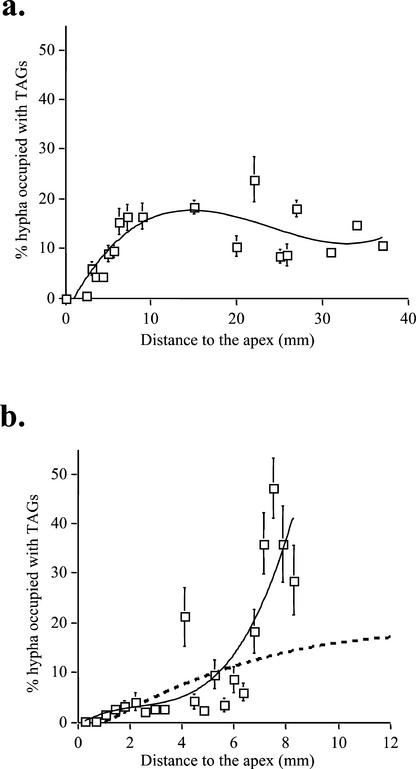
Two-photon laser-scanning microscopy (2PM) observations of Gigaspora margarita external hyphae show that the ERM of this AM fungus has a lipid distribution pattern similar to that of G. intraradices (Fig. 5). Three-dimensional reconstruction of lipid densities in the ERM show that in the 10 mm closest to the apex Gi. margarita runner hyphae contain more storage lipid than G. intraradices, with lipid bodies occupying nearly 50% of the hyphal volume in some areas (Fig. 5a). The gradient of storage lipid bodies along the runner hyphae of each fungus is plotted in Figure 6, from which the difference in total volume of lipid transported per hypha by each species is evident.
Figure 5.
Two-photon microscopy of storage lipids in symbiotic Gi. margarita extraradical hyphae. The same gradient of lipids than in the case of G. intraradices (most abundant in zones closer to the host root [a–c], less abundant approaching hyphal growing front [d–f]) is observed for this AM fungus. Percentages represent the fraction of total hyphal volume occupied by lipid globules ± se. An estimate of total TAGs volume present in each image is also provided.
Figure 6.
The gradient of storage lipid globules along symbiotic hyphae of G. intraradices (a) and Gi. margarita (b) as a result of plotting total storage lipid volume versus distance to the hyphal tip. The dashed line in b represents the expected curve we should obtain for Gi. margarita if applying the same equation curve as for G. intraradices.
Lipid Movement in the ERM
2PM movies of Nile red (Aldrich, Milwaukee, WI) stained ERM of G. intraradices (movie 5) and Gi. margarita (movies 6 and 7) show active cytoplasmic streaming in the thicker runner hyphae. Some of the lipid deposits were irregularly shaped as they moved along the cytoplasmic streams whereas others appeared to be anchored to the hyphal membrane or cell wall (movie 5) or to unknown sites within the cytoplasm (movie 6). In other cases lipid bodies were observed to move against the cytoplasmic streaming (movies 6 and 7). The fastest lipid movement in the ERM was seen along the thick runner hyphae and was calculated as being 4 μm s−1 for G. intraradices (movie 6), and 8 to 11 μm s−1 in Gi. margarita (movies 7 and 8).
Labeling Experiments
Previous labeling experiments using [13C]Glc and 2H2O to follow metabolism in G. intraradices/carrot root monoxenic cultures showed that the fungus makes TAGs in the IRM and moves some of it to the ERM (Pfeffer et al., 1999). In that study no evidence was found for de novo synthesis of storage lipid in the ERM. However this does not exclude the recirculation of lipid from the ERM to the IRM. Microscopy (above) shows that lipid bodies move in both directions in the ERM, but this may only represent movement on short time and distance scales. Therefore, it is important to determine whether significant quantities of carbon do in fact move from the ERM to the IRM in the symbiotic state.
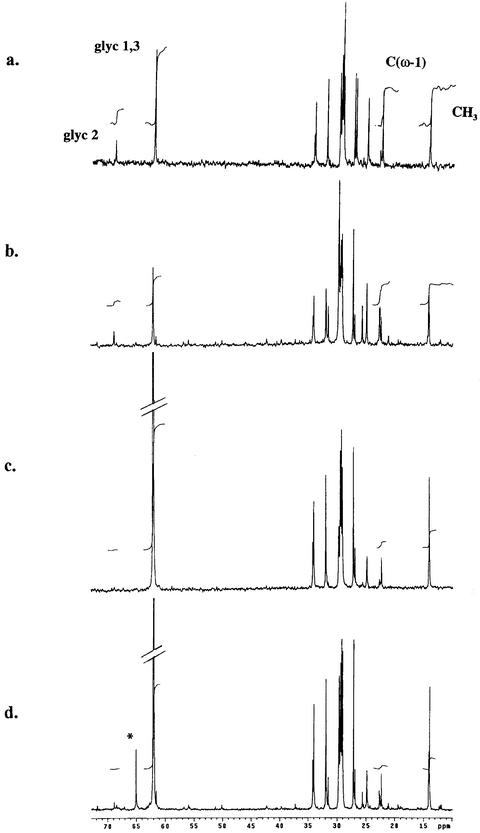
13C-NMR spectra of lipid extracts (Fig. 7) are dominated by the signals of TAGs. In the ERM (Fig. 7, a and c) almost all the fatty acid (FA) moieties are C16:1c11, whereas in the mycorrhizal roots (Fig. 7, b and d), both fungal and plant TAG containing mainly C18:2c9,11 are present (Pfeffer et al., 1999). Such spectra allow the extent of 13C labeling to be determined in different carbon positions of storage lipids from each mycelial phase (either intra- or extraradical). The host and fungal FAs give separate signals in the double bond region of the NMR spectrum, allowing labeling in each to be separately observed (Pfeffer et al., 1999). When [13C1,3]glycerol ([13C1,3]Glyc) was added to the ERM, spectra of TAGs extracted from it (Fig. 7a) and from mycorrhizal roots of the same cultures (Fig. 7b) show labeling in the glyceryl but not the FA moieties of TAGs. In these experiments the labeling level in the glyceryl moieties of the TAG in the ERM is 2.7% ± 0.5 (here and elsewhere, mean ± se, n = 3), and 1.7% ± 0.3 in lipid extracted from the mycorrhizal roots. When [13C1,3]Glyc was added to the mycorrhizal root compartment, TAGs in both ERM (Fig. 7c) and mycorrhizal roots (Fig. 7d) became highly labeled, with 13C enrichment in glyceryl moieties of TAGs in both compartments being 42% ± 4.0 in the ERM and very similar in the IRM (ratio of labeling [ERM/IRM] = 1.1). In the FA moieties of TAGs, labeling was 1.5% ± 0.1 in the mycorrhizal roots and 1.9% ± 0.3 in the ERM. Incubation of G. intraradices germinating spores (asymbiotic fungus) with [13C1,3]Glyc (not shown) resulted in incorporation of the label to the glyceryl moiety, but not in the FA moieties. When [13C2]acetate was used to label either ERM or germinating spores, no labeling of TAG was detected in either glyceryl or FA moieties (spectra not shown), despite efficient entry of the acetate into anabolism via the glyoxylate cycle (Bago et al., 1999a; Lammers et al., 2001).
Figure 7.
13C-NMR spectra of neutral lipids extracted from the ERM of G. intraradices (a and c) and from transformed carrot roots colonized by G. intraradices (b and d) after incubation with [13C1,3]Glyc, which was added to make a final concentration of 10 mm either to the ERM in the fungal compartment (a and b) or to the mycorrhizal root compartment (c and d). Peaks from 13C at different molecular positions of TAG are labeled: glyc1,3 the spectroscopically equivalent C1 and C3 carbons of the glyceryl moiety of TAG, glyc2 the C2 carbon of the glyceryl moiety, CH3 the terminal carbon of the FA moieties, C(ω-1) the penultimate carbon of the FA moieties. The fungal TAG contains almost entirely a shorter chain length FA, C16:1c_11, whereas the host TAG is C18:2c_11. Natural abundance signals from unlabeled positions give integrals that are proportional to the number of carbons of each type. Thus CH3= C(ω-1) when FAs are unlabeled and C1,3 = 2 × C2 when the glyceryl is unlabeled. Incorporation of label from [13C1,3]Glyc results in increased intensity at C1,3 and at CH3, but not at the (ω-1) position since labeling is almost entirely at even-numbered carbons.
Identification of Genes Related to Lipid Metabolism
Sequencing of 420 randomly selected clones of a cDNA library from germinating spores of G. intraradices (Lammers et al., 2001) yielded partial sequence with high homology to known acyl-CoA dehydrogenases. The full-length sequence for the acyl-CoA dehydrogenase-like sequence was obtained (see methods) and the deduced amino acid sequence aligned with acyl-CoA dehydrogenases from other species is shown in Figure 8. The C-terminal tripeptide sequence is AKL, which has been shown in other species, including yeast, to be a peroxisomal targeting sequence for targeting proteins to peroxisomes/glyoxysomes (Hettema et al., 1999). A similar tripeptide sequence was found at the end of the amino acid sequence of a glyoxylate cycle enzyme, malate synthase, from G. intraradices (Lammers et al., 2001).
Figure 8.
Multiple alignments of acyl-CoA dehydrogenase sequences from G. intraradices, Neurospora crassa, Pseudomonas aeruginosa, and Homo sapiens. Noteworthy are the substantial N-terminal extension and the C-terminal PTS-1 type peroxisomal/glyoxysomal tripeptide targeting sequence (in bold) that G. intraradices shares with the Neurospora sp. sequence but not with the bacterial or human sequences.
The N-terminal 100 amino acids of the fungal acyl-CoA dehydrogenases contain a putative heme-binding domain that does not align with the majority of acyl-CoA dehydrogenases in the databases. Similar N-terminal heme domains are found in a number of oxidoreductases, such as plant and fungal nitrate reductases (Campbell and Kinghorn, 1990), sulfite oxidase (Guiard and Lederer, 1977), yeast flavocytochrome b2 (Xia and Mathews, 1990), and plant cytochrome B5/acyl lipid desaturase fusion protein (Sperling et al., 1995). As shown in Figure 8, this domain is conserved in the related Neurospora sp. sequence but missing in the human and Pseudomonas sp. acyl-CoA dehydrogenase sequences.
Examination of 291 expressed sequence tag (EST) sequences from germinating spores of G. intraradices also revealed a sequence (GS 193; GenBank accession no. AY033937) with strong homology to FA CoA ligase (acyl-CoA synthase) proteins as well as to several polyketide synthases. The amino acid identity between the G. intraradices sequence and the Mycobacterium tuberculosis acyl-CoA synthase sequence was 37% over a 339-amino acid stretch. Thus, the protein product of this sequence may be responsible for coupling coenzyme A to FAs released from TAGs in G. intraradices. Other EST sequences from G. intraradices with putative metabolic functions assigned by sequence similarity can be viewed at http://darwin.nmsu.edu/∼plammers/glomus/.
Expression of Putative Acyl-CoA Dehydrogenase
Expression of the putative acyl-CoA dehydrogenase was demonstrated in germinating spores and ERM of G. intraradices using quantitative real time reverse transcriptase (RT)-PCR (see “Materials and Methods”). Table I shows the number of copies of transcripts for β-tubulin, acyl-CoA dehydrogenase, and rRNA in equal quantities of total RNA from each tissue. Five times as much total RNA was isolated per mg of ERM tissue than from germinating spore tissue. This is probably because germ tubes account for only a small proportion of the mass of germinating spore samples whereas the ERM was harvested from plates before extensive sporulation had taken place.
Table I.
Transcript copy numbers for genes expressed in extra radical mycelium and germinating spores of G. intraradices
| Gene | Extraradical Mycelium
|
Germinating Spore Tissue
|
||
|---|---|---|---|---|
| −RT | +RT | −RT | +RT | |
| Acyl-CoA Dehydrogenase | <50 | 8,300 (870) | <50 | 2,700 (310) |
| β-Tubulin | <50 | 6,700 (2,600) | <50 | 2,800 (710) |
| RRNA | 55 (3) | 6.6 (2.0) × 108 | 102 (6) | 5.6 (0.74) × 108 |
Triplicate real-time RT-PCR were performed on 6 ng of DNAse I-treated total RNA isolated from the two tissues. Table values list transcript copy numbers reported as means and (sd).
Acyl-CoA dehydrogenase and β-tubulin transcript levels were expressed at similar, significant levels. Both were higher in RNA from ERM tissue compared with germinating spores (by factors of 3- and 2.4-fold, respectively) despite similar levels of rRNA per nanogram of total RNA in the two tissues. The results for measurements without RT (data columns labeled −RT in Table I) show very low copy numbers, meaning that DNA contamination of total RNA extractions was insignificant.
DISCUSSION
The Nature and Distribution of Lipid Bodies in AM Fungi
Nile red selectively stains neutral lipids (Greenspan et al., 1985), and our observations indicate that AM fungi translocate their storage lipids in lipid bodies, also called “oleosomes,” or “lipid globules.” These consist of a central core of insoluble lipid (usually TAG) surrounded by a monolayer of phospholipid, into which some proteins involved in TAG metabolism are often inserted (Murphy, 1991; Kamisaka and Noda, 2001). The protein coating renders the lipid bodies stable, and this is consistent with our observation that they were not disrupted or amalgamated despite being exposed to shearing and frictional forces during cytoplasmic streaming.
In AM fungal germ tubes (hyphae growing out of germinating spores), we observed more lipid bodies near the spore and progressively fewer approaching the germ-tube tip. Similarly, in the ERM there are more lipid bodies near the root and progressively fewer further out in the mycelium. These distributions are consistent with the results of previous labeling studies showing that storage lipids are synthesized in the IRM and utilized, but not synthesized by both the ERM (Pfeffer et al., 1999) and germinating spores (Bago et al., 1999a) and with the evidence for expression of an acyl CoA dehydrogenase and a acyl-CoA synthase.
Mechanisms and Rates of Lipid Translocation
Most lipid bodies were observed to move with the cytoplasmic stream, but some were stationary, apparently bound to the cell membrane or hyphal wall. Others appear to move opposite to the prevailing direction of cytoplasmic streaming. Observations indicating that translocation along hyphae is not simply governed by cytoplasmic streaming have also been made on the motion of nuclei in AM and other fungi (Aist, 1995; Bago et al., 1998c, 1999b). Movement of lipid bodies independently of cytoplasmic streaming may be associated with the fungal cytoskeleton, perhaps on microtubular arrays, which have been shown to form “tracks” along AM fungal hyphae (Åström et al., 1994; Timonen et al., 2001). A β-tubulin sequence has been reported, and its expression has been demonstrated by Butehorn and coworkers (1999) and recently sequences putatively encoding α-tubulin, an actin, actin-related protein, and dynein have been found (J. Jun, P. J. Lammers, and Y. Shachar-Hill, unpublished data). These, together with previous microscopic observations, indicate that AM fungi have a full array of cytoskeletal proteins including those associated with movement of cytoplasmic constituents.
The total flux of storage lipid mass out of the IRM along an AM runner hypha may be estimated by multiplying the average speed of lipid bodies along the hypha by the fraction of cytoplasmic volume that they occupy, by the cross-section area of the cytoplasm, and by the density of TAG. The velocity (even direction) of translocation varies between AM fungi and within the same fungal colony, and this makes the range of estimated transfer rates large. However, by setting upper bounds on the rates of lipid translocation, such estimates allow one to test whether the observed rates of lipid movement could account for the known carbon fluxes from plant to fungus and for the likely carbon needs of the fungal mycelium. If we take the density of the TAGs translocated by G. intraradices and Gi. margarita to be similar to that of Glyc tripalmitate (d = 0.88 g L−1) we see that a maximum of 0.26 μg TAG per hour are transported through a main extraradical runner hyphae by symbiotic G. intraradices. The corresponding value for Gi. margarita is 1.34 μg TAG h−1. The difference between the estimates for these two fungi may reflect differences between AM fungal isolates in respect to their C demand from the host plant. But before such a conclusion can be drawn one must also take into account the number of runner hyphae per root length and the total colonized root length. This varies widely between different plant/fungus combinations and under different growth conditions (Jakobsen et al., 1992; for review, see Smith and Read, 1997).
One may ask whether the observed lipid flux can account for the amount of lipid that accumulates in the ERM? In the monoxenic cultures of G. intraradices/transformed carrot roots after sporulation in the external compartment is complete, there is 1.7 ± 0.1 mg of TAG per plate (determined by weighing lipid extracted from the fungal compartment). All the TAG exported to the ERM in the fungal compartment must flow through the hyphae that cross the barrier separating the two compartments, so the flux per hypha times the number of crossovers is the total export to the ERM. There are typically about 10 thick runner hyphae crossing the barrier so that in 2 months, (10 hyphae) × (0.26 μg h−1 hypha−1) × (24 h d−1) × (60 d) = 3.7 mg are exported. Thus the observed rates of lipid movement are sufficient to account for all the lipid accumulated in the ERM, because some of the exported lipid is consumed (Pfeffer et al., 1999; Lammers et al., 2001) and some is recirculated to the IRM (see below).
These fluxes may be useful in estimating how much carbon is exported from colonized roots in the form of lipid from plants. Such calculations would require estimates of the number of active hyphae connecting each root system to the ERM, and this again varies widely (Smith and Read, 1997).
Labeling Experiments
The high degree of labeling measured in glyceryl moieties of TAGs in both mycorrhizal roots and ERM when [13C1,3]Glyc was supplied to the root compartment (Fig. 7, c and d) is consistent with previous experiments showing active synthesis of lipid in the IRM and export to the ERM (Pfeffer et al., 1999). The labeling in glyceryl moieties of TAG in the ERM was much less when label was supplied to the hyphal compartment (Fig. 7a). This and the absence of labeling in the FA moieties in those experiments (or when [13C2]acetate was applied to the ERM) are consistent with earlier findings that showed no sign of significant de novo FA synthesis in the ERM (Pfeffer et al., 1999). The low level of labeling of the glyceryl moieties of TAG when [13C]Glyc was supplied to the ERM is not due to a failure of this substrate to enter metabolism (specifically to label dihydroxyacetone phosphate) in the ERM, because trehalose in the ERM of cultures labeled in this way becomes highly enriched (Lammers et al., 2001). The labeling pattern obtained from both ERM and germinating spores when [13C1,3]Glyc is supplied to them seems therefore to be due to a relatively slow turnover of the glyceryl moieties of TAG in the ERM, presumably by transacylation; and to the absence of significant synthesis of FA moieties in the ERM and in germinating spores.
The labeling of the glyceryl moiety of TAG in the mycorrhizal roots when 13C-Glyc is supplied to the ERM is low compared with the level reached when the substrate is supplied to the colonized roots (compare Fig. 7, b and d). But compared with the labeling in the glyceryl of TAG in the ERM it is very significant (compare Fig. 7, b and a) so that the ratio of labeling in TAGs from mycorrhizal roots to labeling in TAG from ERM is 0.6 ± 0.1 when label was supplied to the ERM. This observation suggests that there is very substantial recirculation of lipid globules from the ERM back to the IRM.
Alternatively, it might arise if label supplied as Glyc to the ERM is translocated back to the IRM in a form other than neutral lipid and then incorporated into TAG in the IRM. When labeled Glyc is supplied to the ERM, trehalose becomes highly labeled (Lammers et al., 2001) with substantial scrambling due to the operation of the pentose phosphate pathway. Thus, if this or another carbohydrate were to play any role in C translocation from ERM to IRM, then one would expect substantial scrambling to other positions of the glyceryl moieties and/or labeling of the FA moieties of TAG. Neither was observed. One would also expect labeling of TAGs extracted from colonized roots when labeled acetate is provided to the ERM because labeling patterns and levels after supplying [13C2]acetate or [13C1]acetate show that the entry point of Glyc into central metabolism, dihydroxyacetone phosphate, is highly labeled by acetate (Pfeffer et al., 1999; Lammers et al., 2001). However no labeling of lipid in colonized roots was observed when acetate was supplied to the ERM (spectra not shown).
Therefore we conclude that the labeling of TAG in the IRM when the ERM is labeled is due to substantial recirculation of lipids from the ERM back to the IRM. These experiments do not indicate whether the TAG returns to the same colonized root from which it came or to the IRM in another root. It seems likely, given the relatively long branched coenocytic hyphae along which the lipid bodies move, that the movement from ERM to IRM can be to other roots than the root from which each lipid body originally came, and this provides a mechanism for the well-known transfer of carbon from the roots of one plant to those of another via their common mycorrhizal network.
Implications of Gene Identification and Deduced Amino Acid Sequences
The fact that small-scale random sequencing yielded putative sequences for two key genes involved in the breakdown of storage lipid suggests significant levels of expression of lipid utilization genes in germinating spores. A lipase is necessary for lipid breakdown and lipase activity has been identified in AM fungi (Gaspar et al., 1997). The putative FA-CoA synthase identified in the collection of EST sequences and the acyl-CoA dehydrogenase provide evidence and sequence tags for the next two steps in lipid breakdown. These findings are consistent with the idea that the diminishing level of lipids along hyphae at greater distances from the spore or IRM is due to metabolic consumption of the translocated lipid.
Acyl-CoA dehydrogenase catalyzes the first step of β-oxidation of FAs to acetyl-CoA. β-oxidation in animals takes place in the mitochondrion via acyl-CoA dehydrogenase, and also in the peroxisomes via acyl-CoA oxidase (in which case hydrogen peroxide is produced) whereas in plants and fungi, most or all of β-oxidation takes place in the peroxisomes/glyoxysomes (Mannaerts and vanVeldhoven, 1996). In higher plants and in the yeasts, β-oxidation is via acyl-CoA oxidase, but in at least two filamentous fungi, N. crassa (Thieringer and Kunau, 1991) and Aspergillus niger (Baltazar et al., 1999) acyl-CoA oxidation in the peroxisomes/glyoxysomes is apparently catalyzed mainly or entirely by dehydrogenase rather than oxidase. The acyl-CoA dehydrogenase identified here has high homology to the sequence of dehydrogenase from Neurospora crassa, which like the G. intraradices sequence has a peroxisomal/glyoxysomal targeting sequence (Fig. 8). The Glomus sp. and Neurospora sp. acyl-CoA dehydrogenase sequences also have an N-terminal extension similar to the heme-binding domains of cytochromes B5. This domain has also been found in plant acyl lipid desaturases (Napier et al., 1999), although the remainder of the sequence is homologous not to desaturases but rather to a series of dehydrogenases. The fungal acyl-CoA dehydrogenase N-terminal heme binding domains may play a functional role in reoxidation of the FADH2 cofactor produced during β-oxidation in glyoxysomes.
Recent labeling and gene identification experiments in G. intraradices show that the glyoxylate cycle is very active in both ERM and germinating spores (Lammers et al., 2001), and the lipid exported to the ERM or stored in the spores seems to be the source of carbon entering this anabolic pathway (Pfeffer et al., 1999). Thus the acyl-CoA dehydrogenase in G. intraradices is probably functioning in the flux of carbon from lipid into anabolism and may also be important in the production of acetyl-CoA destined for oxidation in the tricarboxylic acid cycle.
Measurement of Gene Expression
The finding that the putative acyl-CoA dehydrogenase is expressed at levels similar to β-tubulin, which is known to be expressed significantly during the AM fungal life cycle (Butehorn et al., 1999), is consistent with a role for this enzyme in lipid utilization both in the ERM and during germination. Similarities in the metabolic pathways that are active, lipid body movement and distribution patterns, and gene expression observations suggest that AM fungi may use similar mechanisms of lipid utilization during spore germination and in their ERM.
Conclusions: Why Are Lipids Present at High Levels in AM Fungi?
The large quantities of lipid present in the AM fungal hyphae means that the Glomales qualify as “oleogenic” fungi, that is, they can accumulate over 25% of their dry weight as lipid (Jabaji-Hare, 1988; Murphy, 1991). Known oleogenic fungi accumulate these large amounts of lipid when carbon is available but other nutrients—particularly N—limit growth. In these fungi the accumulated lipid is used later, if and when nutritional restrictions are lifted (Murphy, 1991). However, this does not appear to be the pattern in AM fungi, in which synthesis and utilization of lipids are spatially but not always temporally separated. Storage lipids are synthesized in the IRM and utilized, but not synthesized, in the ERM (Pfeffer et al., 1999) and germinating spores (Bago et al., 1999a). Rather, for AM fungi the need for a compact form of carbon for translocation and storage may be the reason for abundant lipid bodies. Indeed it would be impossible for the hyphae to contain carbohydrate at a calorically equivalent density to the TAG levels that were observed in many parts of the fungal hyphae. This fact may also underlie the energetically inefficient strategy of turning hexose acquired within the host root (Shachar-Hill et al., 1995) into lipid in the IRM (Pfeffer et al., 1999) and then exporting it to the ERM, where substantial amounts are turned back into carbohydrate via the glyoxylate cycle (Lammers et al., 2001).
At first sight, the recirculation of lipid bodies that we observed microscopically and confirmed by labeling experiments and NMR spectroscopy might also appear inefficient. Because this seems not to be due to a lack of mechanisms to control the movement of lipid bodies, it may be a strategy to ensure that lipid is distributed throughout the fungal mycelium to be utilized as needed. By this scheme, continuous circulation of lipid bodies ensures the availability of carbon throughout the mycelium, and regulation of lipid utilization would be at the level of transcription and/or post-transcriptional activation of the proteins responsible for directing carbon from lipid bodies into storage, catabolism or anabolism.
MATERIALS AND METHODS
Biological Material
Spores of Gigaspora rosea Nicolson & Schenk (DAOM 194757, Biosystematic Research Centre, Ottawa) were collected from pot cultures by wet sieving and decantation (Gerdemann and Nicolson, 1963) and surface-sterilized (Mosse, 1962). Glomus intraradices spores (DAOM 197198, Biosystematic Research Centre) were collected from monoxenic cultures (Bécard and Fortin, 1988; St-Arnaud et al., 1996) under sterile conditions. Spores of these two species were cultured (25°C, darkness) in sterile water-agar medium (0.8% [w/v] Agar Bacto Difco in distilled water) and, once germinated, were transferred to special microscope observation chambers (Chambered Coverglass, NUNC InterMed, Naperville, IL) as previously described (Bago et al., 1998c). The chambers were filled with 1 mL of autoclaved water-agar medium to which 0.05 μg/mL of the specific neutral lipid dye Nile red (Greenspan et al., 1985; Butt et al., 1989) was added. Three pregerminated spores were transferred per chambered coverglass, and a total of five replicates were prepared. Spores were cultured at 25°C in the dark for 6 d. In the case of Gi. rosea the chambers were placed on a slope of approximately 70° to encourage the negatively geotropic germ tubes of this fungus (Watrud et al., 1978) to grow as close as possible to the coverglass.
AM monoxenic cultures were established as described by Bécard and Fortin (1988) and St-Arnaud et al. (1996). Briefly, clones DC-2 and DC-1 of carrot (Daucus carota) Ri-T DNA transformed roots were cultured, respectively, with G. intraradices or Gigaspora margarita (BEG 34 was kindly provided by Dr. Gillaume Bécard, Toulouse, France) in regular Petri dishes (Gi. margarita) containing M medium (Chabot et al., 1992) or two-compartmented Petri dishes (G. intraradices). For G. intraradices, cultures were initiated in one compartment (“root compartment”) of each plate, which contained M medium. Fungal hyphae, but not roots, were allowed to grow over to the second (“hyphal”) compartment, which contained M medium lacking Suc (St-Arnaud et al., 1996).
Choice of Developmental Stage of the Cultures
All Gi. rosea and G. intraradices spores transferred to the chambered coverslips regerminated, and their germ-tubes developed showing normal morphogenic features. After 6 d of axenic culture, fungal tips showed no retraction of cytoplasm and were growing actively.
Gi. margarita and G. intraradices monoxenic cultures had extensively developed in the culture medium after 2 and 3 months of culture, respectively. Gi. margarita cultures showed numerous runner hyphae, auxiliary cells, BAS (Bago et al., 1998b), and actively growing apices; no spores had been yet formed. G. intraradices had extensively colonized the root compartment, and its ERM profusely developed in the hyphal compartment. Numerous runner hyphae showing BAS at regular intervals could be observed in this hyphal compartment, and few spores had formed. The morphogenic features shown by both fungi correspond to the described “assimilative ” phase of ERM development (Bago et al., 1998a).
Choice of Dye Concentration for in Vivo Microscopy
Nile red inhibited germination of Gi. rosea spores when present in the medium at 0.2 μg/mL or above; 0.5 μg/mL inhibited G. intraradices germination (data not shown). Both spore germination and germ-tube development was reduced at dye concentrations of 0.1 μg/mL for Gi. rosea and of 0.2 μg/mL for G. intraradices. However at 0.05 μg/mL no effect on spore germination or germ-tube growth was observed for either fungus (percent germination: Gi. rosea, 32.3 [control] versus 18.8 [Nile red]; G. intraradices, 53.9 [control] versus 59.0 [Nile red]. Germ-tube development [mm]: Gi. rosea, 164.2 ± 80 [control] versus 177 ± 99 [Nile red]; G. intraradices, 39.3 ± 20.6 [control] versus 35.44 ± 15.96 [Nile red]). After incubation of fungal material in 0.05 μg/mL, lipid globules were clearly observable by conventional fluorescence microscopy (using a fluorescein filter), although fluorescence faded very rapidly (5–10 s) in agreement with previous reports (Butt et al., 1989). However under multiphoton microscopy (two-photon microscopy, 2PM), photobleaching was much slower, so that enough fluorescence for imaging was observed for over 10 min of observation.
In Vivo Observation of Lipids
Several hours before microscopic observations a sterile solution of the fluorescent dye Nile red was poured over the media containing the AM monoxenic cultures to reach a final concentration of 0.05 μg/mL.
Images of Nile red-stained lipid globules in the cultures were acquired with a homebuilt two-photon laser-scanning microscope (2PM). Imaging was carried out at using 840-nm mode-locked excitation (80-fs pulses; 80 MHz repetition rate) obtained from an argon-pumped Ti:Sapphire laser (Tsunami, Spectra Physics, Mountain View, CA). The beam was attenuated and directed into a retrofitted confocal scanning box (MRC-600, Bio-Rad, Hercules, CA) aligned to a fixed stage upright microscope (AX-70, Olympus, Tokyo). A 60x/0.90W water immersion objective (Olympus) was used by directly immersing the lens in the culture media flooded with water. The specimens were exposed to illumination intensities of 5 to 10 mW. Because multiphoton fluorescence excitation is intrinsically localized (Denk, et al., 1990; Williams, et al., 1994), confocal detection optics are unnecessary and fluorescence detection was accomplished external to the confocal scanbox. The fluorescence was separated from the excitation light by a 680-nm long pass dichroic mirror (Chroma Technology Corp., Brattleboro, VT), filtered with a 2-mm-thick BGG22 blue glass filter (380–530 nm, Chroma Technology Corp.) and monitored with a photo multiplier tube (HC125–02, Hamamatsu, Bridgewater, NJ) placed at the back of the objective lens. The signal from the external photomultiplier was sent to the external input of the confocal scanning box.
Images of lipid globules were acquired as serial optical sections (z series) with steps varying from 1 to 4 μm. Images were three-dimensional-projected using the Confocal Assistant 3.1 software (Bio-Rad). Lipid movement was recorded as time-lapse series of images (t series) consisting of multiple (1 s) frames acquired at 1- to 5-s intervals using the Bio-Rad confocal software. Quantification of the lipid to hyphae ratio was accomplished using the interactive graphics manipulation language IDL (Research Systems Inc., Boulder, CO). Image “objects” were automatically selected using an object recognition protocol in which the image to be analyzed was defined by its luminosity compared with a defined background threshold level by an amount given by the average background pixel value plus three sds (3ς) of this background. The resulting objects were then sequentially eroded and dilated by a 3- × 3-pixel kernel to remove isolated noise islands. The threshold to determine lipid droplets was set at 3ς above the surrounding hyphae region pixel values. The threshold to determine hyphae regions was set at 3ς above the general image background. The identified lipid and hyphae “object” areas were summed for every image within the three-dimensional image stack to determine lipid to hyphae volume ratios.
Isotopic Labeling and NMR Analysis
Labeling experiments were carried out as previously described (Bago et al., 1999a; Pfeffer et al., 1999). Briefly, 10 mm [13C1,3]Glyc or [13C2]Glyc was added to the hyphal compartment of divided Petri plates of G. intraradices monoxenic cultures (St-Arnaud et al., 1996) 1 to 2 weeks after fungal crossover. The cultures were then grown for a further 6 to 7 weeks for full mycelial development and sporulation to occur. [13C1,3]Glyc (10 mm) was added to germinating G. intraradices spores in liquid M medium lacking Suc and grown for 2 weeks.
Extraction of water-soluble metabolites was carried out with 70:30 (v/v) methanol:water and of lipids was with isopropyl alcohol as previously described (Pfeffer et al., 1999). The extracts were evaporated to dryness and dissolved in deuterated water or chloroform for NMR analysis. 13C enrichments were measured from NMR spectra taken at 400 MHz for 13C as previously described (Pfeffer et al., 1999).
RNA Extraction
G. intraradices tissues were obtained from the same cultures described above for NMR and microscopy experiments. Total RNA was isolated from 11-d-old germinating G. intraradices spores using a modified hot phenol/SDS method (Wan and Wilkins, 1994) followed by CsCl ultracentrifugation (Sambrook et al., 1989). Tissues were harvested and ground to a fine powder with sand under liquid nitrogen. The ground tissue was resuspended in a 1:2 mixture of hot (65°C) phenol and lysis buffer (0.1 m Tris [tris(hydroxymethy) aminomethane], 0.1 m LiCl, 5 mm EDTA, 0.1 m NaCl, 0.1 m sodium acetate, 1% [w/v] SDS, and β-mercaptoethanol, pH 5.2). The suspension was incubated 10 min at 65°C with occasional vortexing. One-fourth volume of chloroform-isoamyl alcohol (24:1) was added and the aqueous phase was recovered after centrifugation. The aqueous phase was extracted repeatedly with hot phenol (pH 4.5) and phenol:chloroform:isoamyl alcohol (25:24:1) until there was no interphase. To the cleaned aqueous phase, guanidine thyocyanate was added to make 4 m solution and ultracentifuged in the presence of CsCl. The RNA pellet was washed with 70% (v/v) ethanol, dissolved in TE, and stored at −80°C.
cDNA Libraries
A cDNA library from 11-d-old germinating G. intraradices spore total RNA was constructed using the SMART kit (CLONTECH Laboratories, Palo Alto, CA). One microgram of total RNA served for first strand cDNA synthesis, and the resulting single stranded cDNA was amplified by PCR. After digestion with SfiI and size fractionation, the cDNA was ligated into the SfiI-digested λTriplEx2 vector, which contains the asymmetrical SfiI sites (A&B) in the multicloning site. The ligated cDNA was packaged using Gigapack III λ extract (Stratagene, La Jolla, CA). A cDNA library from G. intraradices ERM from the same tissue culture system made by the same approach was the generous gift of Dr. M.J. Harrison (Noble Foundation, Ardmore, OK).
cDNA Sequencing and Analyses
Preparation and random DNA sequencing of a cDNA library constructed from RNA isolated from 10-d germinating spore tissue incubated with 1% (v/v) CO2 was as described previously (Lammers et al., 2001). The sequences are available from National Center for Biotechnology Information with dbEST accession numbers from 5812585 to 5812875 and GenBank numbers BE603746 to BE604036 and also at http://www.chemistry.nmsu.edu/glomus/. The putative acyl-CoA ligase is GenBank accession number AY033937.
Full-length cDNA sequence for the acyl-CoA dehydrogenase gene (GenBank accession no. AY033936) was obtained using the SMART RACE cDNA amplification kit (CLONTECH). Total RNA (250 ng) was used to synthesize each 5′ and 3′ RACE ready cDNA. The sequences of gene-specific primers employed for RACE were based on the sequences of original acycl-CoA dehydrogenase gene fragment. The resulting RACE fragments for each gene were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced with M13 forward and reverse primers.
Sequence analysis of the acyl-CoA dehydrogenase gene was accomplished using the BLAST 2.0 program against the National Center for Biotechnology Information non-redundant database and the Neurospora crassa genomic sequence database (http://www.genome.wi.mit.edu/annotation/fungi/neurospora/). Two putative introns were removed from the best N. crassa homolog based on comparison of the BLAST pairwise analysis between the full-length G. intraradices amino acid sequence and the N. crassa genome using TBLASTN. In each case, canonical 5′-GT and AG-3′ intron boundary dinucleotides were found in the coding regions for the beginning and end of an insertion in the N. crassa sequence relative to the G. intraradices sequence. The unique low-complexity Ala, Pro-rich region of the Neurospora sp. sequence shown in Figure 8 does not appear to be associated with any classical intron boundary sequences. The multiple alignment was constructed using CLUSTAL W.
Real-Time RT-PCR Quantification of Gene Expression
RNA isolation from ERM and germinating spores was performed as described by Lammers et al. (2001). ERM from the fungal compartments of five plates (51 mg) was isolated before the accumulation of large numbers of spores, which yielded 3.05 μg of total RNA. Approximately 50 mg of spores was sieved then germinated for 3.5 d in the presence of 1% (v/v) CO2. Hyphae and spores were harvested yielding 600 ng of total RNA. Gene expression was monitored using an Applied Biosystems PRISM 7700 instrument and “Taqman” assays designed for β-tubulin, acyl-CoA dehydrogenase, and ribosomal RNA. The amplification and probe sequences for each assay are shown below along with amplicon sizes and the region of each cDNA amplified in parentheses. Primers were used at 500 nm and probes at 100 nm final concentrations. Absolute quantification was based on standard curves for each assay. Plasmid DNA containing each amplicon was prepared using Qiagen USA (Valencia, CA) kits and quantified by UV absorbance spectroscopy. Standard curves were determined from duplicate samples at 102, 103, 104, and 106 copies for each assay (not shown).
To prepare template for RT-PCR assays, total RNA was treated with RNase-free DNase-I (DNA-free, Ambion, Austin, TX) for 1 h followed by DNase-I removal as specified by the manufacturer. Triplicate assays used 6-ng aliquots of the DNase-treated RNA pre-incubated for 15 min at 95 C then placed on ice to remove any potential interfering secondary structures. The reverse amplification primer served as the primer for reverse transcription. Each RT-PCR assay was run in 50 μL total volume using One-Step RT-PCR Master mix containing AmpliTaq Gold DNA polymerase to which 12.5 units of MultiScribe enzyme was added (all from Applied Biosystems, Foster City, CA). The reactions were incubated at 48°C for 60 min for reverse transcription followed by a 10-min incubation at 95°C to activate the AmpliTaq Gold polymerase and 45 cycles of 15 s at 95°C, and 1 min at 60°C. MultiScribe enzyme was omitted from the no-reverse transcriptase control reactions.
18S rRNA (142–217) 76 bp
Forward primer, CCGTGAATCATCGAATCTTTGAA, anneals between residues 142 and 164 with a Tm of 60°C. Reverse primer, CACTGACCCTCAAACAGGCATA, anneals between residues 217 and 196 with a Tm of 59°C. Taqman probe, TGCACTCTCTGGCAACCCGGG (21), anneals between residues 172 and 192 with a Tm of 68°C.
β Tubulin (316–397) 82 bp
Forward primer, AGAAAGTCTACCACGGAAAATAGTAGCT, anneals between residues 316 and 343 with a Tm of 59°C. Reverse primer, TTCACGTAATATGATGGCTGCAT, anneals between residues 397 and 375 with a Tm of 58°. Taqman probe, CGGTCAAATATCTTCCATGACGAGGATCG (29), anneals between residues 345 and 373 with a Tm of 69°C.
Acyl CoA-Dehydrogenase (1,204–1,287) 84 bp
Forward primer, GATGTTATTCGTAATAAACTTGCCCATA (28 bp), anneals between residues 1,204 and 1,231 with a Tm of 59°C. Reverse primer, TGTTTGATAAATTAATGACTCCATCCA (27 bp), anneals between residues 1,287 and 1,261 with a Tm of 59°. Taqman probe, CGCGTAAAATTGAGGCAACCCATGC (25 bp), anneals between residues 1,235 and 1,259 with a Tm of 69°C.
ACKNOWLEDGMENTS
The authors thank Dr. D. Douds and J. Brouillette for technical support, Dr. Guillaume Bécard for providing Gi. margarita monoxenic culture in DC-1 transformed carrot roots, and Dr. M.J. Harrison for the ERM cDNA library.
Footnotes
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010466.
LITERATURE CITED
- Aist JR. Independent nuclear motility and hyphal tip growth. Can J Bot. 1995;73:S122–125. [Google Scholar]
- Åström H, Giovannetti M, Raudaskoski M. Cytoskeletal components in the arbuscular mycorrhizal fungus Glomus mosseae. Mol Plant Microbe Interact. 1994;7:309–312. [Google Scholar]
- Bago B, Azcón-Aguilar C, Goulet A, Piché Y. Branched absorbing structures (BAS): a feature of the extraradical mycelium of symbiotic arbuscular mycorrhizal fungi. New Phytol. 1998a;139:375–388. [Google Scholar]
- Bago B, Azcón-Aguilar C, Piché Y. Architecture and developmental dynamics of the external mycelium of the arbuscular mycorrhizal fungus Glomus intraradices grown under monoxenic conditions. Mycologia. 1998b;90:52–62. [Google Scholar]
- Bago B, Pfeffer PE, Douds DD, Jr, Brouillette J, Bécard G, Shachar-Hill Y. Carbon metabolism in arbuscular mycorrhizal spores as revealed by NMR spectroscopy. Plant Physiol. 1999a;121:263–271. doi: 10.1104/pp.121.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Shachar-Hill Y, Pfeffer PE. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 2000;124:949–957. doi: 10.1104/pp.124.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Zipfel W, Williams RC, Chamberland H, Lafontaine J-G, Webb WW, Piché Y. In vivo studies on the nuclear behavior of the arbuscular mycorrhizal fungus Gigaspora r. grown under axenic conditions. Protoplasma. 1998c;203:1–15. [Google Scholar]
- Bago B, Zipfel W, Williams RC, Piché Y. Nuclei of symbiotic arbuscular mycorrhizal fungi as revealed by in vivo two-photon microscopy. Protoplasma. 1999b;209:77–89. doi: 10.1007/BF01415703. [DOI] [PubMed] [Google Scholar]
- Baltazar MF, Dickinson FM, Ratledge C. Oxidation of medium-chain acyl-CoA esters by extracts of Aspergillus niger: enzymology and characterization of intermediates by HPLC. Microbiology UK. 1999;145:271–278. doi: 10.1099/13500872-145-1-271. [DOI] [PubMed] [Google Scholar]
- Bécard G, Fortin A. Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- Beilby JP. Effects of inhibitors on early protein, RNA, and lipid synthesis in germinating vesicular-arbuscular mycorrhizal fungal spores of Glomus caledonium. Can J Bot. 1983;29:596–601. doi: 10.1139/m83-093. [DOI] [PubMed] [Google Scholar]
- Beilby JP, Kidby DK. Biochemistry of ungerminated and germinated spores of the vesicular-arbuscular mycorrhizal fungus Glomus caledonium: changes in neutral and polar lipids. J Lipid Res. 1980;21:739–750. [PubMed] [Google Scholar]
- Bonfante P, Balestrini R, Mendgen K. Storage and secretion processes in the spore of Gigaspora margarita Becker and Hall as revealed by high-pressure freezing and freeze substitution. New Phytol. 1994;128:93–101. doi: 10.1111/j.1469-8137.1994.tb03991.x. [DOI] [PubMed] [Google Scholar]
- Bonfante-Fasolo P. Anatomy and morphology of VA mycorrhizae. In: Powell CL, Bagyaraj DJ, editors. VA Mycorrhiza. Boca Raton, FL: CRC Press; 1984. pp. 5–33. [Google Scholar]
- Butehorn B, Gianinazzi-Pearson V, Franken P. Quantification of beta-tubulin RNA expression during asymbiotic and symbiotic development of the arbuscular mycorrhizal fungus Glomus mosseae. Mycol Res. 1999;103:360–364. [Google Scholar]
- Butt TM, Hoch HC, Staples RC, St. Leger RJ. Use of fluorochromes in the study of fungal cytology and differentiation. Exp Mycol. 1989;13:303–320. [Google Scholar]
- Campbell WH, Kinghorn JR. Functional domains of assimilatory nitrate and nitrite reductases. Trends Biochem Sci. 1990;15:315–319. doi: 10.1016/0968-0004(90)90021-3. [DOI] [PubMed] [Google Scholar]
- Chabot S, Bécard G, Piché Y. Life cycle of Glomus intraradix in root organ culture. Mycologia. 1992;84:315–321. [Google Scholar]
- Cox G, Sanders FE, Tinker PB, Wild JA. Ultrastructural evidence relating to host-endophyte transfer in vesicular-arbuscular mycorrhizas. In: Sanders FE, Mosse B, Tinker PB, editors. Endomycorrhizas. London: Academic Press; 1975. pp. 297–312. [Google Scholar]
- Denk W, Piston DN, Webb WW. Two-photon molecular excitation in laser scanning fluorescence microscopy. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. New York: Plenum Press; 1995. pp. 445–458. [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Douds DD, Pfeffer PE, Shachar-Hill Y. Carbon partitioning, cost and metabolism of arbuscular mycorrhizas. In: Kapulnik Y, Douds DD, editors. Arbuscular Mycorrhizas: Physiology and Function. Dordrecht, The Netherlands: Kluwer Academic Press; 2000. [Google Scholar]
- Francis R, Read DJ. Direct transfer of carbon between plants connected by vesicular arbuscular mycorrhizal mycelium. Nature. 1984;307:53–56. [Google Scholar]
- Gaspar L, Pollero R, Cabello M. Partial purification and characterization of a lipolytic enzyme from spores of the arbuscular mycorrhizal fungus Glomus versiforme. Mycologia. 1997;89:610–614. [Google Scholar]
- Gaspar ML, Pollero RJ, Cabello MN. Triacylglycerol consumption during spore germination of vesicular-arcuscular mycorrhizal fungi. J Am Oil Chem Soc. 1994;71:449–452. [Google Scholar]
- Gerdemann JW, Nicolson JH. Spores of mycorrhizal Endogone species extracted from soil by wet-sieving and decanting. Trans Br Mycol Soc. 1963;46:235–244. [Google Scholar]
- Graham JH. Assessing costs of arbuscular mycorrhizal symbiosis in agroecosystems. In: Podila GK, Douds DD, editors. Current Advances in Mycorrhizae Research. St. Paul, MN: APS Press; 2000. pp. 127–140. [Google Scholar]
- Graves JD, Watkins NK, Fitter AH, Robinson D, Scrimgeour C. Intraspecific transfer of carbon between plants linked by a common mycorrhizal network. Plant Soil. 1997;192:153–159. [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard B, Lederer F. The “B5-Like” domain from chicken-liver sulfite oxidase: a new case of common ancestral origin with liver cytochrome B5 and bakers' yeast cytochrome B2 core. Eur J Biochem. 1977;74:181–190. doi: 10.1111/j.1432-1033.1977.tb11379.x. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Distel B, Tabak HF. Import of proteins into peroxisomes. BBA Mol Cell Res. 1999;1451:17–34. doi: 10.1016/s0167-4889(99)00087-7. [DOI] [PubMed] [Google Scholar]
- Ho I, Trappe JM. Translocation of 14C from Festuca plants to their endomycorrhizal fungi. Nature. 1973;244:30–31. doi: 10.1038/newbio244030a0. [DOI] [PubMed] [Google Scholar]
- Jabaji-Hare S. Lipid and fatty acid profiles of some vesicular-arbuscular mycorrhizal fungi: contribution to taxonomy. Mycologia. 1988;80:622–629. [Google Scholar]
- Jakobsen J, Abbot LK, Robson AD. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. 1. Spread of hyphae and phosphorus inflow into roots. New Phytol. 1992;120:371–379. [Google Scholar]
- Kamisaka Y, Noda N. Intracellular transport of phosphatidic acid and phosphatidilcholine into lipid bodies in an oleaginous fungus, Mortierella ramanniana var. angulispora. J Biochem. 2001;129:19–26. doi: 10.1093/oxfordjournals.jbchem.a002831. [DOI] [PubMed] [Google Scholar]
- Lammers P, Jun J, Abubaker J, Arreola R, Gopalan A, Bago B, Hernández-Sebastiá C, Allen JW, Douds DD, Pfeffer PE, Shachar-Hill Y. The glyoxylate cycle in an arbuscular mycorrhizal fungus. Carbon flux and gene expression. Plant Physiol. 2001;127:1287–1298. [PMC free article] [PubMed] [Google Scholar]
- Mannaerts GP, vanVeldhoven PP. Functions and organization of peroxisomal beta-oxidation, peroxisomes: biology and role in toxicology and disease. Proc NY Acad Sci. 1996;804:99–115. doi: 10.1111/j.1749-6632.1996.tb18611.x. [DOI] [PubMed] [Google Scholar]
- Mosse B. The stablishment of vesicular-arbusculer mycorrhiza under aseptic conditions. J Gen Microbiol. 1962;27:509–520. doi: 10.1099/00221287-27-3-509. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. Storage lipid bodies in plants and other organisms. Prog Lipid Res. 1991;29:299–324. [PubMed] [Google Scholar]
- Napier JA, Sayanova O, Sperling P, Heinz E. A growing family of cytochrome b(5)-domain fusion proteins. Trends Plant Sci. 1999;4:2–4. [Google Scholar]
- Pfeffer PE, Douds DD, Bécard G, Shachar-Hill Y. Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol. 1999;120:587–598. doi: 10.1104/pp.120.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Fitter AH. The magnitude and control of carbon transfer between plants linked by a common mycorrhizal network. J Exp Bot. 1999;50:9–13. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG. Partitioning of intermediate carbon metabolism in VAM colonized leek. Plant Physiol. 1995;108:7–15. doi: 10.1104/pp.108.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. London: Academic Press; 1997. , 605 pp. [Google Scholar]
- Solaiman MD, Saito M. Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol. 1997;136:533–538. doi: 10.1046/j.1469-8137.1997.00757.x. [DOI] [PubMed] [Google Scholar]
- Sperling P, Schmidt H, Heinz E. A cytochrome-b5-containing fusion protein similar to plant acyl lipid desaturases. Eur J Biochem. 1995;232:798–805. [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus G. intraradices in an in vitro system in the absence of host roots. Mycol Res. 1996;100:328–332. [Google Scholar]
- Sward RJ. The structure of the spores of Gigaspora margarita: III. Germ-tube emergence and growth. New Phytol. 1981;88:667–673. [Google Scholar]
- Thieringer R, Kunau WH. The β-oxidation system in catalase-free microbodies of the filamentous fungus Neurospora crassa: purification of a multifunctional protein possessing 2-enoyl-CoA hydratase, l-3-hydroxyacyl-CoA dehydrogenase, and 3-hydroxyacyl-CoA epimerase activities. J Biol Chem. 1991;266:13110–13117. [PubMed] [Google Scholar]
- Timonen S, Smith FA, Smith SE. Microtubules of mycorrhizal fungus Glomus intraradices in symbiosis with tomato roots. Can J Bot. 2001;79:307–313. [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Watrud LS, Heithaus JJ, III, Jaworski A. Geotropism in the endomycorrhizal fungus Gigaspora margarita. Mycologia. 1978;70:274–277. [Google Scholar]
- Williams RM, Piston DW, Webb WW. Two-photon molecular excitation provides intrinsic 3-dimensional resolution for laser-based microscopy and microphotochemistry. FASEB J. 1994;8:804–813. doi: 10.1096/fasebj.8.11.8070629. [DOI] [PubMed] [Google Scholar]
- Xia ZX, Mathews FS. Molecular structure of flavocytochrome b2 at 2.4 A resolution. J Mol Biol. 1990;212:837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]
- Xu C, Webb WW. Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J Opt Soc Am B. 1996;13:481–491. [Google Scholar]
- Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological non-linear microscopy. Proc Natl Acad Sci USA. 1996;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]