Abstract
Because of the use of viral kinetics during polyethylene glycol (PEG)-interferon-ribavirin therapy and the development of specific new anti-hepatitis C virus (anti-HCV) drugs, assessment of the efficacy of anti-HCV drugs needs to be based not on end-point PCR assays but on real-time PCR. The aim of this study was to determine if the two available commercial real-time PCR assays, the Abbott RealTime HCV assay and the Roche Cobas TaqMan HCV assay, can become the standard for HCV RNA quantification. We investigated the prognostic relevance of HCV RNA viral loads at baseline, week 4, and week 12 to a rapid and early virological response to antiviral therapy by using the two assays. Of 59 naïve patients chronically infected by HCV (41 infected with genotype 1) who were treated with ribavirin plus PEG-interferon alfa-2b for 48 weeks, 24 patients (41%) showed a sustained virological response (SVR). With the two assays, viral loads were highly correlated, irrespective of genotype (R2 = 0.94 for all cases). No difference in diagnostic value was found between the Abbott and Roche assays at week 4, with respective negative predictive values (NPVs) of 84% and 78% and positive predictive values (PPVs) of 62% and 56% (not significant), and at week 12, the respective NPVs were 91% and 90% and PPVs were 44% and 46% (not significant). At week 12, 83% (20/24) and 96% (23/24) of patients with SVR tested negative for HCV RNA by the Abbott and Roche assays, respectively (the difference is not significant). In conclusion, the high sensitivities and large dynamic ranges of the Abbott and Roche assays show that a single real-time quantitative PCR assay is fully adequate for clinical and therapeutic management of HCV.
Based on pivotal trials in large multicenter studies, positive and negative predictions of sustained virological response (SVR) using viral load kinetics have been established and are now used for recommendations on antiviral therapy management by the American and European international consensus conference (4, 6, 9, 14). The data for the proposed algorithm for the management of antiviral therapy in patients with chronic hepatitis C were mainly based on measurements of viral loads assessed by end-point PCR assays (NGI Superquant and Cobas Monitor Amplicor HCV 2.0) in centralized laboratories (3, 6, 13). General application may be difficult due to the lack of standardization of previous hepatitis C virus (HCV) RNA quantitation methods (15). The most common methods available for detecting and quantifying HCV RNA are based on PCR and on the signal amplification technique (branched DNA and Versant HCV RNA; Bayer). Two semiautomated PCR assays, the complete bioanalytical system (Cobas Monitor Amplicor HCV 2.0) and the LCx HCV RNA assay (Abbott), are commonly used by many laboratories. To make the quantitation of HCV RNA comparable among the different assays, the National Institute for Biological Standards and Controls and the World Health Organization (WHO) developed and certified a uniform standard for measuring HCV RNA in international units (IU) (16). However, this standardization of HCV RNA assays to IU is based on genotype 1 panels. Little is known on the variation in commercially available HCV RNA assays for quantification of other HCV genotypes.
Despite the introduction of IU, discrepancies may occur when patients are monitored using different types of assays (19). In a previous study, false-positive and false-negative results, as well as variations in the HCV RNA level of >1 to 2 log IU, were observed. These results may have an impact on the management of patients receiving interferon therapy, for example, when treatment decisions are made using a single HCV RNA determination (11). Two standardized HCV RNA quantitative assays based on real-time PCR were recently developed (Abbott RealTime HCV and Roche Cobas TaqMan HCV). Both have a high sensitivity (based on the lower limit for detecting HCV RNA) and a great dynamic range and were standardized using the second WHO panel. The principle of real-time PCR techniques is to detect amplicon synthesis and to deduce the number of viral genomes in the starting clinical sample during the PCR rather than at the end (15). The main advantage of these tests is that each has been standardized, which makes results by different laboratories comparable through reporting in IU. Because of the use of viral kinetics during polyethylene glycol (PEG)-interferon-ribavirin therapy and the development of specific new anti-HCV drugs, assessment of the efficacy of anti-HCV drugs needs to be based not on end-point PCR assays but on real-time PCR.
To determine if the new real-time PCR-based assays can become the standards for HCV RNA detection and quantitation, we investigated the prognostic relevance of HCV RNA viral loads at baseline, week 4, and week 12 to the virological response to antiviral therapy. Also, we compared the performances of the two commercially available real-time PCR assays.
MATERIALS AND METHODS
Patients.
A total of 59 consecutive naïve patients were included in the study (34 men; mean age, 50 ± 13 years) just before starting interferon and ribavirin therapy for chronic HCV infection in two hepatology and gastroenterology units (Arnault Tzanck Institute, St. Laurent du Var, and Saint Joseph Hospital, Marseille, France).
All tested positive for HCV RNA and negative for human immunodeficiency virus. Other causes of chronic hepatitis were excluded by appropriate serological testing and liver histology. The HCV genotype was determined by sequence analysis using the HCV genotyping Trugene assay (Bayer, Puteaux, France) (12). There were 41 genotype 1, 5 genotype 2, 10 genotype 3, and 3 genotype 4 patients. All patients were treated with 800 mg ribavirin per day orally plus PEG-interferon alfa-2b (1.5-μg/kg subcutaneous injection once a week) for 48 weeks.
The virological response was assessed by a qualitative HCV RNA assay with a lower limit of sensitivity of 50 IU/ml (HCV Amplicor 2.0; Roche Diagnostics, Meylan, France). According to the qualitative HCV RNA results, patients were defined as virologic sustained responders (HCV RNA negative 6 months after the end of therapy) or nonresponders.
Sera for HCV RNA analysis.
A total of 177 serum samples were obtained from the 59 patients. Each of the specimens was divided into four aliquots and frozen to −80°C within 2 h of collection (10). Each aliquot was used for HCV RNA quantitation by the following assays: Cobas TaqMan HCV (Roche Diagnostics, Meylan, France), with a detection limit of 15 IU/ml (7), and the Abbott RealTime HCV quantitative assay (Abbott Laboratories, Rungis, France), with a detection limit of 10 IU/ml.
The HCV RNA viral load was measured at the following three time points: baseline and 4 and 12 weeks after the beginning of therapy. SVR was determined 6 months after the end of therapy with the Amplicor qualitative assay (Roche Diagnostics, Meylan, France). Values for HCV RNA are reported in IU/ml. All assays were done at Alphabio Laboratory.
Roche Taqman HCV assay.
The Cobas TaqMan HCV test (Roche Molecular Systems, Inc., Branchburg, NJ) is a real-time nucleic acid amplification assay for quantitative detection of HCV RNA in human serum or plasma. Like the TaqMan HCV analyte-specific reagent (TaqMan HCV ASR; Roche Molecular Systems), this assay was developed for use with the recently introduced Cobas TaqMan 48 analyzer (CTM 48; Roche Molecular Systems). Amplification and detection were performed according to the manufacturer's instructions for TaqMan HCV with a CTM 48 analyzer and Amlilink software, v. 3.0.1 (Roche Diagnostics, Meylan, France). The lower limit of detection is 15 IU/ml, with ≥95% probability, using 1 ml of serum (7).
Abbott RealTime HCV assay.
The RealTime HCV assay (Abbott Molecular Inc., Des Plaines, IL) was performed at Abbott Laboratories according to the manufacturer's specifications. Total nucleic acids were extracted from 0.5 ml serum or plasma by magnetic microparticle technology, using an Abbott m1000 automated sample preparation system. The internal control (IC), derived from the hydroxypyruvate reductase gene of the pumpkin plant, Cucurbita pepo, was introduced as armored RNA into the sample lysis buffer. The IC was processed simultaneously with each sample. RNA was captured by magnetic microparticles, washed to remove unbound sample components, and eluted. A second step consisted of adding master mix (HCV oligonucleotide reagent including primers and probes, thermostable rTth polymerase enzyme, and activation reagent) to extract nucleic acid samples into 96-well optical reaction plates. Plates were sealed and placed on an Abbott m2000rt instrument (m2000rt; Abbott Molecular Inc.) for reverse transcription, PCR amplification, and detection/quantitation.
The HCV probe is a short linear oligonucleotide labeled with a fluorophore at the 5′ end and a quencher at the 3′ end. The PCR primers and probe target conserved regions of the 5′ untranslated region of the HCV genome. In the absence of an HCV target, the fluorescence is quenched. In the presence of the HCV target sequence, the HCV probe preferentially hybridizes to the target sequence, allowing fluorescence detection. A second set of primers and a probe labeled with a different fluorophore are used for amplification and detection of the internal control. The IC probe is a single-stranded DNA oligonucleotide with a fluorophore at the 5′ end and a quencher at the 3′ end. IC probe fluorescence is quenched in the absence of IC target sequences. In the presence of IC target sequences, IC probe hybridization to complementary sequences separates the fluorophore and quencher and allows fluorescence emission and detection. The noncompetitive internal control is detected at all HCV levels.
The HCV- and IC-specific probes are each labeled with a different fluorophore, thus allowing for simultaneous detection of both amplified products at each cycle. The amplification cycle at which a reactive level of fluorescent signal is detected by the m2000rt instrument is proportional to the log of the HCV RNA concentration present in the original sample. The homogeneous format and sealed PCR tray eliminate contamination by amplified products.
The assay meets the Second WHO International Standard for HCV RNA. The lower limit of detection is 12 IU/ml, with ≥95% probability. The dynamic range of the assay extends from 12 to 100,000,000 IU/ml. The Abbott RealTime HCV assay detects and quantitates genotypes 1 to 6.
Statistical analysis.
Values are expressed as medians or means (with 95% confidence intervals). Correlations between assays were evaluated by linear regression. The chi-square test or Wilcoxon test was used for data comparison. P values of <0.05 were considered significant.
RESULTS
Reverse transcription-PCR (RT-PCR) viral load comparison.
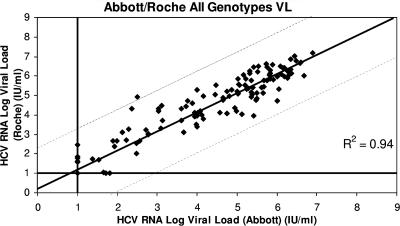
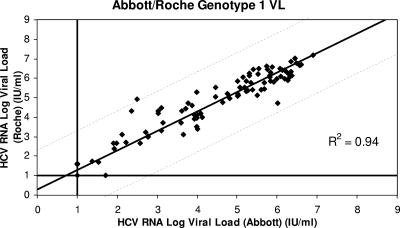
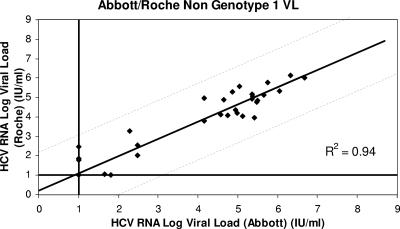
Figure 1 shows the distribution of viral loads measured by each assay in the 59 patients at baseline, week 4, and week 12. One hundred seventy-seven viral loads were measured for each assay. The viral loads measured by the Abbott and Roche assays were highly correlated (R2 = 0.94). Figure 2 shows the distribution of viral loads for genotype 1 patients measured by the Abbott and Roche assays; both assays were again highly correlated (R2 = 0.94), as they were for non-genotype 1 patients (R2 = 0.94) (Fig. 3).
FIG. 1.
Correlation between the results for 59 HCV-infected patients tested by Abbott RealTime HCV and Roche Cobas TaqMan HCV, irrespective of the genotype. Results shows a good correlation between the two assays (R2 = 0.94).
FIG. 2.
Correlation between the results for genotype 1 patients tested by the Abbott and Roche assays (R2 = 0.94).
FIG. 3.
Correlation between the results for non-genotype 1 patients tested by the Abbott and Roche assays (R2 = 0.94).
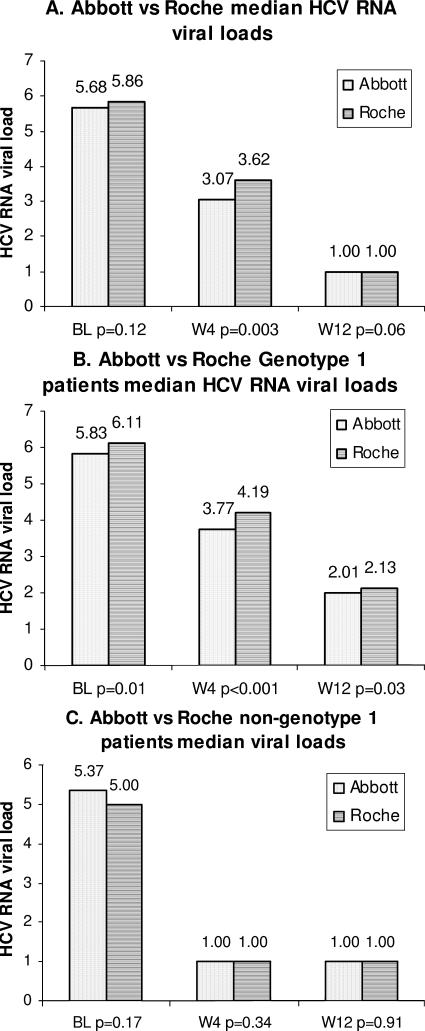
Median viral loads measured by the Abbott and Roche RT-PCR assays at baseline, week 4, and week 12 are shown in Fig. 4A for all genotypes and in Fig. 4B and C for genotype 1 and non-genotype 1 patients, respectively. Significant differences in medians were observed between the Abbott and Roche assays at week 4 when all genotypes were considered (P = 0.003); all medians were significantly different at baseline, week 4, and week 12 for genotype 1 patients; and no medians were significantly different at baseline, week 4, and week 12 for non-genotype 1 patients.
FIG. 4.
Histograms of median viral loads measured by the Abbott and Roche assays for all genotypes (A), genotype 1 patients (B), and non-genotype 1 patients (C). Bars with dots stand for the Abbott assay, and bars with horizontal lines stand for the Roche assay.
Table 1 shows mean interassay differences between the Abbott and Roche RT-PCR assays. With the Roche assay, quantification of genotype 1 patients was slightly higher (−0.28), and quantification of genotypes 2 and 4 was slightly lower (0.22 and 0.34). Quantification of genotype 3 patients was similar for the two assays (−0.03).
TABLE 1.
Mean interassay differences between Abbott and Roche RT-PCR assays
| Parameter | Value for genotype
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Mean interassay difference (log IU/ml) | −0.28 | 0.22 | −0.03 | 0.34 |
Viral kinetics in response to antiviral therapy. (i) RVR.
No differences in negative predictive values (NPVs) and positive predictive values (PPVs) were found between the Abbott and the Roche assays: the respective NPVs were 84% and 78%, and the respective PPVs were 62% and 56%. Among the 20 genotype 1 patients with viral load drops of <2 log IU/ml at week 4 (rapid viral response [RVR]) in the Abbott assay, 18 were nonresponders (NPV = 90%), compared with 19 of 23 for the Roche assay (NPV = 83%) (not significant).
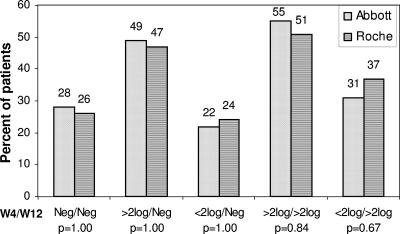
Figure 5 shows that there were no differences between the Abbott and Roche assays in viral load detection and viral load decline after 4 weeks and 12 weeks of treatment. The percentages of patients who became HCV RNA negative within 4 weeks and remained negative after 12 weeks were similar for both assays (P = 1.00) (Fig. 5). With the Abbott assay, all patients (14/14) who had RVR at week 4 were HCV RNA negative at week 12, and with the Roche assay, 1/15 patients who had RVR at week 4 was HCV RNA positive at week 12 (not significant).
FIG. 5.
Histograms of differences between the Abbott and Roche assays in viral load detection and viral load decline after 4 weeks and 12 weeks of treatment.
(ii) EVR.
No differences in PPV and NPV values were found between the Abbott and Roche assays for the early virological response (EVR): NPVs were 91% and 90%, respectively, and PPVs were 44% and 46%, respectively.
For genotype 1 patients, we found no EVR difference in PPV and NPV values between the Roche and the Abbott assays: the NPV was 100% for both, and PPVs were 24% and 29%, respectively. All genotype 1 patients with viral load drops of <2 log IU/ml at week 12 (EVR) were nonresponders (NPV = 100%) (eight patients for the Abbott assay and seven for the Roche assay). At treatment week 12, 83% (20/24) and 96% (23/24) of patients with SVR tested negative for HCV RNA by the Abbott and Roche assays, respectively (not significant).
(iii) Qualitative and quantitative RT-PCR comparison.
For a subsequent population of patients with low viral loads, we compared the absolute viral loads at week 12 measured by both the qualitative HCV Amplicor 2.0 (Roche) assay and the two real-time RT-PCR assays. The quantitative assay diagnostic values were slightly higher than the qualitative assay values but were not significantly different (Table 2).
TABLE 2.
Diagnostic values for quantitative and qualitative assays after 12 weeks of treatment
| Parameter | Value (%) for:
|
P value
|
|||
|---|---|---|---|---|---|
| Quantitative assays
|
Qualitative assay (HCV Amplicor 2.0 [Roche]) | RealTime HCV vs HCV Amplicor 2.0 | Cobas TaqMan HCV vs HCV Amplicor 2.0 | ||
| RealTime HCV (Abbott) | Cobas TaqMan HCV (Roche) | ||||
| NPV | 94 | 94 | 82 | 0.72 | 0.69 |
| PPV | 53 | 55 | 54 | 0.89 | 0.89 |
| Specificity | 46 | 49 | 32 | 0.40 | 0.27 |
| Sensitivity | 95 | 96 | 92 | 0.84 | 0.97 |
DISCUSSION
The results show that real-time PCR using the two available commercial assays is adequate for the management of therapeutic responses to chronic hepatitis C. Considering the viral kinetics and the rapid and early virological responses, a very good correlation was observed between the Abbott and Roche assays, irrespective of the HCV genotype. For a decision of treatment discontinuation on the basis of the 2-log-decline rule at week 12 in genotype 1 patients, the two assays would serve to make the correct decision for all sustained responders. EVR NPVs and PPVs for the two assays were in the same range, and their means were compatible with clinical use (the respective Abbott and Roche NPVs were 91% and 90%, and the respective PPVs were 44% and 46%). For patients with high baseline HCV RNA levels, the week 12 NPV was 100% for the two assays (six patients for the Abbott assay and four patients for the Roche assay; results are similar to those of Ferenci et al. [5]). To explain this finding, a recent study using real-time PCR reported that the presence of detectable residual hepatitis C viremia is associated with a significantly increased risk of relapse after completion of therapy and could therefore be a valuable marker to predict treatment outcomes (2).
Regarding the absolute values at baseline, week 4, and week 12, the two assays differed significantly, without any consequence on the virological response. Considering the linearity of quantification of the different genotypes using the two assays, we observed an excellent correlation for all genotypes (R2 = 0.94), regardless of the genotype. Previous data have reported that the Cobas Taqman assay cannot provide a linear correlation with genotypes 2 and 3 compared with the Cobas Amplicor Monitor 2.0 assay (18). A recent study reported that this phenomenon can be corrected by a modified extraction procedure with linear quantification of genotypes 1 to 6 (7).
End-point-PCR-based assays (Cobas Monitor and LCx) and the signal amplification assay (Versant; Bayer) should now be called “old-generation HCV RNA quantitation,” as opposed to “new-generation real-time PCR” assays, for at least the following three reasons: (i) the absence of complete automation, (ii) the need to dilute for quantification above the upper range (8), and (iii) the fact that the former assays were standardized with the first WHO panel, and due to the different secondary standards used for calibration, the IUs measured by these assays are different. In a recent comparative study (11), we demonstrated that the absolute values obtained with these assays are different and that the HCV RNA viral load decline, assessed by different assays during antiviral therapy, can give different results regardless of the use of international units.
Novel therapeutic strategies against HCV are mainly focused on the development of agents that inhibit the specific steps of the life cycle of the virus. Protease and polymerase inhibitors require the highest HCV RNA sensitivity and dynamic range, because results are mainly based on their ability to reduce the viral load and to reach the largest percentage of patients with SVR (1, 17, 20).
In conclusion, the high sensitivity and large dynamic range offered by the new real-time PCR-based assays must definitively close the discussion about the use of a qualitative or quantitative assay for HCV clinical and therapeutic management. This was confirmed by our findings related to the slightly higher NPVs and PPVs observed with real-time PCR assays than with the qualitative HCV Amplicor 2.0 assay (Table 2). These observations emphasize the need for commercial HCV RNA quantification assays with a broader range of linear quantification, such as real-time PCR-based assays. One other topic is the cost of real-time PCR technology. National health departments might review their respective reimbursement policies, as real-time PCR assay costs are equivalent to those of the older generation of assays.
The 2002 NIH consensus conference (14) gave recommendations for the use of molecular tests, in addition to enzyme-linked immunosorbent assays, for diagnosis. These recommendations concern the decision to treat, optimal treatment schedules, and assessment of the viral response to antiviral therapy. For all of these indications, we propose that a real-time quantitative PCR assay is fully adequate for clinical and therapeutic management of chronic HCV infection without additional costs.
REFERENCES
- 1.Afdhal, N., M. Rodriguez-Torres, and E. Lawitz. 2005. Enhanced antiviral efficacy for valopicitabine (NM283) plus peg-interferon in hepatitis C patients with HCV genotype-1 infection: results of a phase IIa multicenter trial. J. Hepatol. 42:39-40. [Google Scholar]
- 2.Bergk, A., C. Sarrazin, G. Teuber, P. Buggisch, H. Klinker, V. Weich, B. Wiedenmann, and T. Berg. 2005. Detection of minimal residual hepatitis C viremia at treatment week 12 is associated with a high probability of relapse, abstr. 1188. Hepatology 42(Suppl. 1):663A-664A. [Google Scholar]
- 3.Davis, G. L., J. B. Wong, J. G. McHutchison, M. P. Manns, J. Harvey, and J. Albrecht. 2003. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38:645-652. [DOI] [PubMed] [Google Scholar]
- 4.European Association for the Study of the Liver. 1999. EASL International Consensus Conference on Hepatitis C. Paris, 26-28, February 1999, consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 5.Ferenci, P., M. W. Fried, M. L. Shiffman, C. I. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, M. Chaneac, and K. R. Reddy. 2005. Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin. J. Hepatol. 43:425-433. [DOI] [PubMed] [Google Scholar]
- 6.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 7.Germer, J. J., W. S. Harmsen, J. N. Mandrekar, P. S. Mitchell, and J. D. Yao. 2005. Evaluation of the COBAS TaqMan HCV test with automated sample processing using the MagNA pure LC instrument. J. Clin. Microbiol. 43:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourlain, K., A. Soulier, B. Pellegrin, M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and J. M. Pawlotsky. 2005. Dynamic range of hepatitis C virus RNA quantification with the Cobas Ampliprep-Cobas Amplicor HCV Monitor v2.0 assay. J. Clin. Microbiol. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 10.Halfon, P., H. Khiri, V. Gerolami, M. Bourliere, J. M. Feryn, P. Reynier, A. Gauthier, and G. Cartouzou. 1996. Impact of various handling and storage conditions on quantitative detection of hepatitis C virus RNA. J. Hepatol. 25:307-311. [DOI] [PubMed] [Google Scholar]
- 11.Halfon, P., G. Penaranda, M. Bourliere, H. Khiri, M. F. Masseyeff, and D. Ouzan. 2006. Assessment of early virological response to antiviral therapy by comparing four assays for HCV RNA quantitation using the international unit standard: implications for clinical management of patients with chronic hepatitis C virus infection. J. Med. Virol. 78:208-215. [DOI] [PubMed] [Google Scholar]
- 12.Halfon, P., P. Trimoulet, M. Bourliere, H. Khiri, V. de Ledinghen, P. Couzigou, J. M. Feryn, P. Alcaraz, C. Renou, H. J. Fleury, and D. Ouzan. 2001. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J. Clin. Microbiol. 39:1771-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 14.National Institutes of Health. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C: 2002—June 10-12, 2002. Hepatology 36:S3-S20. [DOI] [PubMed] [Google Scholar]
- 15.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 122:1554-1568. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 17.Reesink, H., S. Zeuzem, and A. Van Vliet. 2005. Initial results of a phase 1B, multiple-dose study of VX-950, a hepatitis C virus protease inhibitor. Abstr. Dig. Dis. Week, 14 to 19 May 2005, Chicago, Ill., abstr. 527.
- 18.Sarrazin, C., B. Gaertner, D. Sizmann, R. Babiel, U. Mihm, P. Hofmann, M. Von Wagner, and S. Zeuzem. 2005. HCV RNA quantification of HCV genotypes 1 to 5: comparison of 4 different assays. Hepatology 41:abstr. AASLD 591.
- 19.Shiffman, M. L., A. Ferreira-Gonzalez, K. R. Reddy, R. K. Sterling, V. A. Luketic, R. T. Stravitz, A. J. Sanyal, C. T. Garrett, M. De Medina, and E. R. Schiff. 2003. Comparison of three commercially available assays for HCV RNA using the international unit standard: implications for management of patients with chronic hepatitis C virus infection in clinical practice. Am. J. Gastroenterol. 98:1159-1166. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, X. J., N. H. Afdhal, and E. Godofsky. 2005. Pharmacokinetics and pharmacodynamics of valopicitabine (NM283), a new nucleoside HCV polymerase inhibitor. Results from a phase I/II dose-escalation trial in patients with HCV-1 infection. J. Hepatol. 42:229. [Google Scholar]