Abstract
The COBAS AMPLICOR hepatitis B virus assay targets a conserved region of the genome and is widely used to monitor treatment of hepatitis B in order to identify emerging resistance. However, the assay failed to recognize increasing viremia levels when YMDD mutations were paralleled by mutations in the segment targeted by the COBAS AMPLICOR probe.
The aim of treating chronic hepatitis B virus (HBV) infection is to prevent the emergence of liver cirrhosis or hepatocellular carcinoma. Currently, treatment is given either as injections of pegylated interferon or as oral doses of antivirals, such as lamivudine or adefovir. Lamivudine therapy has the advantages of documented efficiency, lower cost, and minimal side effects but carries an annual 15 to 20% risk of emergence of resistant mutants, which may cause severe exacerbations of hepatitis. Lamivudine resistance is typically due to a mutation in the YMDD motif of the polymerase (M204I or M204V) (7) and is recognized as increases in HBV DNA levels (by more than 1 log).
Here we describe a case where the COBAS AMPLICOR assay (Roche Diagnostics, Branchburg, NJ) failed to reveal resistance in a 56-year-old male Vietnamese patient with HBV cirrhosis.
The patient had arrived in Sweden in 1999, and his chronic HBV infection was discovered in 2001 by a general practitioner (positive HBsAg and anti-HBc results obtained by use of AxSYM [Abbott, Illinois]). The patient was admitted to the infectious disease clinic, where he presented signs of active cirrhosis with a low platelet count (52 × 109/liter), increased prothrombin time (international normalized ratio, 1.7), and an aspartate aminotransferase level of 2.5 times the upper limit of normal. HBeAg (tested by use of AxSYM [Abbott]) was not detected in serum, but the HBV DNA level was high, above 10 million copies/ml, as detected by use of COBAS AMPLICOR. Hepatitis C antibodies (tested for with AxSYM [Abbott]) were not detected. The patient was put on long-term lamivudine therapy (100 mg daily), which resulted in a 3-log reduction in HBV DNA to around 20,000 copies/ml as measured by COBAS AMPLICOR after 3 months. Thereafter the HBV DNA levels did not decrease further but appeared to remain at levels between 20,000 and 50,000 copies/ml. The aminotransferase levels also remained moderately elevated. A sample drawn after 21 months of treatment was included (by chance) in a study comparing COBAS AMPLICOR and COBAS TaqMan (Roche) (4), which revealed a 2-log underestimation of viremia by COBAS AMPLICOR. This finding led us to further investigate the emergence of mutations in the YMDD motif as well as in the region targeted by the COBAS AMPLICOR assay.
Viremia was measured by two different systems, the COBAS AMPLICOR Monitor (Roche) (5) and an in-house real-time PCR targeting a conserved region of the HBsAg gene. COBAS AMPLICOR testing was performed as instructed by the manufacturer. The S-region real-time PCR assay included DNA extraction by use of Magnapure LC (Roche) with a total nucleic acid isolation kit and 45 cycles of two-step amplification on an ABI 7500 instrument (Applied Biosystems). The amplification was carried out in a 50-μl reaction mixture volume, including 25 μl of universal master mix (Applied Biosystems), 10 μl of extracted DNA, and 0.3 μM concentrations of primers (254F, ACTCGTGGTGGACTTCTCTCAA; 432R, AAGAAGATGAGGCATAGCAGCA). The amplicon was detected by a TaqMan probe (0.2 μM; 6-carboxyfluorescein-TGGATGTGTCTGCGGCGTTTTATCAT-6-carboxytetramethylrhodamine). The HBV DNA concentrations of samples were calculated by use of a quantitation standard consisting of cloned full-length HBV DNA (genotype C).
For direct sequencing, the regions of HBV spanning from nucleotides (nt) 1606 to 2058 (core promoter, precore, and core) and 593 to 796 (S and polymerase regions) were amplified. Using the same primers as those used in PCR, a cycle sequencing reaction was then done with fluorescent stop nucleotides. The sequences were then recorded by an ABI 310 sequence reader (Applied Biosystems).
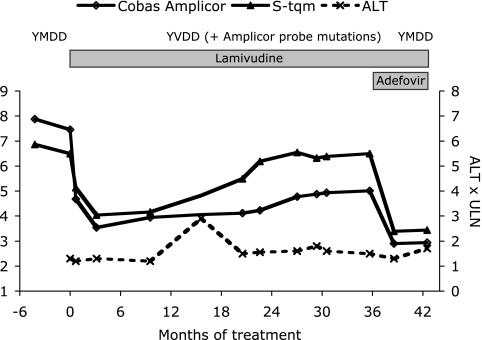
Figure 1 shows the HBV DNA levels before and during lamivudine therapy, as obtained by use of COBAS AMPLICOR and the in-house real-time PCR. Initially, there was a reduction of viremia that was documented similarly by both assays. After 21 months of lamivudine therapy, the HBV DNA level had increased to 300,000 copies/ml, and it then gradually increased further to 3 million copies/ml. However, this was observed only by the in-house real-time PCR and not by COBAS AMPLICOR, which showed values 1.5 logs lower. During this period the aminotransferase levels remained elevated, liver function did not improve, and gastroscopy showed esophageal varices. When it was determined that the viremia levels had increased (by the COBAS TaqMan evaluation mentioned before [4]), adefovir was added to the lamivudine treatment regimen, and viremia levels then decreased (as assessed by both real-time PCR and COBAS AMPLICOR) to around 1,000 copies/ml after a few months. Despite this, the aspartate aminotransferase level remained at 2.5 times the upper limit of normal. Based on a Child-Pugh score of 8 to 9, an elective liver transplantation was done 7 months after the initiation of adefovir treatment, when the HBV DNA level was around 1,000 copies/ml. Five months after transplantation, the patient was alive, the adefovir and HBV immunoglobulin therapy was being continued, and there were no signs of reactivation.
FIG. 1.
Viremia levels during antiviral therapy detected by COBAS AMPLICOR compared to those detected by S-region TaqMan PCR (S-tqm). ULN, upper limit of normal; ALT; alanine aminotransferase.
Sequencing of the polymerase region showed a gradual emergence and takeover by a strain with L180M and M204V mutations from the 21st to the 35th month of treatment (Fig. 1 and Table 1). After 3 months of adefovir treatment, this YVDD mutant had once again been replaced by a YMDD strain at a low concentration. After transplantation, HBV DNA was no longer detectable.
TABLE 1.
Appearance of mutations in four genomic segments as detected by direct sequencing
| Length of therapy (mo) | HBV DNAa (log copies/ml) | Therapyb | Base at indicated nt in the following segment:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymerasec
|
Core promoter
|
Precore
|
Core
|
||||||||
| 667 (180) | 739 (204) | 1757 | 1775 | 1837 | 1846 | 1909 | 1913 | 1915 | |||
| −4 | 6.9 | C | A | A | G | A | A | T | C | G | |
| 0 | 4.0 | Lam | C | A | A | G | A | A | T | C | G |
| 10 | 4.2 | Lam | C | A | A | G | A | A | T | C | G |
| 21 | 5.5 | Lam | A | G | A | A | G | T | C | A | G |
| 27 | 6.5 | Lam | A | G | A | A | G | T | C | A | G |
| 31 | 6.4 | Lam | A | G | G | A | G/A | T | C/T | A | G/C |
| 36 | 6.5 | Lam | A | G | G | A | G/A | T | C/T | A | C |
| 39 | 3.4 | Lam + Adv | C | A | A | A | G | T | C/T | A | G |
Measured by S-region TaqMan PCR.
Lam, lamivudine; Adv, adefovir.
For polymerase segment nt positions, the numbers in parentheses indicate the corresponding amino acid positions.
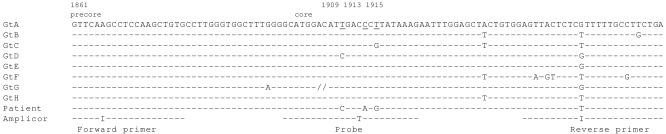
As summarized in Table 1, sequencing of the core promoter-precore-core region showed several mutations. The strain was a C-1858 variant of genotype C, a variant that is relatively common in Southeast Asia and in which the G1896A stop codon mutation rarely evolves (1). Some mutations, including C-1753 and T-1762/A1764 in the core promoter, had emerged before the initiation of lamivudine therapy. Others had emerged in parallel with the M204V mutation. As shown in Fig. 2, three mutations in the amino-terminal part of the core (nt 1909, 1913, and 1915) were located to a segment targeted by the COBAS AMPLICOR probe. One of these mutations (A1913) resulted in an amino acid shift (P5T).
FIG. 2.
Alignment of COBAS AMPLICOR primers and probe to representative sequences of each genotype as well as to the sequence of HBV from the patient (after 2 years of treatment). In addition to showing the mismatches between the COBAS AMPLICOR probe sequence and the patient's HBV sequence, this alignment demonstrates that COBAS AMPLICOR is not accurate for the detection of genotype F (GtF) (due to reverse primer mismatches) or genotype G (due to a 33-nt insertion in the probe region, marked by //). Underlining in the sequence of genotype A indicates positions 1909, 1913, and 1915.
The main mechanism for selection of the mutant strain was probably the antiviral resistance conferred by mutations in the polymerase region (L180M and M204V) (7). The selective advantage of the coemerging mutations in the precore and core is uncertain. G1837 changes the X gene stop codon from TAA to TGA, but neither G1837 nor T1846 induces any amino acid shift of the precore protein. Possibly, these mutations might alter the structure of the 5′ or the 3′ pregenomic RNA loop, although none of them are included in the stem of the epsilon signal.
The core mutations at nt 1909, 1913, and 1915 are of special interest because they are localized to a segment targeted by the COBAS AMPLICOR probe. The hybridization in the COBAS AMPLICOR assay is performed after PCR at a relatively low temperature, and therefore it is probably more tolerant to mismatches than the TaqMan hybridization, which is used in many real-time PCR systems. Still, this case clearly shows that if several mutations are concentrated to the probe region, then the HBV DNA level may be underestimated by COBAS AMPLICOR by as much as 1.5 logs. This observation is of apparent clinical importance, particularly since COBAS AMPLICOR is widely used for monitoring response to antiviral treatment. Of the three mutations, only one results in an amino acid exchange, from proline to threonine at the position corresponding to codon 5 of the core. This P5T mutation has frequently been observed in Asian carriers (2) and may reflect the presence of an epitope encoded by this part of the core (or HBeAg). Alternatively, the impact on the polyadenylation signal (nt 1918 to 1923) might be of relevance for the selection of these mutations.
Whether the precore and core mutations were associated with each other, e.g., by interactions in the secondary structure of the mRNA, is unclear. Such interactions might exist, even though the mutations at nt 1837, 1846, 1909, 1913, and 1915 do not base pair in the epsilon loop. It is also unclear if any of the mutations in the precore-core region may enhance the effects of the lamivudine resistance mutations. Such effects have been described previously for the precore mutation at nt 1896 (3, 6) but not for any of the mutations observed here.
Because the COBAS AMPLICOR assay is widely used for monitoring therapeutic response, it is important to clarify the frequency of mutations in the probe region and the way in which they might influence the quantification results. It is noteworthy that the COBAS TaqMan assay, i.e., the successor of COBAS AMPLICOR, was not affected by the core mutations but showed an HBV DNA level similar to that shown by the in-house TaqMan assay (not shown). This result is explained by the fact that the probe in COBAS TaqMan targets a different, neighboring, and probably more conserved (the probe sequence is not yet published) segment of the precore-core region. The case reported here underlines the importance of making sequence data for commercial assays public to allow investigations of unexpected quantification results. For example, it would be of value to investigate whether the COBAS TaqMan probe region is more affected or less affected by emerging mutations than are the conserved parts of the S region that are targeted by other real-time PCR assays (8) and the assay described here. This is of particular importance because real-time PCR methods using probes are in general even more sensitive to sequence variability than the COBAS AMPLICOR assay.
Nucleotide sequence accession numbers. The sequences reported here appear in GenBank with the accession numbers DQ525717 to DQ525732.
Acknowledgments
This study was supported by the Swedish Medical Research Council and the LUA-ALF funds.
REFERENCES
- 1.Alestig, E., C. Hannoun, P. Horal, and M. Lindh. 2001. Phylogenetic origin of hepatitis B virus strains with precore C-1858 variant. J. Clin. Microbiol. 39:3200-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozkaya, H., B. Ayola, and A. S. Lok. 1996. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology 24:32-37. [DOI] [PubMed] [Google Scholar]
- 3.Chen, R. Y., R. Edwards, T. Shaw, D. Colledge, W. E. T. Delaney, H. Isom, S. Bowden, P. Desmond, and S. A. Locarnini. 2003. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology 37:27-35. [DOI] [PubMed] [Google Scholar]
- 4.Lindh, M., and C. Hannoun. 2005. Dynamic range and reproducibility of hepatitis B virus (HBV) DNA detection and quantification by Cobas Taqman HBV, a real-time semiautomated assay. J. Clin. Microbiol. 43:4251-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noborg, U., A. Gusdal, E. K. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tacke, F., C. Gehrke, T. Luedde, A. Heim, M. P. Manns, and C. Trautwein. 2004. Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of lamivudine-resistant mutants. J. Virol. 78:8524-8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tipples, G. A., M. M. Ma, K. P. Fischer, V. G. Bain, N. M. Kneteman, and D. L. Tyrrell. 1996. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology 24:714-717. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger, K. M., E. Wiedenmann, S. Bohm, and W. Jilg. 2000. Sensitive and accurate quantitation of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR). J. Virol. Methods 85:75-82. [DOI] [PubMed] [Google Scholar]