Abstract
Salmonella enterica serovar Typhimurium is a common cause of nontyphoidal salmonellosis in humans and animals. Multidrug-resistant serovar Typhimurium phage type DT104, which emerged in the 1990s, has become widely distributed in many countries. A total of 104 clinical isolates of Salmonella serogroup B were collected from three major hospitals in Taiwan during 1997 to 2003 and were examined by a multiplex PCR targeting the resistance genes and the spv gene of the virulence plasmid. A total of 51 isolates (49%) were resistant to all drugs (ACSSuT [resistance to ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline]), and all contained a 1.25-kb PCR fragment of integron that is part of the 43-kb Salmonella genomic island 1 (SGI1). The second group was resistant to SSu (28%), and the third was susceptible to all five drugs (13%). Fifty-nine isolates were serotyped to be serovar Typhimurium by the tube agglutination method using H antisera. The virulence plasmid was found in 54 (91.5%) of the 59 serovar Typhimurium isolates. A majority (94.1%) of the Salmonella serogroup B isolates with the ACSSuT resistance pattern harbored a virulence plasmid. Phage typing identified three major phage types: DT104, DT120, and U302. Analysis of the isolates by pulsed-field gel electrophoresis showed six genotypes. We found two genotypes in DT104 strains, two in DT120, and the other two in U302. The presence of a monophasic serovar (4,5,12:i:−) has added difficulty in the determination of the serovars of multidrug-resistant Salmonella serogroup B isolates. Nevertheless, the multiplex PCR devised in the present study appears to be efficient and useful in the rapid identification of ACSSuT-type serovar Typhimurium with SGI1, irrespective of their phage types.
Multidrug-resistant ACSSuT-type (resistance to ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline) Salmonella enterica serovar Typhimurium definitive type 104 (DT104) has risen to prominence in Europe and North America (15, 18, 31) but has been reported less in Asia (26, 32). Although the multidrug-resistant serovar Typhimurium was highly prevalent in Taiwan (19), its phage types have not been determined. The infections caused by serovar Typhimurium include gastroenteritis and occasional outbreaks in humans and wild or hatchery animals. The ACSSuT-type serovar Typhimurium DT104 is derived from two separate evolutionary events. One is the integration of a 43-kb Salmonella genomic island 1 (SGI1), which carries the following multiple antimicrobial resistance genes: pse for ampicillin resistance (A), floR for chloramphenicol resistance (C), str or aad for streptomycin resistance (S), sulI for sulfonamide resistance (Su), and tetR or tetG for tetracycline resistance (T) (2, 3, 10). The other is the integration of P22-like phage into the chromosome to form prophage PDT17 or ST104 (25), which encodes 64 open reading frames without antibiotic resistance genes within the 41-kb DNA fragment (30).
In serovar Typhimurium DT104, SGI1 is located between genes thdF and int2 with an imperfect 18-bp direct repeat flanking at the two ends (10). Near the end of SGI1, a 13-kb antibiotic resistance gene cluster responsible for ACSSuT resistance constitutes a complex class 1 integron of the In4 group (4). It has been suggested that the dissemination of SGI1 among Salmonella serovars occurs through a mechanism of mobilization by the IncC plasmid R55 (10). In the presence of a helper plasmid, the SGI1 can spread between different serovar Typhimurium phage types or between serovars. In addition to DT104, other serovar Typhimurium phage types, such as DT204b and U302, with the ACSSuT resistance phenotype, have also been isolated from animals, foods, and humans (20, 24). Before phage typing, the identification of Salmonella serovars using H antisera is important; however, recent studies discovered a monophasic variant of serovar Typhimurium (4,5,12:1:−) from swine, the H antigen profile of which differs from the typical serovar Typhimurium U302 in that it has no phase switch (9). The situation prevents the rapid determination of serovar Typhimurium. Since 1999, molecular diagnostic methods, including multiplex PCR and array systems, have been developed to detect the ACSSuT-type resistance. Both methods include Salmonella-specific genes including sipBC (5) and invA (13) and five previously known multidrug resistance genes as targets. Among serogroup B Salmonella isolates, a molecular marker, the spv operon (7, 14, 16), that is encoded on the 94.7-kb serovar-specific virulence plasmid, can be used to identify serovar Typhimurium because more than 90% of serovar Typhimurium clinical isolates harbor the virulence plasmid (2, 6). In the present study, we developed a multiplex PCR method to detect the ACSSuT-type serovar Typhimurium strains from 104 multidrug-resistant clinical isolates of serogroup B Salmonella. Representative multidrug-resistant serovar Typhimurium strains were further characterized by phage typing and pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
Bacterial strains and antimicrobial susceptibility.
Clinical isolates of Salmonella serogroup B were collected from the clinical microbiology laboratories of Chang Gung Memorial Hospital and Children's Hospital in northern Taiwan from 1997 to 1999 and from Chiayi Chang Gung Memorial Hospital in southern Taiwan from 2000 to 2003. Four isolates were obtained from National Cheng Kung University Hospital in 2000. After examination by using O antisera, these isolates were further characterized for their differences in serovars, phage types, integrons, genotypes, and antimicrobial susceptibility.
The antimicrobial susceptibility was tested by a standard disk diffusion method (23). The antimicrobial agents used were ampicillin (10 μg), ceftriaxone (30 μg), chloramphenicol (30 μg), florfenicol (30 μg), streptomycin (10 μg), trimethoprim-sulfamethoxazole (1.25 and 23.75 μg), sulfisoxazole (250 μg), and tetracycline (30 μg). Susceptible and resistant isolates were defined according to the criteria suggested by the National Committee for Clinical Laboratory Standards (23).
Multiplex PCR detection of resistance genes and the virulence plasmid of serovar Typhimurium.
The ACSSuT-type serovar Typhimurium DT104 possesses multiple resistance genes that mediate resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline. Serovar Typhimurium also contains an endogenous virulence plasmid that encodes a virulence gene, spv. Primer sequences for the detection of these genes are listed in Table 1. A 50-μl reaction mixture contained six different primer pairs of different concentrations, 200 μM deoxynucleoside triphosphates, 1 U of Taq DNA polymerase (Promega), 1.5 mM MgCl2, and 2 μl of DNA templates. An aliquot of overnight bacterial culture (100 μl) was collected and boiled at 100°C for 10 min. The bacterial lysate was centrifuged at 13,000 × g for 10 min, and the supernatant was used as the DNA template. The multiplex PCR was performed for 35 cycles as follows: denaturation at 95°C for 45 s, annealing at 56°C for 45 s, and extension at 72°C for 1 min. A final step of extension at 72°C for 5 min was performed. To further identify the integrons of these isolates, PCR using primers CS-F and CS-R (Table 1) was performed to amplify the class 1 integron resistance gene cassettes according to the method described previously (18). After amplification, the PCR product was sequenced by using an ABI 3730 autosequencer. In all experiments, serovar Typhimurium BN9181, a ACSSuT-type DT104 strain, was used as a control.
TABLE 1.
Primers used in the multiplex PCR
| Primera | Target | Sequence (5′ to 3′) | Nucleotide positionb | Expected amplicon size (bp) | GenBank accession no.c |
|---|---|---|---|---|---|
| Pse-F | pse | GGCAATCACACTCGATGATGCGT | 263→286 | 156 | AY339985.1 |
| Pse-R | GGCTCAATACGGTCTAGACGAGT | 418→396 | |||
| FloR-F | floR | CTTTGGCTATACTGGCGATG | 3066→3085 | 266 | AY339985.1 |
| FloR-R | GATCATTACAAGCGCGACAG | 3331→3312 | |||
| STR-F1 | str | AGACGCTCCGCGCTATAGAAGT | 2202→2242 | 203 | AY339985.1 |
| STR-R1 | CGGACCTACCAAGGCAACGCT | 2404→2384 | |||
| Sul I-F | sulI | CGGATCAGACGTCGTGGATGT | 1468→1488 | 351 | AY339985.1 |
| Sul I-R | TCGAAGAACCGCACAATCTCGT | 1818→1797 | |||
| TetG-F | tetG | AGCAGCCTCAACCATTGCCGAT | 6934→6955 | 391 | AY339985.1 |
| TetG-R | GGTGTTCCACTGAAAACGGTCCT | 7324→7302 | |||
| SpvC-1 | spvC | ACTCCTTGCACAACCAAATGCGGA | 553→573 | 447 | M64295.1 |
| SpvC-2 | TGTCTCTGCATTTCGCCACCATCA | 999→977 | |||
| CS-F | int | GGCATCCAAGCAGCAAG | Variable | AY339985.1 | |
| CS-R | AAGCAGACTTGACCTGA |
Primer name as it appears in the text.
Corresponding nucleotides in the referenced accession number that correspond to the 5′-to-3′ primer sequence.
Sequence in the database used for primer design.
Identification of virulence plasmid by DNA-DNA hybridization.
The virulence plasmid of serovar Typhimurium isolates was checked by Southern blotting hybridization with the probe of spvC DNA fragment amplified by PCR (7, 8, 28). Plasmid DNA was separated by a method described earlier (17). The plasmid DNA was then transferred onto Zeta-Probe membrane (Bio-Rad) with the method recommended by the manufacturer. PCR products of spvC were purified by Wizard SV gel and PCR Clean-Up System (Promega), labeled with digoxigenin-11-dUTP (Roche), and then hybridized to form digoxigenin-labeled probe-target hybrids. After the addition of anti-digoxigenin antibody conjugated with peroxidase, the membrane was reacted with chemiluminescent substrate and then exposed to X-ray film. A virulence plasmid was identified by a positive PCR and DNA-DNA hybridization.
Serotyping and phage typing.
To confirm whether serogroup B ACSSuT-type Salmonella isolates were serovar Typhimurium, these isolates were further examined for their serovars by the tube agglutination test using H antisera (Becton Dickinson Co., Franklin Lakes, NJ). Representative strains of ACSSuT-type Salmonella serogroup B were phage typed at the French National Center for Salmonella (Institut Pasteur, Paris, France) with the method described by Anderson et al. (1). Strains that did not react with any of the typing phages were considered nontypeable.
PFGE.
Genetic variation of the phage-typed serovar Typhimurium isolates was analyzed by PFGE. Bacteria were collected from overnight broth culture and embedded in 1% agarose to form a plug. The plug was treated with 1 mg of proteinase K/ml at 50°C overnight. The plug was washed eight times with distilled water at 50°C and then with T10E1 buffer for 4 h. The plug was then digested with 50 U of restriction endonuclease XbaI. The digested DNA was subsequently separated by CHEF Mapper XA (Bio-Rad) in 0.5× Tris-borate-EDTA at 14°C for 22 h. Due to the conserved characteristic of the Salmonella genome, band patterns that differed more than one band were designated as different genotypes.
RESULTS
Antimicrobial susceptibility and integron analysis.
According to the results of antimicrobial susceptibility testing, three major types of antimicrobial resistance were detected (Tables 2 and 3). The most predominant group was the isolates that were resistant to ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline (ACSSuT), followed by those resistant to streptomycin and sulfisoxazole (SSu), and those susceptible to all five drugs. The three types were evenly distributed in each year during the study period without obvious fluctuation. None of the isolates was resistant to ceftriaxone.
TABLE 2.
Types of antimicrobial resistance among 104 isolates of Salmonella serogroup B during 1997 to 2003
| Type of resistancea | No. of isolates from (yr of isolation):
|
No. (%) of isolates | |||||
|---|---|---|---|---|---|---|---|
| 1997 | 1998 | 1999 | 2000- 2001 | 2002 | 2003 | ||
| ACSSuT | 11 | 15 | 16 | 3 | 3 | 3 | 51 (49.0) |
| ACSSu | 1 | 0 | 0 | 0 | 0 | 0 | 1 (1.0) |
| SSuT | 0 | 0 | 0 | 1 | 1 | 0 | 2 (1.9) |
| ASSu | 0 | 0 | 1 | 0 | 0 | 0 | 1 (1.0) |
| CST | 0 | 1 | 0 | 0 | 0 | 0 | 1 (1.0) |
| SSu | 11 | 5 | 4 | 2 | 6 | 1 | 29 (27.9) |
| Su | 1 | 0 | 0 | 0 | 0 | 1 | 2 (1.9) |
| C | 0 | 0 | 0 | 0 | 0 | 1 | 1 (1.0) |
| S | 0 | 0 | 0 | 1 | 0 | 0 | 1 (1.0) |
| T | 0 | 1 | 0 | 0 | 0 | 0 | 1 (1.0) |
| All susceptible | 2 | 1 | 5 | 3 | 2 | 1 | 14 (13.5) |
A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfonamide; T, tetracycline.
TABLE 3.
Detection of various antimicrobial resistance types by the disk diffusion and multiplex PCR methods and of the virulence plasmid by the PCR
| Type of resistancea | No. of isolates detected by:
|
No. of isolates with the virulence plasmid | |
|---|---|---|---|
| Disk diffusion | Multiplex PCR | ||
| ACSSuT | 51 | 51 | 48 |
| ACSSu | 1 | 2 | 0 |
| CSSuT | 0 | 3 | 0 |
| SSuT | 2 | 0 | 0 |
| ASSu | 1 | 3 | 0 |
| CST | 1 | 0 | 0 |
| SSu | 29 | 26 | 1 |
| Su | 2 | 6 | 0 |
| C | 1 | 1 | 0 |
| S | 1 | 1 | 0 |
| T | 1 | 1 | 0 |
| All susceptible | 14 | 10 | 5 |
A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfonamide; T, tetracycline.
When we compared the two methods (disk diffusion and PCR), a few discrepancies were found (Table 3). There were some false positives in the detection of antibiotic resistance by PCR method, suggesting the existence of DNA fragments from inactive genes.
However, the detection rate of the ACSSuT-type resistance was exactly the same using either method. All of the resistance genes that were located on SGI1 (2, 3, 10) can be efficiently detected by this multiplex PCR method. PCR amplification of the CS region revealed that all of the 51 ACSSuT-type isolates contained a 1.25-kb DNA fragment derived from SGI1. Sequence analysis of the fragment showed that nucleotide sequence and gene order of the PCR product was identical to those of the SGI1 with an aad gene in the CS region.
Multiplex PCR and plasmid analysis.
The multiplex PCR appeared to be reliable for identifying multidrug-resistant serovar Typhimurium with an ACSSuT-type resistance from the many multidrug-resistant serogroup B Salmonella isolates (Fig. 1). To ensure the specificity of the method, clinical isolates were further characterized for their serovars by using H antisera and virulence plasmids using DNA-DNA hybridization. There are some differences in the antigen profiles between typical serovar Typhimurium and serovar Typhimurium var. Copenhagen: the H antigen profile of serovar Typhimurium is i:1,2,7, while that of serovar Typhimurium var. Copenhagen is i:1,2; also, the O antigen profile of serovar Typhimurium is 1,4,5,12, while that of serovar Typhimurium var. Copenhagen is 1,4,12. In the present study, we identified 10 isolates of serovar Typhimurium var. Copenhagen and three isolates of serovar Typhimurium with the monophasic H antigen 4,5,12:i:− (Table 4). On the other hand, the virulence plasmid was detected in the majority of serovar Typhimurium isolates (54 of 59 [91.5%]) (Table 4). Among the 30 phage-typed Salmonella, 96.3% (26 of 27) of the serovar Typhimurium (DT104, n = 21; DT120, n = 2; U302, n = 4) contained a virulence plasmid (Table 5). The remaining non-Typhimurium serogroup B Salmonella isolates (two serovar Derby and one serovar Agona) lacked a virulence plasmid.
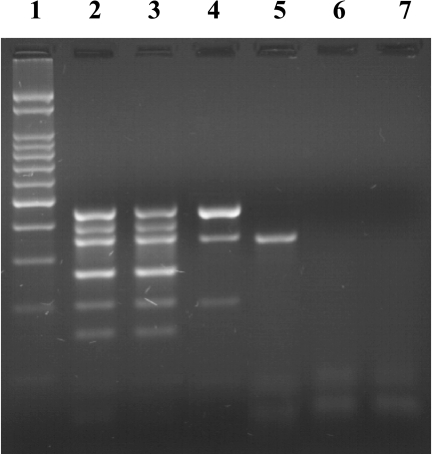
FIG. 1.
Results of the multiplex PCR. The PCR products were analyzed by 2% agarose gel electrophoresis and staining with ethidium bromide. Lanes: 1, 100-bp marker; 2, serovar Typhimurium BN9181 (ACSSuT, DT104 strain); 3, Sal91 (ACSSuT); 4, Sal235 (SSu); 5, Sal42 (Su); 6, Sal14 (susceptible to all antibiotics tested); 7, blank.
TABLE 4.
Plasmid analysis of the 104 clinical isolates of Salmonella serogroup B
| Isolate | No. of isolates:
|
|||
|---|---|---|---|---|
| Total | With the virulence plasmid | With nonvirulence plasmids (>50 kb) | With no plasmid | |
| Serovar Typhimurium | 49 | 47 | 1 | 1 |
| Serovar Typhimurium var. Copenhagen | 10 | 7 | 0 | 3 |
| Others | 45 | 0 | 9 | 36 |
TABLE 5.
Distribution of phage types, virulence plasmid, and type 1 integron among 30 representative isolates with various types of antimicrobial resistance
| Type of resistancea | Phage type | No. (%) of isolates
|
||
|---|---|---|---|---|
| Total | With the virulence plasmid | With the type I integron | ||
| ACSSuT | DT120 | 1 | 1 (100) | 1 (100) |
| DT104 | 21 | 21 (100) | 21 (100) | |
| U302 | 4 | 3 (75) | 4 (100) | |
| Insensitive (serovar Derby) | 2 | 0 (0) | 2 (100) | |
| ACSSu | Insensitive (serovar Agona) | 1 | 0 (0) | 0 (0) |
| SSu | DT120 | 1 | 1 (100) | 0 (0) |
A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfonamide; T, tetracycline.
Phage types and genotypes of the ACSSuT-type serovar Typhimurium isolates.
A total of 26 ACSSuT-type isolates and 1 SSu-type isolate of serovar Typhimurium were phage typed. Phage types DT104 (77.8%) (DT104B, 25.9%; DT104L, 51.9%), DT120 (7.4%), and U302 (14.8%) appeared in Taiwan as early as 1997. Although the number of isolates tested was limited, DT104 seemed to be the most common phage type among ACSSuT-type serovar Typhimurium isolates in Taiwan from 1997 to 2003. The other two phage types with the same ACSSuT type were only found occasionally. All ACSSuT-type isolates contained an integron as expected; however, the SSu-type isolate with a DT120 phage type did not harbor any integron. Based on the difference in the XbaI restriction fragments demonstrated by PFGE, all strains were separated into six genotypes (Fig. 2). We found two genotypes in DT104 strains (A and B), another two in DT120 (C and D), and two in U302 (E and F). Despite the similarity in phage types, serovars, and antibiotic susceptibilities, genomic variation (genotypes) among these predominant ACSSuT-type serovar Typhimurium strains was demonstrated by PFGE (Fig. 2).
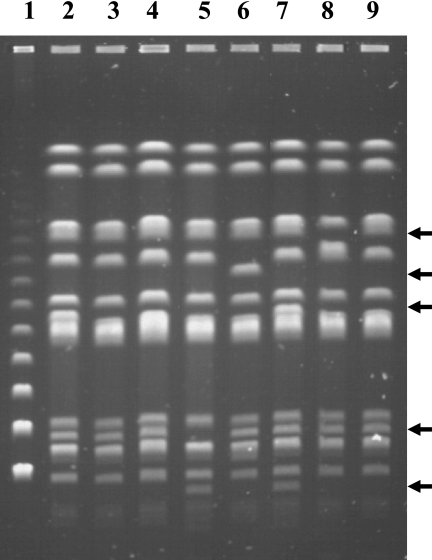
FIG. 2.
Genotypes of selected ACSSuT-type serovar Typhimurium with various phage types by PFGE. Lanes: 1, lambda ladder marker; 2, Sal1B (DT120); 3, Sal21B (DT104); 4, Sal34B (DT104); 5, Sal66B (U302); 6, Sal152B (U302); 7, Sal235B (DT120); 8, CY-S91B (DT104); 9, CY-S142B (DT104). Arrows indicate differences of the DNA fragments.
DISCUSSION
Among the 104 human isolates of serogroup B Salmonella collected from 1997 to 2003, the predominant serovar was serovar Typhimurium; of these, more than 90% of the isolates were ACSSuT-type serovar Typhimurium. Very few serovar Typhimurium isolates were virulence plasmidless, and this justified our multiplex PCR method that includes spvC primers to rapidly identify ACSSuT-type serovar Typhimurium from the multidrug-resistant serogroup B isolates. This PCR method appears to be able to substitute the traditional methods of serotyping and antibiotic susceptibility testing, which have been used widely in epidemiological investigations.
The ACSSuT resistance type has been reported in other serovars of Salmonella, including serovars Agona, Paratyphi A, Albany, and Newport (4, 11, 12, 22). All of these strains contained SGI1 (4, 9, 10, 21). By using PCR to detect class 1 integron in the SGI1, we demonstrated that all ACSSuT-type serovar Typhimurium harbored SGI1. In addition, we also identified SGI1 in multidrug-resistant serovar Derby. Whether the SGI1 of serovar Derby is a new variant warrants further studies.
Serovars of Salmonella are determined by O and H antigen profiles using antisera. The flagellar (H) antigen is expressed in the switching state between phase 1 and phase 2 antigens by the recombination event of an inverted repeat flanking the promoter region (27). Recently, monophasic H antigen expression (4,5,12:i:−) of serovar Typhimurium has been reported in phage type DT U302 (9). The mechanism that contributes to such expression change may be the inactivation of hin or one of the structural genes, fliC and fljB. The present study did identify the similar monophasic H antigen profile 4,5,12:i;− in three clinical serogroup B Salmonella isolates. These isolates contained a 94.7-kb virulence plasmid, indicating that they are serovar Typhimurium, although the phage types of these isolates have not been determined yet. The result also implies that such monophasic strains of serovar Typhimurium may not be uncommon in the clinical setting. Recently, nucleotide sequence analysis of the Salmonella flagellin genes fliC, fljB, and flpA showed that the sequences clustered by the antigens they encode and not by locus (21). Alleles encoding the same flagellar antigen were homologous, suggesting that flagellin genes may be useful targets for the molecular determination of flagellar antigen types (21). The multiplex PCR used in the present study that targets the spv-type virulence plasmid, in addition to the multiple resistance genes, of serovar Typhimurium would add to these methods in the molecular determination of the serovars of clinical Salmonella isolates.
Multidrug-resistant serovar Typhimurium DT104 has been reported in Eastern Asia, including Korea and Japan (26, 32). Here we reported the ACSSuT-type serovar Typhimurium phage types DT104, DT120, and DT U302 isolated from human sources in Taiwan since as early as 1997. The most common phage type identified was DT104 (DT104B and DT104L). The result again reflects the severe problem of antimicrobial resistance to Salmonella in Taiwan (29). On the other hand, PFGE analysis showed that genetic variations are present among different phage types, as well as among isolates within the same phage type, suggesting diversification of serovar Typhimurium in Taiwan. Serovar Typhimurium is a serovar with a broad host range. Whether the drastic genetic variations of serovar Typhimurium isolates derive from this organism's wide host range, i.e., each different type of serovar Typhimurium that caused human infections is derived from a different animal host, will require further investigation.
Acknowledgments
This study was funded by grants from Chang Gung Memorial Hospital (CMRPG32033 and CMRPG63011) and National Science Council (NSC93-2314-B-182A-071) of Taiwan.
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. De Saxe, and J. D. De Sa. 1977. Bacteriophage typing designations of Salmonella typhimurium. J. Hyg. 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrego, J. J., D. Castro, M. Jimenez-Notario, A. Luque, E. Martinez-Manzanares, C. Rodriguez-Avial, and J. J. Picazo. 1992. Comparison of epidemiological markers of Salmonella strains isolated from different sources in Spain. J. Clin. Microbiol. 30:3058-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., G. A. Peters, A. Cloeckart, K. Sidi Boumedine, E. Chaslus-Dancia, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serotype Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, S. A., L. F. Bolton, C. E. Briggs, H. S. Hurd, V. K. Sharma, P. J. Fedorka-Cray, and B. D. Jones. 1999. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell Probes 13:213-222. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, C. H., T. Y. Lin, and J. T. Ou. 1999. Prevalence of the virulence plasmids of nontyphoid Salmonella in the serovars isolated from humans and their association with bacteremia. Microbiol. Immunol. 43:899-903. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, C. H., and J. T. Ou. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34:2619-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, C. H., T. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y. Y. Chou, H. S. Wang, and Y. S. Lee. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De la Torre, E., D. Zapata, M. Tello, W. Mejía, N. Frías, F. J. García Peña, E. M. Mateu, and E. Torre. 2003. Several Salmonella enterica subsp. enterica serotype 4,5,12:i:− phage types isolated from swine samples originate from serotype Typhimurium DT U302. J. Clin. Microbiol. 41:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckert. 2005. The Salmonella genomic island 1 is an integrative mobilization element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 11.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublet, B., F. X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier, M., and B. W. Blais. 2004. Cloth-based hybridization array system for the detection of multiple antibiotic resistance genes in Salmonella enterica subsp. enterica serotype Typhimurium DT104. Lett. Appl. Microbiol. 38:265-270. [DOI] [PubMed] [Google Scholar]
- 14.Gulig, P. A., H. Danbara, D. G. Guiney, A. J. Lax, F. Norel, and M. Rhen. 1993. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 7:825-830. [DOI] [PubMed] [Google Scholar]
- 15.Helms, M., S. Ethelberg, K. Molbak, et al. 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg. Infect. Dis. 11:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmuth, R., R. Stephan, C. Bunge, B. Hoog, A. Steinbeck, and E. Bulling. 1985. Epidemiology of virulence-associated plasmids and outer membrane protein patterns within seven common Salmonella serotypes. Infect. Immun. 48:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kado, C., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 174:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai-King, N. G., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauderdale, T. L., F. M. Aarestrup, P. C. Chen, J. F. Lai, H. Y. Wang, Y. R. Shiau, I. W. Huang, C. L. Hung, et al. 2006. Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn. Microbiol. Infect. Dis., 55:140-155. [DOI] [PubMed]
- 20.Liebana, E., L. Garcia-Migura, C. Clouting, F. A. Clifton-Hadley, E. Lindsay, E. J. Threlfall, S. W. McDowell, and R. H. Davies. 2002. Multiple genetic typing of Salmonella enterica serotype Typhimurium isolates of different phage types (DT104, U302, DT204b, and DT49) from animals and humans in England, Wales, and Northern Ireland. J. Clin. Microbiol. 40:4450-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McQuiston, J. R., R. Parrenas, M. Ortiz-Rivera, L. Gheesling, F. Brenner, and P. I. Fields. 2004. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42:1923-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A7, 7th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Pritchett, L. C., M. E. Konkel, J. M. Gay, and T. E. Besser. 2000. Identification of DT104 and U302 phage types among Salmonella enterica serotype Typhimurium isolates by PCR. J. Clin. Microbiol. 38:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross, I. L., and M. W. Heuzenroeder. 2005. Discrimination within phenotypically closely related definitive types of Salmonella enterica serovar Typhimurium by the multiple amplification of phage locus typing technique. J. Clin. Microbiol. 43:1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sameshima, T., M. Akiba, H. Izumiya, J. Terajima, K. Tamura, H. Watanabe, and M. Nakazawa. 2000. Salmonella typhimurium DT104 from livestock in Japan. Jpn. J. Infect. Dis. 53:15-16. [PubMed] [Google Scholar]
- 27.Silverman, M., J. Zieg, M. Hilmen, and M. Simon. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. USA 76:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern, E. M. 1975. Detection of specific sequence among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 29.Su, L. H., C. H. Chiu, C. Chu, and J. T. Ou. 2004. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin. Infect. Dis. 39:546-551. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, K., K. Nishimori, S. I. Makino, T. Nishimori, T. Kanno, R. Ishihara, T. Sameshima, M. Akiba, M. Nakazawa, Y. Yokomizo, and I. Uchida. 2004. Molecular characterization of a prophage of Salmonella enterica serotype Typhimurium DT104. J. Clin. Microbiol. 42:1807-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Threlfall, E. J., B. Rowe, and L. R. Ward. 1993. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol. Infect. 111:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, S. J., K. Y. Park, S. H. Kim, K. M. No, T. E. Besser, H. S. Yoo, S. H. Kim, B. K. Lee, and Y. H. Park. 2002. Antimicrobial resistance in Salmonella enterica serovars Enteritidis and Typhimurium isolated from animals in Korea: comparison of phenotypic and genotypic resistance characterization. Vet. Microbiol. 86:295-301. [DOI] [PubMed] [Google Scholar]