Abstract
An apparently transient infection by a superimposed torquetenovirus (TTV) in a subject who already carried three different genotypes of the virus is described. The superinfection induced a rapid increase in the plasma TTV load and a decline in immunocomplexed virus. The superinfecting TTV was a novel group 2 genotype.
Anelloviruses are a vast group of unenveloped viruses with a small negative-sense single-stranded circular DNA genome. Due to the lack of tissue culture and serological methods, diagnosis relies exclusively on viral DNA detection. Genome sequencing has shown that anelloviruses are widely diverse. Indeed, based on their heterogeneity, anelloviruses are currently classified into two, possibly three species, each subdivided into numerous genotypes. Thus, torquetenovirus (TTV), the first anellovirus species identified (11), is currently subdivided into approximately 40 genotypes, which cluster in five clearly distinct phylogenetic groups that are designated 1 to 5 (2, 7, 11-13).
One major feature of anelloviruses is their ability to produce chronic viremia. In fact, >80% of people harbor variably high viral loads (VL) of TTV in plasma, regardless of geographic origin, age, and health conditions, and quite often such VL consist of several genetic forms of the virus (1). The present study is the first, to our knowledge, to show that a sudden transient increase of plasma VL can be sustained by an apparently short-lived superinfection by a TTV different from the ones already present in the host.
Starting observation.
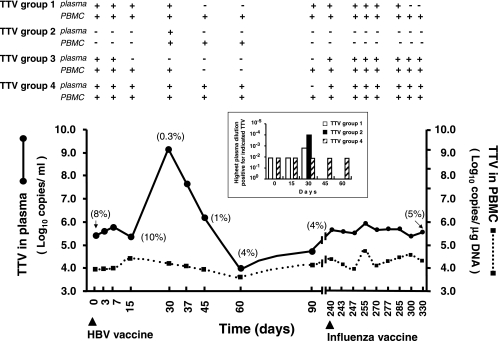
During studies of the consequences of routine immunizations on TTV viremia using a universal quantitative real-time PCR (9), one study subject (MON) manifested a 3.75-log increase in plasma VL, along with the development of mild flu-like symptoms at 30 days after hepatitis B vaccination. The increase was followed by a rapid decline that brought VL first to a brief dip and then back to prevaccination levels (ca. 5.5 × 106 DNA copies per ml). Subsequently, despite immunization with a second vaccine, the VL remained stable over the remaining observation period (Fig. 1). Because the transient increase of TTV replication observed in MON was much greater than that seen in the rest of the vaccinated study group (0.4 ± 0.1 log10, mean ± the standard deviation), we considered it worthy of further investigation.
FIG. 1.
Dynamics of TTV in the plasma and PBMC of the study subject. The arrowheads under the time scale indicate administration of HBV (day 0) and influenza vaccines (day 240). In parentheses, the percent TTV immunocomplexed is shown. At the top, the results of the group-specific PCRs, performed on selected samplings, are shown. The group 2-specific PCR reported was the one with expanded breadth modified to amplify ViPi04, as well as the other genotypes in the group. The results with the group 5-specific PCR are not shown because no sample reacted positively throughout the observation period. The inset shows semiquantitative data obtained by examining plasma collected at the times indicated with group-specific PCRs.
In-depth analysis of plasma viremia in the study subject.
The TTV(s) carried by MON was further investigated by amplifying plasma with five distinct PCRs, each specific for one of the TTV groups, and then typed by sequencing the resulting amplicons exactly as described previously (10). At the start of the study, MON was found to be infected with three distinct TTVs that belonged to genotype 6 of group 1 (80% nucleotide identity), genotype 27 of group 3 (79% identity), and genotype 21 of group 4 (97% identity). Importantly, this TTV profile was consistent throughout the observation period, except for the occasional finding of undetectable levels of one or another genotype and for a 2.5-month eclipse of genotype 27 around the time of the VL peak (Fig. 1), changes that were most likely due to fluctuations in the respective VL. Furthermore, in quadruplicate analyses of 10-fold dilutions of the plasma samplings obtained just before and at VL peak using group 1- and 4-specific PCRs (a similar analysis of group 3 TTV could not be performed due to the previously described undetectability of this genogroup in such specimens) showed that the preexisting TTVs had increased only by approximately 1 log (genotype 6) or not at all (genotype 21), thus failing to explain the large VL increase observed. Selected plasma samples were also examined for immunocomplexed TTV by immunoprecipitation (8). Briefly, plasma was mixed with 4 volumes of a 1:10 dilution of rabbit anti-human immunoglobulin G (Dade Behring, Marburg, Germany), incubated at 4°C overnight, and centrifuged at 14,000 × g for 10 min. The precipitate, washed once and resuspended in 140 μl of sterile saline, and the supernatant were then stored at −80°C until tested for TTV content. The proportion of TTV immunocomplexed was 8 to 10% at baseline, decreased dramatically in the VL peak and immediately subsequent samplings, and then stabilized at 4 to 5% (Fig. 1).
Identification of a novel group 2 TTV genotype.
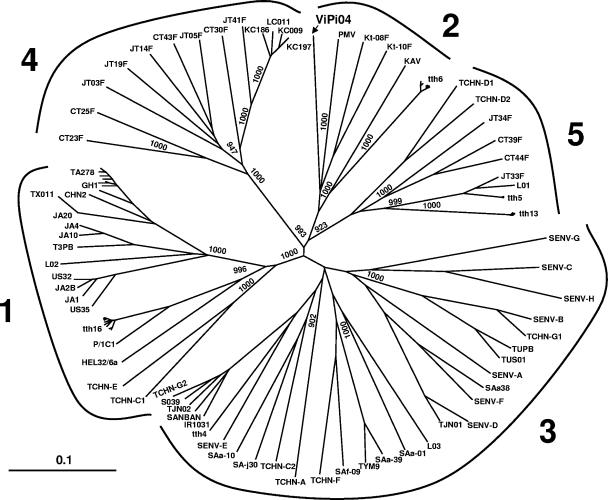
Collectively, the findings presented above suggested that the cause of the plasma VL burst observed in MON was superinfection by a TTV amplified by universal PCR but not by group-specific PCRs. To verify this possibility, we sequenced the TTV dominant in plasma at peak VL (day 30) using various sets of primers designed to this purpose (Table 1). This led us to identify a novel TTV genotype, designated ViPi04, which segregated within group 2 but diverged 35 to 39% from the other viruses in this group, thus representing the first member of a distinct group 2 genotype (Fig. 2). We used the ViPi04 sequence to design new primers for the group 2-specific PCR (Table 1), so that it detected ViPi04 as effectively as the others in the group and yet had a specificity and sensitivity similar to the original assay (4,000 DNA copies per ml of plasma), and then retested MON plasma samples with this improved assay. The only positive plasma sample was the one at peak VL, indicating that ViPi04 had persisted briefly in plasma (Fig. 1). However, semiquantitative analyses showed that, at this time, ViPi04 was by far the dominant genotype, exceeding genotypes 6 and 21 by at least 1 and 2 logs (Fig. 1, inset), respectively, and that it mostly circulated as free virus, being much less immunocomplexed than the other TTVs (data not shown).
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′-3′) | Position (nt)a | PCR product length (bp) |
|---|---|---|---|
| ViPi04 amplificationb | |||
| PCR 1 | |||
| CLONS | AGTTTTCCACGCCCGTCCGC | 114-133 | |
| P1AS | ACGCACTCCTCGCTTACGCTCG | 3075-3095 | |
| UNIVSc | TCAAGGGGCAATTCGGGCT | 205-223 | |
| P3ASc | CTTACGCTCGGAGTGCTTAGTG | 3063-3083 | 2,879 |
| PCR 2 | |||
| MONS1 | TATCACCTACACGCCCCCATTATCCA | 2781-2806 | |
| MONAS2 | TCGCCTCCATCTCCATCGCCAC | 647-668 | |
| MONS3c | GACTGGAAAGAGGAACAGGATG | 2870-2891 | |
| MONAS4c | TTGGGAGCGGGCAGAGCGGG | 550-569 | 1,474 |
| ViPi04 sequencing | |||
| MONSEQ1 | CTGGAGATGGTGGTGGAGACGC | 617-638 | |
| MONSEQ2 | GCTTTCTTCTTCTGAGGTTTGCC | 2626-2648 | |
| MONSEQ3 | AAAGACACTCAAATAACAAG | 1149-1168 | |
| MONSEQ4 | AAATCTAAAGACTATGGGT | 2174-2192 | |
| MONS5 | CGTCACGGCAGCCATTTTA | 3407-3425 | |
| MONAS6 | TACAGCCCAGGAAATGAATC | 54-73 | |
| Group 2 specificd | |||
| G2new1 | TAGACGCCCCAGAGTAAGGAGA | 719-740 | |
| G2new2 | AGTTTTGTTGGTGAAATATGC | 2206-2226 | |
| G2tth2 | GTGAAATAGGCAGATGTACCA | 2196-2216 | |
| G2new3 | CCTGCGGTTTCCGTTCTG | 1361-1378 | 866e |
Nucleotide (nt) positions are according to isolate ViPi04.
Overlapping PCR fragments encompassing the entire sequence of isolate ViPi04 were obtained by two distinct PCR reactions.
Primers also used for sequencing isolate ViPi04.
Genogroup 2-specific PCR was performed by using G2new1, G2new2, and G2tth2 in the first step and G2new3, G2new2, and G2tth2 in the second step, respectively.
Size of PCR products ranging from 856 to 866 bp depending on the binding of G2tth2 or G2new2 to the target sequence, respectively.
FIG. 2.
Phylogenetic analysis of TTV ViPi04 based on full-length or near-full-length sequences. The branching pattern of the trees was obtained by the FastME algorithm included in DAMBE program (version 4.2.13). Bootstrap resampling was used to test the robustness of the tree, and bootstrap values of >900 out of 1,000 replicates are shown at the branch points. The tree was drawn by using Treeview (version 1.6.6). ViPi04 (indicated in boldface; accession number DQ361268) is 3,774 nucleotides in size and has a genetic organization (i.e., UTR length, presence of conserved motifs, number and presumed transcriptional profile of open reading frames) similar to what reported for other group 2 TTV isolates (5). The numbers in boldface around the tree indicate TTV genogroups according to Biagini et al. (2). The scale bar represents a 10% genetic difference.
Analysis of PBMC.
Since frozen peripheral blood mononuclear cells (PBMC) were available for most MON samplings, we examined how plasma viremia changes had impacted the TTV contents of these cells. As shown by Fig. 1, the PBMC VL fluctuated very moderately throughout the observation period. Moreover, TTVs detected mirrored only partly those in plasma: in particular, the undetectability of genotype 27 around the time of peak VL lasted less than in plasma, and ViPi04 was detected not only at day 30 but also at days 45 and 60.
Prevalence of group 2 TTV in the population with the upgraded PCR.
The redesigned group 2-specific PCR was used to examine stored plasma from 48 adults, among healthy individuals and patients with various diseases, and 11 pairs of nasal swabs and plasma from children with acute respiratory disease, all known to be TTV positive. The only one proving to be group 2 positive was one nasal swab, and this, on sequencing, yielded a TTV clearly distinct from ViPi04 and 98% identical to the previously described PM virus isolate (5).
Conclusions.
This report describes an apparently self-limited infection by a superimposed TTV genotype in an individual who already carried three different genetic forms of the virus. The superinfection showed up as a rapid increase in plasma VL, and the superinfecting virus was recognized as a novel group 2 TTV genotype, designated ViPi04. Studies (3-5, 10) have shown that group 2 TTV is much less common than other TTV groups (e.g., 35-fold relative to group 3 in reference 10). Testing clinical specimens with group 2-specific PCR primers redesigned to detect the new, as well as the other, genotypes in the group showed that ViPi04 is as uncommon as other group 2 TTV genotypes. The reasons for the widely uneven prevalence rates for the five TTV groups are unknown, but a clear possibility is that, at least in part, they reflect differences in the ability to persist. Thus, it may not be a coincidence that ViPi04 was detected in the study subject plasma only very briefly. PBMC, the only other specimen of the subject available, tested ViPi04 positive for only 15 additional days, possibly indicating complete eradication of the superinfecting genotype. The circumstance that the subject was already carrying three different genotypes and—as revealed by the substantial aliquot of the preexisting VL that was immunocomplexed—possessed antibodies to these viruses may have helped clearing the superimposed virus. That preformed antibodies may be more effective at controlling fresh than established infection is well known (6). The serological relationships between TTV groups are still completely unexplored.
REFERENCES
- 1.Bendinelli, M., M. Pistello, F. Maggi, C. Fornai, G. Freer, and M. L. Vatteroni. 2001. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of man. Clin. Microbiol. Rev. 14:98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biagini, P., D. Todd, M. Bendinelli, S. Hino, A. Mankertz, S. Mishiro, C. Niel, H. Okamoto, S. Raidal, B. W. Ritchie, and G. C. Teo. 2004. Anellovirus, p. 335-341. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A Ball (ed.), Virus taxonomy, 8th report of the International Committee for the Taxonomy of Viruses. Elsevier/Academic Press, New York, N.Y.
- 3.Biagini, P., P. Gallian, J. F. Cantaloube, H. Attoui, P. de Micco, and X. de Lamballerie. 2006. Distribution and genetic analysis of TTV and TTMV major phylogenetic groups in French blood donors. J. Med. Virol. 78:298-304. [DOI] [PubMed] [Google Scholar]
- 4.Devalle, S., and C. Niel. 2004. Distribution of TT virus genomic groups 1-5 in Brazilian blood donors, HBV carriers, and HIV-1-infected patients. J. Med. Virol. 72:166-173. [DOI] [PubMed] [Google Scholar]
- 5.Hallett, R. L., J. P. Clewley, F. Bobet, P. J. McKiernan, and C. G. Teo. 2000. Characterization of a highly divergent TT virus genome. J. Gen. Virol. 81:2273-2279. [DOI] [PubMed] [Google Scholar]
- 6.Hangartner, L., R. M. Zinkernagel, and H. Hengartner. 2006. Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol. 6:231-243. [DOI] [PubMed] [Google Scholar]
- 7.Hino, S. 2002. TTV, a new human virus with single stranded circular DNA genome. Rev. Med. Virol. 12:151-158. [DOI] [PubMed] [Google Scholar]
- 8.Itoh, Y., M. Takahashi, M. Fukuda, T. Shibayama, T. Ishikawa, F. Tsuda, T. Tanaka, T. Nishizawa, and H. Okamoto. 2000. Visualization of TT virus particles recovered from the sera and feces of infected humans. Biochem. Biophys. Res. Commun. 279:718-724. [DOI] [PubMed] [Google Scholar]
- 9.Maggi, F., E. Tempestini, L. Lanini, E. Andreoli, C. Fornai, S. Giannecchini, M. Vatteroni, M. Pistello, S. Marchi, P. Ciccorossi, S. Specter, and M. Bendinelli. 2005. Blood levels of TT virus following immune stimulation with influenza or hepatitis B vaccine. J. Med. Virol. 75:358-365. [DOI] [PubMed] [Google Scholar]
- 10.Maggi, F., E. Andreoli, L. Lanini, C. Fornai, M. Vatteroni, M. Pistello, S. Presciuttini, and M. Bendinelli. 2005. Relationships between total plasma load of torquetenovirus (TTV) and TTV genogroups carried. J. Clin. Microbiol. 43:4807-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto, H., and M. Mayumi. 2001. TT virus: virological and genomic characteristics and disease associations. J. Gastroenterol. 36:519-529. [DOI] [PubMed] [Google Scholar]
- 13.Peng, Y. H., T. Nishizawa, M. Takahashi, T. Ishikawa, A. Yoshikawa, and H. Okamoto. 2002. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch. Virol. 147:21-41. [DOI] [PubMed] [Google Scholar]