Abstract
The studies that correlate the results obtained by different typing methodologies rely solely on qualitative comparisons of the groups defined by each methodology. We propose a framework of measures for the quantitative assessment of correspondences between different typing methods as a first step to the global mapping of type equivalences. A collection of 325 macrolide-resistant Streptococcus pyogenes isolates associated with pharyngitis cases in Portugal was used to benchmark the proposed measures. All isolates were characterized by macrolide resistance phenotyping, T serotyping, emm sequence typing, and pulsed-field gel electrophoresis (PFGE), using SmaI or Cfr9I and SfiI. A subset of 41 isolates, representing each PFGE cluster, was also characterized by multilocus sequence typing (MLST). The application of Adjusted Rand and Wallace indices allowed the evaluation of the strength and the directionality of the correspondences between the various typing methods and showed that if PFGE or MLST data are available one can confidently predict the emm type (Wallace coefficients of 0.952 for both methods). In contrast, emm typing was a poor predictor of PFGE cluster or MLST sequence type (Wallace coefficients of 0.803 and 0.655, respectively). This was confirmed by the analysis of the larger data set available from http://spyogenes.mlst.net and underscores the necessity of performing PFGE or MLST to unambiguously define clones in S. pyogenes.
Typing methods are major tools for the epidemiological characterization of bacterial pathogens, allowing the determination of the clonal relationships between isolates based on their genotypic or phenotypic characteristics. Recent technological advances have resulted in a shift from classical phenotypic typing methods, such as serotyping, biotyping, and antibiotic resistance typing, to molecular methods such as restriction fragment length polymorphism (8), pulsed-field gel electrophoresis (PFGE) (25), and PCR serotyping (4). With the availability of affordable sequencing methods, another shift occurred towards sequence-based typing methods such as multilocus sequence typing (MLST) (18) and emm sequence typing (2). Sequence-based methods have a wide appeal since they provide unambiguous data and are intrinsically portable, allowing the creation of databases that, if publicly available through the internet, enable the comparison of local data with those of previous studies in different geographical locations. Ideally, an analysis of each typing method, in terms of discriminatory power, reproducibility, typeability, feasibility, and other characteristics as suggested by Struelens (31), should be performed to better determine which method is appropriate in a given setting.
Several molecular epidemiology studies of clinically relevant microorganisms provide a characterization of isolates based on different typing methods (6, 8, 20, 23). Frequently these studies focus on a comparison between the assigned types of different typing methods, from a qualitative point of view, i.e., indicating correspondences between the types of the different methods. Although this may be useful for the comparison of the genetic backgrounds of the particular set of isolates under study, it does not allow for a broader view of how the results of the different typing methods are related.
As more bacterial genomes are completed, novel typing methods will appear based on the new information available. Comparisons of these new methods to those currently available should be complemented by a quantitative measure of how much information is gained from a new method in terms of discriminatory power, type assignment, or even phylogenetic information about the isolates. It is conceivable that less-sophisticated molecular methods can recover levels of information about the relationships between the isolates that are similar to those obtained with newer sequence-based methods. Since typing schemes analyze different phenotypic or genotypic properties of bacteria, if some congruence between the methods is found, it suggests that a phylogenetic signal is being recovered by both methods, allowing greater confidence about the evolutionary hypothesis or clonal dispersion of the strains under study. These quantitative comparisons should allow the informed choice of which typing method is more appropriate in a given clinical or microbiological research setting, also taking into account other factors, such as the ability to identify isolates of interest, execution time, cost-effectiveness, or ease of interpretation of the results. A great diversity of typing methods is used to characterize bacterial isolates, rendering the comparison of the various studies difficult. If one could infer the missing information from the available data provided in each study, one could overcome this problem. In order to do this, a method offering a quantitative assessment of the confidence of predicting an unknown character from another typing method or set of methods is needed.
Streptococcus pyogenes or group A streptococci (GAS) are known to cause infections ranging from mild manifestations, such as pharyngitis, to severe invasive infections, such as streptococcal toxic shock syndrome and necrotizing fasciitis (7). These human pathogens provide a good case study for mapping relationships between typing schemes, since multiple typing methods have been used in their characterization, including T and M serotyping, antibiotic resistance typing, PFGE, restriction fragment length polymorphism (vir typing), emm sequence typing, and MLST (9, 17, 21). Although all these methods have proven useful for the characterization of GAS isolates, phenotypic methods have declined in popularity and the mainstream methods are now emm sequence typing, PFGE, and MLST. In the study that defined the MLST scheme for GAS, the authors compared MLST and emm sequence typing and concluded that the majority of emm types define clones or clonal complexes (9). This conclusion, together with the existence of extensive data on serological M types that are directly comparable to emm sequence type data and a technically simpler and more economic determination of emm types as opposed to the characterization by MLST, led to the frequent use of emm typing as the main typing technique for GAS clone definition. Notwithstanding the advantages of sequence-based methods, GAS virulence has been related to the presence of phages and to horizontal transfer of large fragments of DNA (1, 32). These observations suggest that techniques, like PFGE, that probe genomic organization could be more discriminative than sequence-based methods, since phage insertions can alter band positions in an agarose gel and, consequently, create more diversity within PFGE types.
In this paper we propose the use of measures of clustering concordance—Adjusted Rand (15, 24) and Wallace (34) coefficients—to compare type assignments, allowing a quantitative approach for exploring the concordance between typing methods. The proposed methods were applied to a set of 325 macrolide-resistant GAS for which extensive typing information was available and, when possible, we generalized the conclusions based on this data set by using typing data available from the MLST database. The proposed framework also allows the evaluation of possible gains in discriminatory power obtained by using different methods or any combination of typing schemes and the identification of which of the typing methodologies used will be more informative in clone definition. Ultimately, this framework may allow a mapping of type equivalences between typing methods.
MATERIALS AND METHODS
Strain collection.
A collection of 325 macrolide-resistant S. pyogenes isolates recovered from throat swabs associated with a diagnosis of tonsillopharyngitis, from the period between 1998 and 2003 in Portugal, was analyzed. Results of antimicrobial susceptibility testing, T typing, macrolide-resistant phenotyping and genotyping, and emm typing were reported previously (29). Eleven T types were identified (1, 2, 4, 5/27/44, 6, 9, 12, 13, 25, 28, and B3264). Since twelve isolates were nontypeable by this method, the typeability of this method in our collection was 97%. All isolates were analyzed by PFGE using SfiI and either SmaI or Cfr9I endonucleases. Twelve emm sequence types were identified (1, 2, 4, 6, 9, 11, 12, 22, 28, 75, 77, and 89). Forty-one strains were chosen for MLST analysis by selecting at least one isolate from each SmaI/Cfr9I cluster. Ten sequence types (ST) were found (20, 28, 36, 38, 39, 45, 46, 52, 75, and 150) (28).
Gel analysis.
A database of PFGE patterns was created in Bionumerics version 4.5 from Applied Maths (Sint-Martens-Latem, Belgium).
The gel digital photos acquired and stored in a Kodak EDAS 290 system were imported into a Bionumerics database, as inverted 8-bit grayscale TIF images.
For each image, spectral analysis included in the software was used, to determine the disk size that should be used in “rolling disk” background subtraction (Background scale) and the cutoff threshold for least-squares filtering (Wiener cutoff scale). Furthermore, a median filter was used in the image to further smooth the densitometric curves.
After this image preprocessing, intergel and intragel normalizations of PFGE runs were done using a Lambda PFGE molecular marker (New England Biolabs, Ipswich, Ma.). All the gels had three markers in the first, middle, and last lanes. Ten lambda bands were used from 48.5 kb to 485 kb.
On all gel images, band assignment was manually curated after automatic band detection. Bands ranging from 22.8 kb to 608 kb were considered in this study.
The settings used for comparing the strains' PFGE patterns were 1.0% optimization and 1.5% band tolerance.
Diversity indices.
Hunter and Gaston (16) proposed the use of Simpson's index of diversity (30) to measure the discriminatory ability of typing systems. This index indicates the probability of two strains sampled randomly from a population belonging to two different types. Grundmann et al. (13) proposed a method for determining confidence intervals (CIs) of Simpson's index, thereby improving the objective assessment of the discriminatory power of typing techniques. The formulas of Simpson's Index (D) and the CI are presented in the following equations:
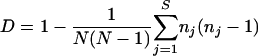 |
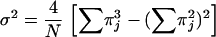 |
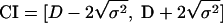 |
where N is the total number of strains in the sample population, S the total number of types described, nj is the number of strains belonging to the jth type, and πj is the frequency nj/N.
Other diversity indices exist, such as the Shannon-Wiener index (27) and others from the Hill family of indices, of which Simpson's index is a special case (14). Notwithstanding, the ease of interpretation of Simpson's index of diversity as a probability and the possibility of calculating a confidence interval justifies the choice of this index in our study.
Clustering comparison coefficients: Rand, Adjusted Rand, and Wallace.
In molecular epidemiology studies, the term cluster is frequently used to describe a group of isolates sharing similar characteristics according to a given typing method. Frequently, the clusters are obtained by hierarchical methods, such as the unweighted-pair group method with arithmetic means (UPGMA), providing further detail on the relationships of the isolates within clusters. In these cases, the definition of types relies on partitioning the resulting dendrogram at a given similarity value. In this paper, the terms partition, cluster, and type will be used interchangeably to identify a group of isolates sharing similar characteristics according to a given typing method.
To compare two sets of results of different microbial typing methods, an objective measure of agreement is needed. Several measures were developed for comparing two sets of partitions (10, 15, 22, 24, 34), taking different approaches to how partitions should be compared. For their ease of interpretation, in this study we use Adjusted Rand's index and Wallace coefficient. Rand (24) and Adjusted Rand (15) are symmetric coefficients, i.e., they do not take into consideration which partition is considered the standard, while others, like the ones proposed by Wallace (34), do. It is also important to note in this context that none of the partitions tested are considered the “correct” partition in terms of microbial typing.
Given two partition schemes of the same data set, P and P′, all these coefficients are calculated based on the fact that a pair of points (in microbial typing, a pair of points is a pair of isolates under study) from the data set will fall into one of the following conditions: a, the number of point pairs that are in the same cluster in P and P′; b, the number of point pairs that are in the same cluster in P but not in P′; c, the number of point pairs that are in the same cluster in P′ but not in P; or d, the number of point pairs that are in different clusters in P and P′.
The coefficients can then be defined as shown in Table 1.
TABLE 1.
Clustering comparison coefficients: Rand, Adjusted Rand, and Wallace
| Coefficient | Formula (s)a |
|---|---|
| Rand | 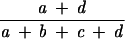 |
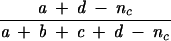 |
|
| Adjusted Rand | 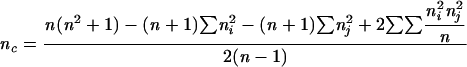 |
| Wallace | 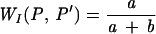 |
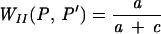 |
See the text for definitions of a, b, c, and d; n represents the number of strains under study, ni and nj represent the numbers of strains in types i and j of partitions P and P′, respectively.
Rand's index represents the proportion of agreement for both matches (a) and mismatches (d). An acknowledged limitation of this coefficient is that, when comparing two random partitions, the expected value of the Rand's index does not take a null value (indicating nonagreement). To address this issue, Hubert and Arabie (15) assumed a hypergeometric distribution as the random model, adding a correction factor designed to take into account the presence of chance agreement. The Hubert and Arabie's Adjusted Rand index, here referred to simply as Adjusted Rand, allows a better quantitative evaluation of the global congruence between the two partitions.
Wallace proposed two coefficients, based on Fowlkes and Mallows ' coefficient (10). They are easy to interpret since they represent the probability that a pair of points which are in the same cluster under P are also in the same cluster under P′ and vice versa.
Wallace's coefficients provide an estimate of, given a typing method, how much new information is obtained from another typing method. A high value of Wallace's coefficient indicates that partitions defined by a given method could have been predicted from the results of another method, suggesting that the use of both methodologies is redundant.
To facilitate the use of these indices in studies conducted by others, we have made available a Bionumerics script that calculates these indices from any two sets of data generated by different typing methods. The script can be downloaded from http://biomath.itqb.unl.pt/ClusterComp.
Visual representation of cluster congruence.
To facilitate the interpretation and representation of the comparisons between partitions, we developed a visual method where all the clusters and cases under comparison are represented in a figure similar to a sequence dot plot (12, 19). The strains are ordered by type and cluster size for each of the typing methods under comparison. A dot is then plotted at the intersection of the position of each strain. Vertical and horizontal lines delimit the clusters in the figure.
Examples of this visual representation are shown in Fig. 2D and in Fig. SA1 in the supplemental material.
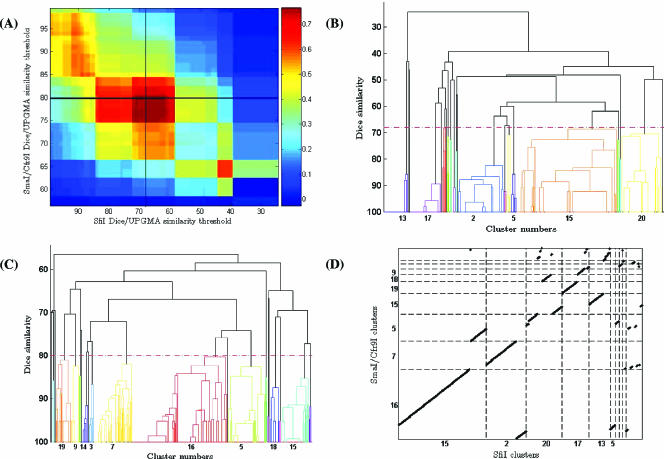
FIG. 2.
Panel A. Adjusted Rand values for all possible cutoff values in each of the SmaI/Cfr9I and SfiI dendrograms (in panels B and C). Panel B. Dice/UPGMA dendrogram for SfiI band patterns of the 325 isolates. Panel C. Dice/UPGMA dendrogram for SmaI/Cfr9I band patterns of the 325 isolates. Panel D. Visual representation of cluster congruence between SmaI/Cfr9I and SfiI clusters at the cutoff levels indicated in panels B and C.
RESULTS
Pulsed-field gel electrophoresis.
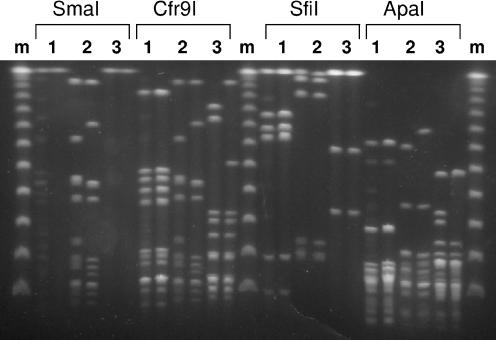
For this study, 325 isolates of S. pyogenes were analyzed by PFGE, using two restriction enzymes: SfiI and either one of two isoschizomers—SmaI or Cfr9I. The use of Cfr9I was necessary since the majority of the 150 isolates presenting macrolide resistance phenotype M were refractory to cleavage with SmaI, in agreement with previous studies (21). As expected, DNA of isolates that were digested with SmaI presented identical band patterns when digested with Cfr9I (Fig. 1). In this way the use of Cfr9I increased the typeability of PFGE to 100%, compared to 54% (175/325) estimated for the use of SmaI alone, in our collection.
FIG. 1.
PFGE profiles of three pairs of macrolide-resistant isolates generated after digestion with either SmaI, Cfr9I, SfiI, or ApaI. The isolates in pair 1 were refractory to cleavage with SmaI similarly to what was previously reported in the literature for isolates presenting the M phenotype (21) and showed identical profiles upon digestion with Cfr9I, SfiI, and ApaI. The isolates in pair 2 were digested by both SmaI and Cfr9I and, as expected, the profile of each isolate was identical with either endonuclease. The isolates in pair 2 showed different SmaI/Cfr9I and ApaI profiles but were indistinguishable by SfiI. The isolates in pair 3 were refractory to cleavage with SmaI and presented different profiles upon digestion with Cfr9I (four-band difference). These isolates presented identical SfiI profiles but exhibited different profiles with ApaI. m, lambda ladder PFG marker (New England Biolabs, Beverly, Ma.).
In previous studies, SfiI is the endonuclease most frequently used to characterize M isolates by PFGE (21), although a few studies use ApaI (3). There was a concern that SfiI patterns did not allow sufficient discrimination, since these patterns had only 3 to 10 bands, compared to SmaI/Cfr9I patterns that presented 8 to 17 bands (Fig. 1). This hypothesis was tested using Simpson's index of diversity and Adjusted Rand to determine the threshold that better defines types compared to those defined by SmaI/Cfr9I.
The Adjusted Rand index was calculated for each possible combination of partitions given by varying the threshold cutoffs for each UPGMA/Dice dendrogram using SfiI and SmaI/Cfr9I endonucleases similarly to what was previously described (5). The threshold value that produced the maximum coefficient value was determined, and the results are displayed in Fig. 2.
The maximum value of Adjusted Rand of 0.771 was found at a threshold level of 77% similarity in the Dice/UPGMA dendrogram of SmaI/Cfr9I and at a 68% similarity in the SfiI dendrogram.
An 80% similarity value was previously shown to be useful and concordant with proposed visual comparison criteria when defining types by Dice/UPGMA dendrograms of SmaI profiles of Streptococcus pneumoniae (5, 11, 26, 33). At this commonly used similarity threshold cutoff, a maximum Adjusted Rand value of 0.765 was found, corresponding to a threshold value of 68% in the SfiI dendrogram. Due to its acceptance as the cutoff value to define clusters in SmaI Dice/UPGMA dendrograms and the small difference observed in the value of Adjusted Rand at these two threshold levels, we opted to use the 80% cutoff for SmaI/Cfr9I and the 68% cutoff for SfiI in the remainder of the analysis.
At these cutoff values, 21 clusters were defined by either SmaI/Cfr9I or SfiI. Simpson's index of diversity calculations for the partitions found at these threshold levels for either endonuclease were of the same value of 0.81 (95% CI, 0.78-0.84), resulting in equal discriminatory power for SmaI/Cfr9I and SfiI (Table 2).
TABLE 2.
Number of types and Simpson's index of diversity of each typing method
| Typing method | No. of types found | Simpson's index of diversity (95% CI) | No. of isolates typed (% typeability) |
|---|---|---|---|
| T typing | 12 | 0.72 (0.68-0.77) | 316 (97)a |
| emm typing | 12 | 0.77 (0.74-0.81) | 325 (100) |
| T + emm typing | 33 | 0.81 (0.77-0.84) | 325 (100) |
| SmaI/Cfr9I 80% | 21 | 0.81 (0.78-0.84) | 325 (100) |
| MRPb | 3c | 0.51 (0.50-0.53) | 325 (100) |
| SfiI 68% | 21 | 0.81 (0.78-0.84) | 325 (100) |
| MLST | 10 | —d | 41 (100) |
Twelve strains were nontypeable. For the purpose of calculating the Simpson's index of diversity, they were all considered to belong to the same group.
MRP, macrolide resistance phenotyping.
Three macrolide resistance phenotypes were considered: M, cMLSB, and iMLSB.
—, Simpson's index of diversity was not calculated for the 41 isolates since it would not be comparable to the remaining results calculated for the entire 325-isolate data set.
Comparing typing methods.
Simpson's index of diversity provides a measure of the discriminatory power of the different typing methods as applied to our study data. Table 2 summarizes this coefficient for the methods used: T typing, emm typing, a combination of both these methods, PFGE typing defined at an 80% cutoff value on the SmaI/Cfr9I Dice/UPGMA dendrogram, macrolide resistance phenotyping, PFGE typing defined at a 68% cutoff value on the SfiI Dice/UPGMA dendrogram, and MLST data.
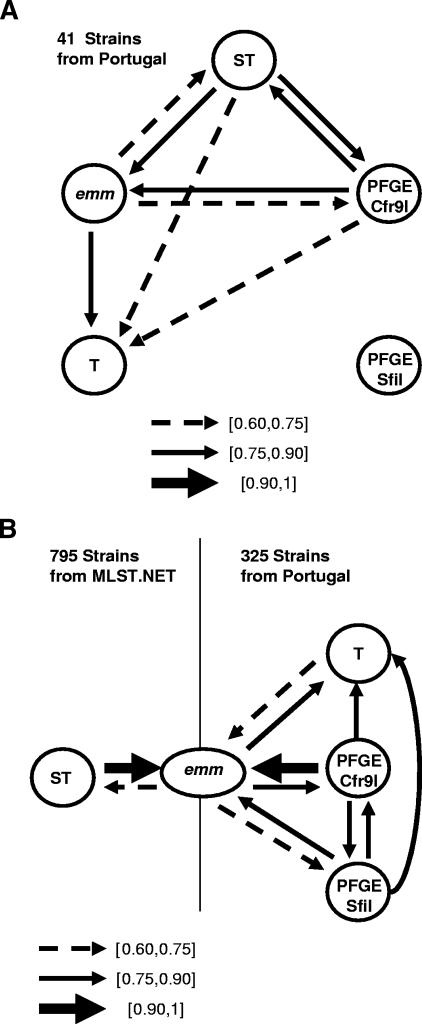
For comparing the congruence between type assignments of the different typing methods, adjusted Rand and Wallace coefficients were calculated for the subset of 41 isolates for which the results of all typing methods were available. These isolates are a diverse collection representing most of the types defined by the various methods in the entire collection of 325 isolates. The results are shown in Fig. 3A and in Tables SA1 and SA2 in the supplemental material. The data indicate a strong correlation between the information provided by SmaI/Cfr9I PFGE, MLST, and emm typing. It is interesting to note that there was a robust bidirectional correspondence between SmaI/Cfr9I PFGE types and ST. In contrast, a strong correspondence was found only in the direction of emm type for both ST and SmaI/Cfr9I PFGE types but not in the reverse direction (Fig. 3A).
FIG. 3.
(A) Representation of correspondences between the typing methods used in the 41-strain subset, calculated by Wallace coefficients. The arrows represent Wallace coefficients of >0.60.
(B) Representation of correspondences between the typing methods used in the 325-strain data set and on the 795 strains from www.mlst.net, calculated by Wallace coefficients. The arrows represent Wallace coefficients of >0.60.
To increase the robustness of the values of the Adjusted Rand and Wallace coefficients for the correspondences between the various typing methods utilized to characterize GAS, we excluded the MLST data and used the entire collection of 325 isolates for which information regarding all other typing methods was available. The results are shown in Tables 3 and 4. The values of Rand and Wallace coefficients were consistently higher when considering the 325 isolates. This was only expected if the smaller data set of 41 isolates already reflected the true correspondences between the various typing methods.
TABLE 3.
Adjusted Rand coefficients for the methods used to characterize the 325 macrolide-resistant GAS
| Typing method | Typing method
|
|||||
|---|---|---|---|---|---|---|
| T typing | emm typing | T + emm typing | MRPa | SmaI/Cfr9I 80% | SfiI 68% | |
| T typing | 1.000 | |||||
| emm typing | 0.693 | 1.000 | ||||
| T + emm typing | 0.768 | 0.905 | 1.000 | |||
| MRPa | 0.268 | 0.462 | 0.397 | 1.000 | ||
| SmaI/Cfr9I 80% | 0.566 | 0.837 | 0.764 | 0.399 | 1.000 | |
| SfiI 68% | 0.514 | 0.724 | 0.663 | 0.382 | 0.762 | 1.000 |
MRP, macrolide resistance phenotyping.
TABLE 4.
Wallace coefficients for the methods used to characterize the 325 macrolide-resistant GAS
| Typing method | Typing method
|
|||||
|---|---|---|---|---|---|---|
| T typing | emm typing | T + emm typing | MRPa | SmaI/Cfr9I 80% | SfiI 68% | |
| T typing | 1.000 | 0.697 | 0.697 | 0.723 | 0.559 | 0.528 |
| emm typing | 0.861 | 1.000 | 0.861 | 0.989 | 0.803 | 0.724 |
| T + emm typing | 1.000 | 1.000 | 1.000 | 0.988 | 0.802 | 0.725 |
| MRPa | 0.415 | 0.459 | 0.395 | 1.000 | 0.392 | 0.386 |
| SmaI/Cfr9I 80% | 0.818 | 0.952 | 0.817 | 1.000 | 1.000 | 0.812 |
| SfiI 68% | 0.764 | 0.848 | 0.731 | 0.972 | 0.802 | 1.000 |
MRP, macrolide resistance phenotyping.
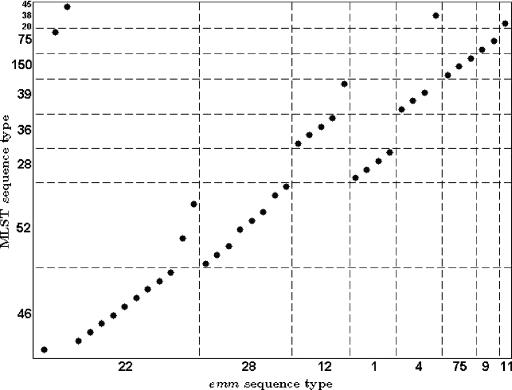
Previous publications indicated that emm types defined clones as assessed by MLST (9). Our limited data suggested a more complex relationship between emm typing and MLST (Fig. 4), resembling the comparison between emm and SmaI/Cfr9I PFGE types for our entire data set (see Fig. SA1 in the supplemental material).
FIG. 4.
Relationship between MLST and emm sequence typing for the 41 strains. In this example, the figure illustrates the fact that the majority of strains with emm type 22, the most frequent emm type in this subset, presented ST 46, and two isolates presented ST 52. However, the majority of the strains with ST 52 have emm sequence type 28.
To better evaluate the discriminatory power of MLST, we calculated Simpson's index of diversity for the 795 strains that had unambiguous information about both ST and emm type in the S. pyogenes MLST database (http://spyogenes.mlst.net; with a total of 847 isolates on 29 August 2005). Likewise, the same index was calculated for emm type to provide a more global view than that afforded by our data set. The results are shown in Table 5. The same could not be done for T types, since only 90 strains had T type information; 23 of those were nontypeable, and the majority of the 67 remaining strains had ambiguous information (two types). The distribution in this data set of emm sequence types per ST, and vice versa, as well as the overall concordance between emm typing and MLST among the strains in the S. pyogenes MSLT database can be found in the supplemental material (Fig. SA2 and SA3).
TABLE 5.
Diversity indices and Wallace coefficients for emm typing and MLST data available from the online MLST database
| Typing method | No. of types found | Simpson's index of diversity (95% CI) | Wallace coefficient
|
|
|---|---|---|---|---|
| MLST | emm typing | |||
| MLST | 274 | 0.985 (0.982-0.988) | 1.000 | 0.952 |
| emm typing | 155 | 0.978 (0.975-0.981) | 0.655 | 1.000 |
The Adjusted Rand for the comparison of the clustering by MLST and emm typing is 0.77, indicating a good overall match between partitions. The Wallace coefficient provides more information: considering ST as the standard for comparison, the value of Wallace's index is 0.952, i.e., the probability of two strains having the same ST also sharing the same emm type is 95%. However, the probability of two isolates that share the same emm type sharing the same ST is only 66% (Wallace's index is 0.655) (Table 5).
The correspondences between the various typing methods defined by using these expanded data sets are graphically represented in Fig. 3B.
DISCUSSION
The primary objective of this report was to provide a framework for the quantitative assessment of correspondence between type assignments obtained by different microbial typing methods. This quantification is achieved by the use of Simpson's index of diversity, Hubert and Arabie's Adjusted Rand, and the Wallace coefficient and is complemented by the visualization of the congruence between partitions generated by different typing methods.
An important application of the proposed framework is in evaluating if clusters generated by a given typing method could have been predicted by another methodology, allowing the evaluation of the usefulness of using several typing methods to characterize the same collection of isolates. This is also important in benchmarking the novel information offered by new typing schemes and in establishing if one can infer unknown typing information for a given isolate from other known characters.
While Simpson's index was previously used in this context (16) and it allows for a measure of the discriminatory power of a typing method, it does not evaluate the degree of equivalence between type assignments of two distinct typing methods. This goal is achieved here by the Adjusted Rand index, which provides an overall measure of the congruence between two typing methods. On the other hand, Wallace's coefficient is more informative and offers a clear interpretation since it represents the probability that a pair of strains which are assigned to the same type by one method are also classified in the same type by the other method. To facilitate the use of these indices in studies conducted by others, we have made available a Bionumerics script that calculates these indices from any two sets of partitions generated by different typing methods. To test the validity of the framework proposed, we applied it to a collection of macrolide-resistant GAS characterized by T serotyping, emm sequence typing, PFGE using two different endonucleases, and MLST.
Although intuitively we could expect that the PFGE band patterns generated after SfiI digestion would be less discriminatory than those of SmaI/Cfr9I, since the former presented fewer bands, this was not the case. The clusters defined by either SmaI/Cfr9I or SfiI, at the cutoff levels showing the highest agreement, showed the same Simpson’s index value, namely, 0.81 (95% CI, 0.78 to 0.84).
In spite of the similar discriminatory powers, SmaI/Cfr9I and SfiI assigned a significant number of isolates to different types (Adjusted Rand, 0.765). The value of the Wallace index was almost the same in either direction and indicated that as many as one out of every five pairs of isolates classified in the same cluster by an endonuclease are in separate clusters using the other endonuclease. This is represented in Fig. 2D, where it can be seen that isolates that belong to a single cluster when using one endonuclease were scattered into at least two other clusters when using the other.
In view of this data, which endonuclease is more suitable for typing macrolide-resistant S. pyogenes? A weaker correspondence between SfiI PFGE and the other typing methods is shown in Fig. 3A, where no line connects SfiI PFGE and T types whereas multiple correspondences are associated with SmaI/Cfr9I PFGE types. When using the full data set of 325 isolates this was not so pronounced, but the correspondences established between SmaI/Cfr9I PFGE and the other typing methods were consistently stronger than those observed for SfiI PFGE (Fig. 3B). We would therefore argue that the groups defined by SmaI/Cfr9I, by being more concordant with other typing methods, are more likely to accurately represent the true relationships between the isolates, justifying the use of these endonucleases in typing GAS isolates by PFGE. The use of the SmaI isoschizomer Cfr9I was essential in increasing the typeability of macrolide-resistant GAS, since the majority of the isolates presenting the M phenotype were refractory to cleavage by SmaI but were susceptible to digestion with Cfr9I (Fig. 1). A combination of the two endonucleases is therefore recommended for the characterization of macrolide-resistant GAS.
If two isolates belong to the same cluster according to SmaI/Cfr9I, one can predict confidently that they will also share the same emm sequence type (Wallace, 0.952). The reverse correspondence, on the other hand, cannot be made with the same certainty since only four out of five pairs of strains sharing the same emm sequence type were also grouped into the same SmaI/Cfr9I cluster (Wallace, 0.803). Even considering that SmaI/Cfr9I PFGE was comparable to emm sequence typing in terms of discriminatory power (Table 2), it was more informative given its predictive power over emm sequence typing (Fig. 3B).
T typing alone, although presenting a Simpson's index of diversity similar to that with emm sequence typing, separated the isolates into groups that could not be mapped confidently in any of the other typing methods, indicating that this methodology is poorly congruent with any of the others (Fig. 3). The combination of T typing and emm sequence typing showed a discriminatory power comparable to that of SmaI/Cfr9I (Table 2), but it did not improve the mapping relative to emm typing alone as demonstrated by Adjusted Rand and Wallace coefficients (Table 3 and Table 4). A drawback of using the combination of these two methods was the reduced typeability of T typing compared with all other typing methods and the necessity of maintaining a set of specific sera for T typing.
When comparing the ST of the 795 isolates referenced in the MLST database, what had started to emerge with our limited data set of 41 isolates became clear: the majority of emm types subdivide themselves into two or more ST (see Fig. SA3 in the supplemental material). This is supported by a Wallace coefficient of 0.655 when comparing emm types using MLST as a standard. In the reverse comparison, a Wallace coefficient of 0.952 implies that when two strains have the same ST we will only misclassify them according to their emm types once out of every 20 pairs. These results show that clone definition by MLST is more consistent than by emm sequence typing.
The correspondences between the various typing methods illustrated in Fig. 3B argue that performing either PFGE using SmaI and Cfr9I endonucleases or MLST is sufficient to predict the emm type of the isolates with less than 5% error but that one cannot accurately predict ST or SmaI/Cfr9I PFGE types from emm data.
A comprehensive comparison between SmaI/Cfr9I PFGE and MLST is outside the scope of this paper. The limited data available from the smaller data set of 41 isolates for which we had MLST information suggested that there is a strong mapping between SmaI/Cfr9I PFGE types and MLST (Fig. 3A; also see Tables SA1 and SA2 in the supplemental material). This was also supported by the similar relationship of each of these methods with emm typing (Fig. 3B). In spite of the role of bacteriophages and of the horizontal exchange of large fragments of genomic DNA in the evolution of virulent GAS strains (1, 32), these observations argued in favor of equally good results when using SmaI/Cfr9I PFGE or MLST to characterize GAS. However, the choice of isolates for which MLST was determined reflects the SmaI/Cfr9I PFGE type assignment, so further studies are necessary to clarify which of the two typing methods would provide a more discriminatory and informative clone definition.
Our data set represented a diverse group of GAS as documented by the use of the various typing methods; for instance, 21 SmaI/Cfr9I PFGE types were defined. However, it could be argued that these isolates do not accurately represent the global diversity of S. pyogenes since they are restricted to macrolide-resistant GAS recovered in Portugal during a limited time period. This would prevent the generalization of the results presented. Although it is certainly true that there was limited diversity in our collection, the expected clonal structure of such a geographically and temporally limited population would reinforce the correspondences between the different typing methods, increasing the values of adjusted Rand and Wallace coefficients. This was not observed for all methods and Wallace coefficients showed strong asymmetries depending on the directionality, suggesting that the results did not reflect a particular clonal composition of the studied population but are general properties of the typing methods used. This was further supported by the analysis of the more extensive data available from the MLST online database that strengthened the conclusions emerging from the study of our data set regarding the relationship between emm typing and MLST.
When using PFGE to characterize macrolide-resistant GAS, the results were in favor of the use of SmaI, complemented by its isoschizomer Cfr9I to circumvent the resistance to cleavage of M isolates as documented previously (21), against the alternative endonuclease SfiI. The analysis also highlighted the importance of using PFGE and MLST, in addition to emm typing, in the characterization of GAS due to the poor predictive value of the latter over the groups defined by the former.
As the data compiled in online databases (such as www.mlst.net) increases, the framework of methods presented will provide further insights into the relationships between isolates, eventually enabling a generic mapping between the different typing methods. The congruence of results between typing methods suggests that a phylogenetic signal is indeed being recovered by the typing data generated by different methods. Accordingly, the progressive identification of mapping functions, such as the probability matrices for agreement of type assignments represented in the supplemental material (Fig. SA4.1 and SA4.2), indicates that a consensus assessment of the relationships between the different types will soon be at hand. Such a tool would allow not only for comparisons of typing results obtained by different methods but would also facilitate the joint analyses of multiple typing methods.
Supplementary Material
Acknowledgments
J. A. Carriço and F. R. Pinto were supported by grants SFRH/BD/3123/2000 and SFRH/BD/6488/2001, respectively, both from the Fundação para a Ciência e Tecnologia, Portugal.
Partial support for this work was provided by PREVIS (LSHM-CT-2003-503413 from the European Community awarded to J. S. Almeida, H. de Lencastre, and J. Melo-Cristino) and by a grant from the Fundação Calouste Gulbenkian awarded to J. Melo-Cristino and M. Ramirez.
This publication made use of the Multi Locus Sequence Typing website (http://www.mlst.net) at Imperial College London developed by David Aanensen and Man-Suen Chan and funded by the Wellcome Trust.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aziz, R. K., R. A. Edwards, W. W. Taylor, D. E. Low, A. McGeer, and M. Kotb. 2005. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., R. R. Facklam, J. A. Elliott, A. R. Franklin, T. Hoenes, D. Jackson, L. Laclaire, T. Thompson, and R. Viswanathan. 1998. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J. Med. Microbiol. 47:893-898. [DOI] [PubMed] [Google Scholar]
- 3.Billal, D. S., M. Hotomi, K. Yamauchi, K. Fujihara, S. Tamura, K. Kuki, R. Sugita, M. Endou, J. Mukaigawa, and N. Yamanaka. 2004. Macrolide-resistant genes of Streptococcus pyogenes isolated from the upper respiratory tract by polymerase chain reaction. J. Infect. Chemother. 10:115-120. [DOI] [PubMed] [Google Scholar]
- 4.Brito, D. A., M. Ramirez, and H. de Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carriço, J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazão, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coenye, T., T. Spilker, A. Martin, and J. J. LiPuma. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 13:64-73. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowlkes, E. B., and C. L. Mallows. 1983. A method for comparing two hierarchical clusterings. J. Am. Stat. Assoc. 78:553-569. [Google Scholar]
- 11.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs, A. J., and G. A. McIntyre. 1970. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur. J. Biochem. 16:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, M. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427-432. [Google Scholar]
- 15.Hubert, L., and P. Arabie. 1985. Comparing partitions. J. Classification 2:193-218. [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataja, J., P. Huovinen, A. Efstratiou, E. Perez-Trallero, and H. Seppäla. 2002. Clonal relationships among isolates of erythromycin-resistant Streptococcus pyogenes of different geographical origin. Eur. J. Clin. Microbiol. Infect. Dis. 21:589-595. [DOI] [PubMed] [Google Scholar]
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maizel, J. V., Jr., and R. P. Lenk. 1981. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl. Acad. Sci. USA 78:7665-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malhotra-Kumar, S., C. Lammens, S. Chapelle, M. Wijdooghe, J. Piessens, K. Van Herck, and H. Goossens. 2005. Macrolide- and telithromycin-resistant Streptococcus pyogenes, Belgium, 1999-2003. Emerg. Infect. Dis. 11:939-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan, G. W., and M. C. Cooper. 1986. A study of comparability of external criteria for hierarchical cluster analysis. Multivariate Behav. Res. 21:441-458. [DOI] [PubMed] [Google Scholar]
- 23.Nemoy, L. L., M. Kotetishvili, J. Tigno, A. Keefer-Norris, A. D. Harris, E. N. Perencevich, J. A. Johnson, D. Torpey, A. Sulakvelidze, J. G. Morris, Jr., and O. C. Stine. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rand, W. M. 1971. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66:846-850. [Google Scholar]
- 25.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 26.Serrano, I., J. Melo-Cristino, J. A. Carriço, and M. Ramirez. 2005. Characterization of the genetic lineages responsible for pneumococcal invasive disease in Portugal. J. Clin. Microbiol. 43:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379-423, 623-656. [Google Scholar]
- 28.Silva-Costa, C., M. Ramirez, and J. Melo-Cristino. 2006. Identification of macrolide-resistant clones of Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 12:513-518. [DOI] [PubMed] [Google Scholar]
- 29.Silva-Costa, C., M. Ramirez, and J. Melo-Cristino. 2005. Rapid inversion of the prevalences of macrolide resistance phenotypes paralleled by a diversification of T and emm types among Streptococcus pyogenes in Portugal. Antimicrob. Agents Chemother. 49:2109-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson, E. H. 1949. Measurement of species diversity. Nature 163:688. [Google Scholar]
- 31.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 32.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 33.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace, D. L. 1983. A method for comparing two hierarchical clusterings: comment. J. Am. Stat. Assoc. 78:569-576. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.