Abstract
In the most common C4 pathway for carbon fixation, an NADP-malic enzyme (NADP-ME) decarboxylates malate in the chloroplasts of bundle sheath cells. Isoforms of plastidic NADP-ME are encoded by two genes in all species of Flaveria, including C3, C3-C4 intermediate, and C4 types. However, only one of these genes, ChlMe1, encodes the enzyme that functions in the C4 pathway. We compared the expression patterns of the ChlMe1 and ChlMe2 genes in developing leaves of Flaveria pringlei (C3) and Flaveria trinervia (C4) and in transgenic Flaveria bidentis (C4). ChlMe1 expression in C4 species increases in leaves with high C4 pathway activity. In the C3 species F. pringlei, ChlMe1 expression is transient and limited to early leaf development. In contrast, ChlMe2 is expressed in C3 and C4 species concurrent with stages in chloroplast biogenesis. Because previous studies suggest that NADP-ME activities generally reflect the level of its mRNA abundance, we discuss possible roles of ChlMe1 and ChlMe2 based on these expression patterns.
Plants using the C4 pathway for carbon fixation minimize the energetically wasteful process of photorespiration. The C4 “syndrome” usually includes extensive vascularization of the leaf, with a ring of photosynthetic bundle sheath (BS) cells surrounding each vein and an outer ring of mesophyll (M) cells surrounding the BS (Dengler and Nelson, 1999). CO2 is first fixed in M cells, shuttled to the BS cells as a 4-carbon acid, enzymatically released from the acid, and refixed by Rubisco in the reductive pentose phosphate pathway (e.g. Furbank and Taylor, 1995; Kanai and Edwards, 1999; Leegood and Walker, 1999).
C4 photosynthesis requires enzyme activities in addition to those needed for the C3 pathway. Among these, NAD-malic enzyme (NAD-ME), NADP-ME, or phosphoenolpyruvate (PEP) carboxykinase (depending on the C4 biochemical type) is needed to release CO2 for refixation in BS cells. Genes encoding these supplementary enzymes are present in both C3 and C4 plants. In C3 plants, the metabolic roles played by these enzymes are likely to differ from those in C4 species. For instance, NADP-ME is induced in a non-cell-specific manner in C3 plants in apparent support of defense responses (for review, see Drincovich et al., 2001). In C4 plants of the NADP-ME type, it is light-induced, localized to chloroplasts of BS cells, and acts to release the CO2 and reducing power transferred from M cells in the form of malate (Edwards and Andreo, 1992). The evolution of this C4 pathway required the re-regulation of an NADP-ME gene to allow the enzyme to function in a cell-specific and light-induced manner (Ku et al., 1996; Monson, 1999; Sheen, 1999).
The genus Flaveria (Asteraceae) contains C3 and C4 species and C3-C4 intermediate species, which have C4 photosynthetic capacity ranging from C3- to C4-like (Ku et al., 1991). This diversity makes Flaveria spp. experimentally useful for investigating the molecular events that accompanied the transition from C3 to C4 photosynthesis. The genus has been divided into two phylogenetic lines (Powell, 1978). The 3-4 phyllary line includes all C3 and C4 species in addition to some C3-C4 intermediates, whereas the 5-6 phyllary line consists of only C3-C4 species. Although the phylogenetic relationship between the two lineages is unclear, evidence from genetic hybridization experiments, the number of phyllaries (floral bracts), eco-geographical considerations, and phylogenetic studies support the division (Powell, 1978; Bayraktaroglu, 1993; Kopriva et al., 1996; Marshall et al., 1996). Of these, the recent phylogenetic data have been quite revealing. Both the studies based on chloroplast genomic RFLP (Bayraktaroglu, 1993) and those based on sequence comparison of the gene encoding the H-protein of the Gly cleavage system (gdcsH; Kopriva et al., 1996) revealed a tree in which the C3-C4 species, placed between the basal C3 group and the apical C4 group, are divided into two distinct clades.
In Flaveria spp., NADP-ME genes are expressed in different patterns, depending on the photosynthetic type. Three protein isoforms of molecular masses 62-, 64-, and 72-kD have been detected immunologically in all Flaveria spp. examined (Drincovich et al., 1998; Casati et al., 1999), although the correspondence between the NADP-ME genes and the observed isoforms is unclear. Several NADP-ME genes have been identified in Flaveria spp. The genes Me1 and Me2 (Marshall et al., 1996) encode chloroplast-localized C4 and non-C4 enzymes, respectively. We refer to them here as ChlMe1 and ChlMe2 to distinguish them from the CytMe gene, which encodes a cytosolic NADP-ME (L. Lai, S.L. Tausta, and T. Nelson, unpublished data). All three of these genes are present in C3, C4 and C3-C4 species of Flaveria, in some species as small gene families (Marshall et al., 1996; L. Lai, S.L. Tausta and T. Nelson, unpublished data).
The roles and regulation of NADP-ME genes in C3 and C3-C4 species are of interest for understanding the recruitment of enzymes for the C4 pathway. The ChlMe1 gene, which encodes an abundant BS cell-specific chloroplastic enzyme in leaves of C4 species, was found in the C3 species Flaveria pringlei (Marshall et al., 1996). Although F. pringlei is possibly a degenerate allotetraploid derived from a C3 and a C3-C4 parent, we confirm in this study the coexistence of the ChlMe1 and ChlMe2 genes in the diploid C3 species Flaveria robusta and in six C3-C4 species. We also characterize the expression patterns of ChlMe1 and ChlMe2 by means of RNA quantitation and transgenic reporter studies. ChlMe1 is expressed in leaves of both C3 and C4 plants. We suggest that the enzyme acts in CO2 refixation schemes in both C3 (only in immature leaves) and C4 species. The accumulation of ChlMe2 transcripts is closely correlated with early developmental states of chloroplasts. We suggest that this enzyme functions in both C3 and C4 plants to provide NADPH for chloroplast biogenesis and/or plastid-localized biosynthesis.
RESULTS
Both ChlMe1 and ChlMe2 Are Present in All Flaveria Species
To assay the presence of ChlMe1 and ChlMe2 in Flaveria spp. of all photosynthetic types, we used thermal asymmetric interlaced PCR (TAIL-PCR; Liu et al., 1995) to clone the 3′ regions of both genes, because these 3′ regions exhibited the most sequence differences between the two ChlMe genes in Flaveria spp. (Marshall et al., 1996). Five species, in addition to the three previously assayed (Marshall et al., 1996), were included: two C3 species (F. robusta and F. pringlei), four C3-C4 species (Flaveria linearis, Flaveria floridana, Flaveria brownii, and Flaveria angustifolia), and two C4 species (Flaveria trinervia and Flaveria bidentis). F. linearis, F. floridana, and F. brownii belong to the 5-6 phyllary line, and the remaining species all belong to the 3-4 phyllary line (Powell, 1978). F. robusta is a diploid without any evidence of introgression of C4 traits, whereas F. pringlei is likely to be more complex. Twenty-four sequences were obtained and are available in GenBank (accession nos. AF288896–AF288919). One of these sequences, F. pringlei ChlMe2-1, was obtained by subcloning from a ChlMe2 genomic clone (see “Materials and Methods”).
The 3′ regions characteristic of distinct ChlMe1 and ChlMe2 genes were obtained from all eight Flaveria spp. Multiple clones from each species were sequenced. Despite interspecific differences (i.e. single base mismatches), high overall sequence similarity was observed among species within each phyllary line. An alignment of the 3′-untranslated region (UTR) sequences of ChlMe1 and ChlMe2 genes is shown in Figure 1. The identification of the highly variable regions and the C4-specific nucleotides results from the analysis of more than 20 sequences of each gene. For simplicity, only one representative sequence from each photosynthetic group is presented.
Figure 1.
Alignment of the representative sequences of the ChlMe1 and ChlMe2 3′-UTRs obtained by TAIL-PCR. A, 3′-UTR sequences of ChlMe1. The sequence from F. trinervia was presented for the C4(3-4) group, F. pringlei ChlMe1-2 for the C3(3-4) group, and F. brownii ChlMe1-1 for the C3-C4(5-6) group. B, 3′-UTR sequences of ChlMe2. The sequence from F. trinervia was presented for the C4(3-4) group, F. pringlei ChlMe2-2 for the C3(3-4) group, and F. brownii ChlMe2-3 for the C3-C4(5-6) group. The sequences included were immediately after the stop codon and upstream of the polyadenylation site, as defined by the F. trinervia ChlMe1 and the F. pringlei ChlMe2 cDNAs (Börsch and Westhoff, 1990; Lipka et al., 1994). F. ang(3-4) was the single species examined of the C3-C4(3-4) group. Dashes at the beginning of F. ang(3-4) ChlMe1 sequence represent unknown sequence; the remaining dashes represent deletions. Black bars indicate regions with high variability. Stars in A indicate the C4-specific nucleotides. See text for more details.
The ChlMe1 3′-UTR sequences obtained from the two C4 species F. trinervia and F. bidentis are identical with those reported previously (Börsch et al., 1990; Rajeevan et al., 1991; Marshall et al., 1996). The major differences among the photosynthetic groups and phyllary lines are found in three regions (indicated by bars in Fig. 1A). The sequences in region I are the same within the C3 and C3-C4(5-6) groups (the number in parentheses indicates the phyllary line). Because only one C3-C4(3-4) species, F. angustifolia, was assessed, the extent of region I sequence divergence within this group is unknown. Although the sequences from the two C4 species were highly similar, significant differences were found in region I (F. trinervia = GCG3A7 and F. bidentis = GCG5A5G). Regions II and III harbor the deletions found in the C3-C4(5-6) species and contribute to the dimorphism found between the two phyllary lines; region II is the site of a previously identified length dimorphism (Marshall et al., 1996). Several of the mismatches throughout the 3′-UTR are C4-specific (indicated by stars in Fig. 1A). The functional significance of these C4-specific mismatches and the three regions discussed above is unknown.
The 3′-UTR sequences of ChlMe2 are more conserved among the Flaveria spp. than are those of ChlMe1. The majority of differences are found in three regions (indicated by bars in Fig. 1B). Region I is characterized by varying numbers of TG dinucleotide repeats (four–six repeats). Similarly, region II varies in the length and sequence of an A-rich seqence [(A/T) A(A/G)(A/C)(A/G)2A2–3A0–1(A/G)0–1].The TCAGCT motif of region III is absent in the single copy of F. angustifolia and in several copies of the C3 species. The functional significance of these regions is presently unknown.
ChlMe1 and ChlMe2 Are Single-Copy Genes in C3-C4 and C4 Flaveria Species but Are Small Gene Families in C3 Species
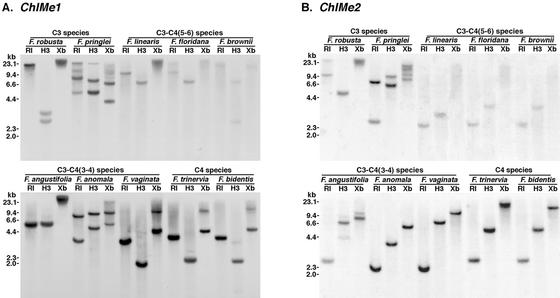
The above study gives an indication of ChlMe1 and ChlMe2 gene copy number. However, the TAIL-PCR approach may not have amplified all ChlMe loci. As an additional measure of gene copy number, genomic-blot analysis was carried out using ChlMe1- and ChlMe2-specific probes (Fig. 2, A and B, respectively). In addition to the species for which we evaluated 3′-UTRs, two more C3-C4(3-4) species, Flaveria anomala and Flaveria vaginata, were included. Although low-stringency hybridization was carried out, the weak signals were obtained in the C3-C4(5-6) lanes. Based on the combined data from the blot analysis and TAIL-PCR of 3′-UTRs, both ChlMe1 and ChlMe2 are single-copy genes in the two C4 species, the two C3-C4(3-4) species F. angustifolia and F. vaginata, and the two C3-C4(5-6) species F. linearis and F. floridana. ChlMe1 and ChlMe2 genes are present as two and one copy, respectively, in the third C3-C4(3-4) species, F. anomala, and as one and three copies, respectively, in the third C3-C4(5-6) species, F. brownii. As expected, there are multiple (two–four) copies of both ChlMe1 and ChlMe2 in the tetraploid C3 species F. pringlei. However, in the diploid C3 F. robusta, two copies of ChlMe1 and three copies of ChlMe2 are present.
Figure 2.
Southern analysis of genomic DNAs from Flaveria spp. using probes specific for ChlMe1 (A) and ChlMe2 (B). The different species are grouped together according to types of photosynthesis. Within each of the C3-C4 groups, the species are placed in order of increasing C4 capabilities (Ku et al., 1991). Note that F. brownii and F. vaginata have been characterized as C4-like. RI, EcoRI; H3, HindIII; Xb, XbaI. Numbers at left indicate sizes in kilobase pairs.
The Two ChlMe Genes Have Distinct Patterns of Expression during Leaf Development in F. pringlei (C3) and F. trinervia (C4)
Because the roles of ChlMe1 and ChlMe2 under various photosynthetic schemes are uncertain, we characterized their expression patterns in representative C3 and C4 species, with particular attention to their responses to leaf development. Studies to date suggest that ME activities in plants directly reflect mRNA abundance, in contrast to the case for other enzymes of the C4 pathway, which are subject to complex regulation at the protein level (for review, see Sheen, 1999). In a previous study (Marshall et al., 1996), ChlMe1 expression was not detected in leaves of the C3 species F. pringlei. However, we reasoned that it might be expressed during a limited stage of leaf development that was not assayed in that study. Therefore, we measured ChlMe1 and ChlMe2 mRNA levels throughout the development of F. pringlei and F. trinervia leaves using relative quantitative RT-PCR.
In F. pringlei (C3) leaves, the level of ChlMe1 mRNA peaked early in development, declined rapidly, and was barely detectable in late stages (Fig. 3A). In the same leaves, the level of ChlMe2 mRNA peaked immediately after that of ChlMe1, then similarly declined. The coincidence of the rapid decrease in ChlMe1 transcripts with the peak of ChlMe2 expression suggests that the two genes might be expressed exclusively of one another, possibly via a negative feedback mechanism (see “Discussion”).
Figure 3.
Changes in the levels of ChlMe1 and ChlMe2 mRNAs during leaf development. A, F. pringlei leaves. B, F. trinervia leaves. Relative quantitative RT-PCR was used to analyze ChlMe1 and ChlMe2 mRNA levels in the same samples obtained from leaves of various lengths. 18S rRNA was used as the internal control in each sample. The results shown were the average of at least two experiments. Error bars represent se.
In developing F. trinervia (C4) leaves, we observed a rise in ChlMe1 mRNA levels coincident with the onset of C4 photosynthetic competency (Fig. 3B), as reported previously for activity levels (Moore et al., 1986; Moore and Edwards, 1988). In contrast, ChlMe2 transcript levels peaked at two stages in development. The first, seen during early development, was similar to that observed in F. pringlei young leaves (Fig. 3A), suggesting that the role ChlMe2 plays in early leaf development may be conserved in C3 and C4 species. The second was coincident with the peak of ChlMe1 expression in this species.
ChlMe Gene Expression in Transgenic F. bidentis (C4)
To evaluate the roles of 5′- and 3′-flanking sequences in the developmental regulation of ChlMe1 and ChlMe2, we monitored the activity of these regions in reporter gene fusion constructs (Fig. 4) in transgenic F. bidentis plants. Although F. pringlei, F. trinervia, and F. bidentis are closely related species, the heterologous nature of the expression studies reported here merits discussion. Because F. trinervia and F. bidentis are related C4 species, we anticipated that the 5′ and 3′ sequences of F. trinervia ChlMe1 would direct β-glucuronidase (GUS) expression in transgenic F. bidentis plants in a pattern highly similar to that of the endogenous ChlMe1 gene. We also anticipated good fidelity of F. pringlei ChlMe2 5′ and 3′ sequences in transgenic F. bidentis plants, because the endogenous ChlMe2 genes in F. pringlei and F. trinervia exhibit only small differences in expression pattern (Fig. 3, A and B). The patterns of GUS expression we observed were consistent among multiple independent transformed lines for each construct.
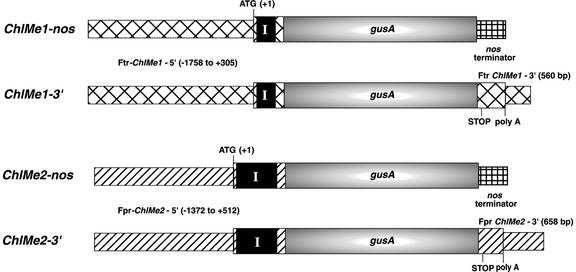
Figure 4.
Translational fusion constructs using 5′ and 3′ regions of F. trinervia ChlMe1 and F. pringlei ChlMe2 for F. bidentis transformation. The gusA gene encoding GUS was used in all constructs as a reporter. The lengths of the 5′ regions are specified with the start codon as +1. The black box labeled I represents the first intron. The length of each region is shown in relative proportion. See “Materials and Methods” for more details. Ftr, F. trinervia; Fpr, F. pringlei.
The non-uniform arrangement of cells relative to veins in Flaveria spp. leaves requires care in interpreting staining patterns. A cauliflower mosaic virus 35S-GUS-nos control construct was expressed at high levels throughout cotyledons and leaves in a non-cell-specific manner. At high concentrations of potassium ferricyanide and potassium ferrocyanide to slow product formation and diffusion during staining (5 versus 0.5 mm), only the tip portions of young, developing leaves showed staining, suggesting some level of developmental regulation. As a result of the preferential positioning of palisade M cells over veins in F. bidentis, GUS staining appeared to be concentrated around veins in paradermal view. At higher magnifications, however, non-cell-specific staining was evident in both BS and all palisade M cells.
The F. trinervia ChlMe1 Promoter Is Sufficient to Direct High-Level, BS-Preferential GUS Expression in F. bidentis Seedlings
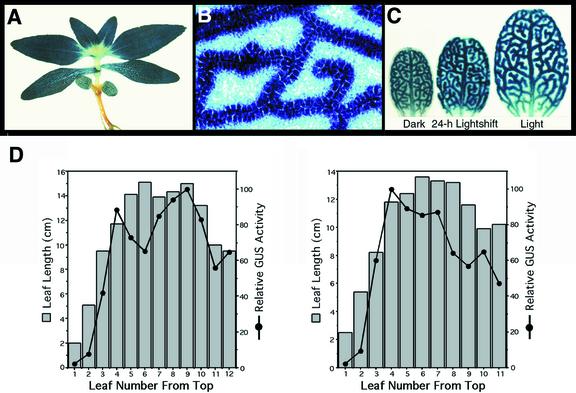
F. bidentis seedlings transformed with the ChlMe1-nos construct exhibited high-level expression in the cotyledons and true leaves (Fig. 5A). Intense GUS staining was concentrated in the BS cells (Fig. 5B). A low level of staining was also apparent in M cells of true leaves, but not in cotyledons. Furthermore, a basipetal increase in GUS staining was evident in developing leaves (youngest leaves in Fig. 5A) and cotyledons of very young seedlings (not shown). In regions with vascular differentiation and morphologically distinct BS cells, BS-preferential staining was observed. The ChlMe1 promoter activity corresponded to the extent of cellular development in the leaf because BS-specific staining increased dramatically with further BS maturity, culminating in very high levels and high specificity toward the tip of the leaf (Fig. 5C, light-grown cotyledon). Note that the cell specificity of staining, which was evident to an observer at the microscope, is less evident in a single photograph, because the background is additive in single exposures of whole mounts. We found that quantitative measurement of GUS activity in separated cells, even with controls, adds different caveats and artifacts associated with the cell separation process.
Figure 5.
Histochemical localization of GUS activity in leaves and cotyledons of F. bidentis seedlings transformed with the construct ChlMe1-nos. A, A 4-week-old seedling showing strong staining in both cotyledons and true leaves. B, High-magnification, paradermal view of a cleared seedling true leaf showing intense blue staining around veins. C, One-week-old cotyledons grown under different light conditions. Illumination enhanced staining somewhat in BS cells and suppressed staining in M cells. D, Relative GUS activity in consecutive leaves taken from a single branch of a greenhouse-grown plant transformed with ChlMe1-nos. Shown are data for two independent transformants. Gray bars represent the length of each leaf measured from the tip to the end of the petiole. Black points show GUS activity obtained from each leaf, plotted relative to the maximal point.
The addition of a 0.5-kb 3′ region of F. trinervia ChlMe1 to the ChlMe1 promoter construct (ChlMe1-3′) did not enhance the high level and BS cell specificity we observed with the F. trinervia promoter alone (data not shown). This observation differs from the results of a previous study (Marshall et al., 1997) that high-level expression from the F. bidentis ChlMe1 promoter required the presence of a 5.9-kb 3′-flanking region from the F. bidentis ChlMe1 gene. To identify potentially significant differences in the ChlMe1 promoters from the two species, we compared their sequences by dot matrix analysis (data not shown). The promoters were collinear, with the exception of the regions from −2,045 to −1,970 and from −1,757 to −1,184 in the F. bidentis promoter, which are absent in the F. trinervia promoter. The latter region exhibits a high AT content, which suggests the possible insertion of a transposable element. Such insertional events have been correlated with modification of gene expression (Wessler, 1998).
To characterize the activity of the F. trinervia ChlMe1 promoter throughout development, we examined GUS histochemical staining and in vitro activity in staged leaves of mature ChlMe1-nos plants. For histochemical staining, we used plants grown to maturity (having 10–11 nodes) on agar medium to overcome the low permeability to GUS substrate of soil-grown plants. In young developing leaves, we observed up-regulation of the ChlMe1 promoter in a basipetal fashion (not shown), similar to that observed in seedling leaves. Strong staining persisted until senescence.
To correlate ChlMe1 promoter activity with leaf developmental stage, we measured GUS activity as a function of leaf length among leaves collected from single branches of greenhouse-grown ChlMe1-nos plants. Consecutive, expanding leaves exhibited a regular increase in GUS activity from the young leaves at the top to older leaves at the bottom of the branch (Fig. 5D). Maximal levels were apparent just before full leaf expansion and dropped steadily by one-half at the time of visible senescence. The pattern of ChlMe1-nos expression corresponds to the expression pattern obtained for endogenous ChlMe1 in F. trinervia developing leaves (Fig. 3B). Because the ChlMe1-nos construct includes only the ChlMe1 promoter, this finding suggests that ChlMe1 expression in F. trinervia is primarily regulated by the 5′ region of the gene.
Light Plays a Role in M Suppression and BS Enhancement of ChlMe1 Expression in Cotyledons
Immunolocalization and in situ hybridization studies have indicated that light is essential for the cell-specific and high-level expression of C4 genes, including ME genes (Berry et al., 1985; Langdale et al., 1988). We therefore assayed how illumination affects ChlMe1 promoter activity in transgenic plants. Because GUS is a highly stable protein, experiments employing dark adaptation of light-grown plants failed to lower GUS levels in the dark within an experimentally useful time. However, we found that cotyledons of dark-germinated seedlings were a suitable substitute, as has been reported for the C4 dicot Amaranthus hypochondriacus (Wang et al., 1993).
In 1-week-old ChlMe1-nos seedlings grown in the dark, GUS staining in the mature regions of cotyledons was BS preferential, albeit at a much lower level than that found in light-grown cotyledons (Fig. 5C). In addition, the ratio of M to BS staining was higher than that found in light-grown cotyledons. After 24 h of illumination, GUS staining increased significantly in BS cells but only slightly in M cells (Fig. 5C), thereby increasing the specificity for BS cells. The BS specificity is likely to be higher than observed because of the likely stability of residual GUS in M cells. The effect of light quality was also examined in 1-week-old ChlMe1-nos seedlings. There was no apparent difference in cell-specific expression of ChlMe1 in seedlings grown in white, blue, red, and far-red light, although much lower levels were observed in far-red light (data not shown). Negative regulation of the C4 NADP-ME gene by far-red light has also been observed in maize (Casati et al., 1998). Together with results from experiments using dark-grown seedlings, we concluded that BS-preferential expression driven by the ChlMe1 promoter is linked to the development of BS cells and that light plays a role in enhancing the expression in these cells while suppressing expression in M cells.
The Expression of ChlMe2 in Transgenic F. bidentis Leaves Is Linked to Cell Differentiation and Light Levels
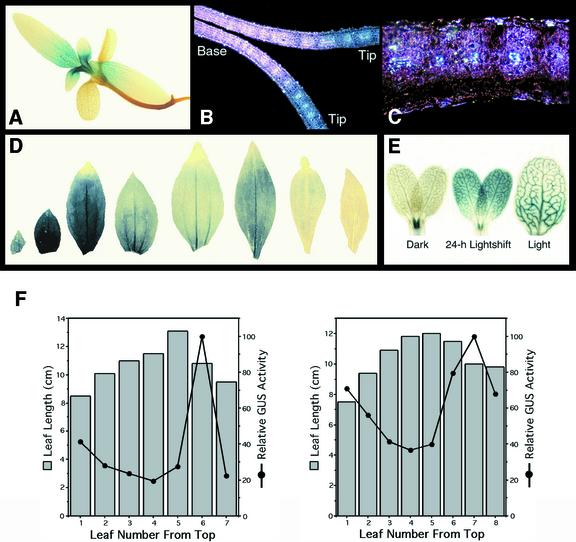
The expression pattern of the ChlMe2 promoter in transgenic F. bidentis was distinct from that of the ChlMe1 promoter. In ChlMe2-nos seedlings, GUS expression was uniform throughout emerging leaves and unexpanded cotyledons, then down-regulated basipetally as these organs matured (Fig. 6A). To view the cell-type specificity and comparative level of promoter activity as a function of the tip to base gradient of maturity in developing leaves, we longitudinally sectioned young leaves approximately 1.5 mm in length after histochemical staining. In the undifferentiated basal region of the leaf, staining was non-cell-specific (Fig. 6B). In regions with active vascular and BS differentiation, GUS staining was stronger in cells with high chloroplast number (Fig. 6C). These were BS cells and M cells immediately adjacent to BS cells (Stockhaus et al., 1997), and the higher GUS activity in these cells gave the appearance of BS-specific staining. In the more mature distal regions, GUS staining was completely absent (Fig. 6B). Thus, the ChlMe2 promoter appears to respond to a developmental gradient within seedling leaves, in a fashion opposite to that of the ChlMe1 promoter.
Figure 6.
Histochemical localization of GUS activity in leaves and cotyledons of F. bidentis seedlings and mature plants transformed with the construct ChlMe2-nos. A, A 3-week-old seedling showing basipetal down-regulation of GUS expression in leaves. B, Dark-field view of a longitudinal section of a primary leaf (approximately 1.5 mm in length). Under dark-field illumination, the crystalline GUS product appears as bright pink spots. Staining was uniform in the basal, non-differentiated region of the leaf and increased in region proximal to developing veins. C, High magnification of a region in (B) with strong staining. Staining was restricted to cells with high chloroplast number (i.e. BS cells and M cells surrounding BS cells). D, Consecutive leaves of a mature plant (youngest leaf at left). Note the absence of basipetal down-regulation of GUS expression. Staining was strongest in young leaves and down-regulated in older leaves. E, One-week-old cotyledons grown under different light conditions. Illumination was required for GUS expression in cotyledons but not in the first true leaves. F, Relative GUS activity in consecutive leaves taken from a single branch of a greenhouse-grown plant transformed with ChlMe2-nos. Shown are data for two independent transformants. Gray bars represent the length of each leaf measured from the tip to the end of the petiole. Black points show GUS activity obtained from each leaf, plotted relative to the maximal point.
In leaves from mature plants, however, the ChlMe2 promoter exhibited uniform activity at all stages of development. Figure 6D shows the staining pattern in consecutive leaves of plants grown to maturity on agar medium. The level of staining reflected the stage of development of the individual leaves. GUS activity increased significantly between the youngest leaf and the second leaf, and then decreased steadily as the leaves reached full expansion. Just before senescence, the expression increased briefly and then became undetectable in senescing leaves. The switch from the seedling to the adult staining pattern appeared to occur during the development of the third pair of true leaves (data not shown). To quantitate the changes in activity of the ChlMe2 promoter in developing leaves of mature plants, consecutive leaves at different stages of expansion were collected from individual greenhouse-grown plants and assayed quantitatively for GUS activity (Fig. 6F). Here, the youngest leaves assayed were probably more mature than the youngest leaf in Figure 6D, so the relative GUS activity curves started with a decline. The pattern of changes in GUS activity was similar to the pattern of staining shown in Figure 6D.
When the 3′ region of F. pringlei ChlMe2 was added to the ChlMe2 construct (ChlMe2-3′; data not shown), we observed some variation in expression pattern. First, GUS activity was somewhat higher in ChlMe2-3′ than in ChlMe2-nos leaves. Second, the peak of GUS activity observed after full leaf expansion in adult ChlMe2-nos plants (Fig. 6F, approximately leaf 6) was absent in ChlMe2-3′ plants. Because older leaves beyond full expansion were not included in the analysis of the mRNA levels of ChlMe2 (Fig. 3) as in the analysis of ChlMe2-nos and ChlMe2-3′ expression, it is not known whether this presenescence peak of GUS activity obtained with ChlMe2-nos is a trait of ChlMe2 expression. In any case, although modest in extent, the differences observed suggest that additional elements controlling the expression of ChlMe2 are present in the 3′-flanking region.
Light influenced ChlMe2 expression in cotyledons in an age-dependent manner. When seedlings containing the construct ChlMe2-nos were germinated in the dark, GUS was only detected at low levels in the veins of the cotyledons and was present uniformly in the developmentally arrested first true leaves (Fig. 6E). Light induction of GUS expression in dark-grown cotyledons appeared to be dependent on the age of the cotyledons. Illumination resulted in GUS expression in 1-week-old (Fig. 6E) but not in 5-week-old (not shown) cotyledons. During the first 24 h of greening, light had no effect on ChlMe2 expression in the first leaves (Fig. 6E), which continued to expand only after illumination. When T1 ChlMe2-nos seeds were germinated under different light qualities, results obtained with red and blue light were similar to that found with white light, whereas far-red light gave results identical to that obtained with dark-grown seedlings (data not shown).
ChlMe1 and ChlMe2 Have Distinct Expression Patterns in Non-Photosynthetic Organs
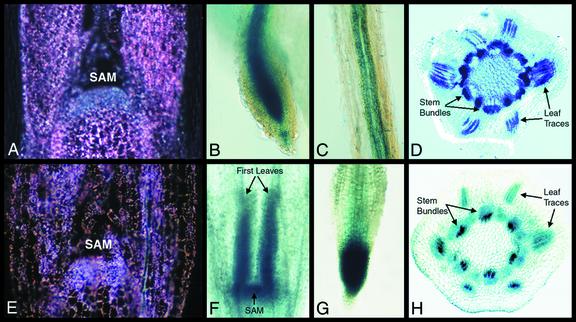
To further distinguish the roles of ChlMe1 and ChlMe2, we examined reporter gene expression patterns in non-photosynthetic tissues of transgenic F. bidentis. In the shoot apical meristem (SAM) of dark- and light-grown plants, ChlMe1 was not expressed although low levels of expression were observed in the surrounding tissues of light-grown plants (Fig. 7A). Root tips of seedlings and mature plants showed GUS staining in the columnellar cells and the vascular tissues flanking the non-staining root apical meristem (Fig. 7B). In more mature regions of the root, however, expression was limited to the phloem (Fig. 7C). Stem cross-sections also showed staining in the vascular tissue, especially in the phloem (Fig. 7D).
Figure 7.
Histochemical localization of GUS activity in non-photosynthetic tissues of F. bidentis seedlings transformed with the constructs ChlMe1-nos (A–D) and ChlMe2-nos (E–H). A, Dark-field view of a longitudinal section through the SAM, where staining was absent. Under dark-field illumination, the crystalline GUS product appears as bright pink spots. B, A root tip showing staining in the vascular bundle and columnellar cells, but not in the root meristem. C, An upper portion of a root showing GUS staining in the phloem. D, A cross-section of a stem showing strong staining in the phloem. E, Dark-field view of a longitudinal section through SAM showing similar staining as the surrounding tissues. F, An apical portion of a dark-grown seedling showing strong staining in the arrested first leaves and SAM. G, A root tip of soil-grown seedling showing strong staining throughout the whole tip, including the root meristem. H, A cross-section of a stem showing staining in the vascular bundles, particularly in the xylem.
In contrast, ChlMe2 transgenics showed strong staining in the SAM, with less staining in the surrounding tissue (Fig. 7E). The SAM and the developmentally arrested first leaves (made up of mostly meristematic tissue) of dark-grown seedlings were also stained strongly (Fig. 7F). The expression of ChlMe2 in seedling root tissues, however, was ambiguous. When grown on agar medium with kanamycin as selection, no GUS staining was detected in roots of ChlMe2 seedlings. However, the root tips of soil-grown seedlings showed strong staining in both the cortical and vascular tissues as well as the root apical meristem (Fig. 7G). The lack of root staining in agar-grown seedlings was unexpected, because selected seedlings grown to maturity on agar without kanamycin yielded GUS staining in roots. In the latter case, significant staining was observed in all root tissues even in the more mature regions (data not shown). Similar to ChlMe1, ChlMe2 was also expressed in the vascular tissues of stems. However, ChlMe2 was expressed preferentially in the xylem instead of in the phloem (compare Fig. 7, D and H).
DISCUSSION
ChlMe Genes in C3 Flaveria Species
The analysis of the 3′-UTR sequences and the copy numbers of the ChlMe genes in various Flaveria spp. uncovered some interesting features. First, the large difference between the sequences of the ChlMe1 genes in the 3-4 and 5-6 phyllary lines revealed a clear divergence between the two lineages of Flaveria spp. Second, we found that the C3 species F. robusta and F. pringlei possess multiple ChlMe genes, in contrast to the single-copy genes we found in all of the C4 and C3-C4 intermediate species assayed (except for F. anomala and F. brownii). Similar observations have been reported for gdcH (encoding the H-protein of Gly decarboxylase) and CytMe (encoding the cytosolic isoform of ME) in Flaveria spp. (Kopriva and Bauwe, 1995; Kopriva et al., 1996; L. Lai, S.L. Tausta, and T. Nelson, unpublished data). In both of these cases, a higher gene copy number was found in the C3 species relative to the C3-C4 and C4 species. Although these findings suggest that the Flaveria C3 species are degenerate (i.e. C4 to C3 reversal), phylogenetic studies (Bayraktaroglu, 1993; Kopriva et al., 1996) support the ancestral status of all C3 species.
Regulation of ChlMe1 in F. trinervia and F. bidentis
We observed properties of the C4-pathway-specific ChlMe1 gene of F. trinervia that differ from those of the ChlMe1 gene from F. bidentis, as described in other studies (Marshall et al., 1997; Ali and Taylor, 2001). In the F. bidentis study, the BS-specific expression of a transgenic reporter construct depended on a 5′-flanking region, but high-level expression required a 3′-flanking region that included the 240-bp 3′UTR plus an additional 135 bp. We did not observe such a 3′ requirement for high-level, BS-specific expression of the F. trinervia gene, although we tested transgenic constructs that included the 253-bp 3′-UTR plus 263 bp of additional downstream sequences (and 42 bp before the STOP codon). The F. bidentis and F. trinervia sequences are identical in this 3′ region, with the exception of region I, as described in “Results”. The F. bidentis and F. trinervia 5′ regions used in the two studies also differed in possibly important ways. The F. bidentis constructs extended to include 24 bp of exon 2, whereas the F. trinervia constructs extended to 89 bp of exon 2. In addition, although the F. bidentis promoter region used in transgenic constructs was longer than the F. trinervia (2,121 versus 1,758 bp before the START codon), dot-matrix analysis revealed two internal non-colinear regions present only in F. bidentis. If only colinear regions are considered, the F. trinervia constructs actually start 0.3 kb farther upstream than the F. bidentis promoter constructs.
These sequence comparisons for the 3′ and 5′ regions from the two species suggest the following: (a) the difference in region I of the 3′-UTR is responsible for the different behavior of the two constructs (i.e. the G-rich sequence in the F. bidentis 3′-UTR is activating), and (b) elements responsible for high-level expression of the F. trinervia construct are found in the distal 0.3 kb of the 5′ region. We speculate that the modification in region I of the F. bidentis 3′-UTR compensated for a loss of high-level expression because of an apparent insertional event in the 5′ region. This is consistent with the observation that the region I of F. trinervia 3′-UTR is more similar to the corresponding sequences in C3 and C3-C4 Flaveria spp. than is that of F. bidentis. A further possibility is that internal sequences contribute to differences in transcription or translation level, because our constructs included approximately two-thirds of the plastid transit peptide coding sequence, whereas the F. bidentis constructs included about one-third.
Roles of ChlMe1 and ChlMe2
The ChlMe1 and ChlMe2 genes are expressed in both C3 and C4 Flaveria spp. in patterns that provide a basis for hypotheses regarding both the roles of the corresponding enzymes and the origin of the C4 pathway in Flaveria spp. The relative levels in fully expanded leaves as well as the overall profiles of mRNAs suggest that in the course of C4 evolution, the expression pattern of ChlMe2 remained constant, whereas that of ChlMe1 changed greatly. In C4 species, ChlMe1 is expressed non-specifically early in leaf development, and becomes BS specific as leaves mature. The later expression corresponds to the well-characterized role of the BS-specific NADP-ME in the C4 pathway. In C3 species the ChlMe1 gene is expressed early in leaf development at times when the organ is likely to be heterotrophic and active in respiration. This expression is consistent with a role for the ChlMe1 product in a scheme to refix respired CO2, perhaps in concert with a cytosolic phosphoenolpyruvate carboxylase (PEPCase). To prevent loss of CO2 to the environment, PEPCase condenses it with glycolysis-generated PEP to form oxaloacetate (OAA). OAA is then shuttled into the tricarboxylic acid (TCA) cycle. Because OAA is supplied by PEPCase, the malate generated by the TCA cycle is not needed to regenerate OAA, and therefore malate is diverted into the chloroplasts. We suggest that CHLME1 then releases CO2, which might be refixed in the reductive pentose phosphate pathway using the NADPH formed by the same reaction. Based on the early non-cell-specific pattern of expression of ChlMe1 in developing leaves of C4 species, the same scheme might operate before C4 photosynthetic maturity.
The proposal that NADP-ME is a component of a CO2 refixation scheme in sink leaves is novel, although there are numerous examples of such a role in other sink tissues, such as fruits. We base this proposal on the observation that the transcription of the rbcS gene begins significantly earlier than that of ChlMe1 in undifferentiated leaf cells. It is possible that the reassimilation of CO2 in the chloroplasts at this time does not go through the entire RPP cycle, as a result of metabolic repression or of limitations in availability of ATP and NADPH. CO2 reassimilation might stop after the ribulose bisphosphate carboxylase reaction, which produces PGA, a useful metabolite with several potential roles, including an exchanging currency with other cellular compartments for metabolites such as malate. Acting this way, the respired CO2 would not be lost but would act in a shuttle importing useful metabolites into developing chloroplasts. Regarding the exchange of malate, the major malate transporter known to be present on the chloroplast envelope is the malate/OAA counter-exchanger, characterized in the M cell chloroplasts of C4 plants. However, other malate translocators have been characterized. Studies of malate uptake by BS chloroplasts have revealed a new malate transport system that is activated by Asp and pyruvate (Kanai and Edwards, 1999). In addition, reports on uptake of metabolites into mitochondria and immature chloroplasts in the first few hours subsequent to illumination suggest that the transport systems in these compartments are different from those present in mature chloroplasts (Wellburn, 1984). Therefore, it is possible that this minor translocator is used during chloroplast morphogenesis in all plants and that it was recruited for C4 photosynthesis in BS chloroplasts in C4 plants.
The expression of ChlMe2 appears to be correlated with chloroplast development in both C3 and C4 species. The role of ChlMe2 might be to provide a burst of NADPH (and pyruvate) for protein and lipid biosynthesis needed during chloroplast biogenesis or other plastid-localized biosynthesis, possibly using malate generated by the TCA cycle as the substrate. Metabolic cooperation between mitochondria and developing chloroplasts is plausible, because the differentiation of chloroplasts is accompanied by a high rate of mitochondrial activity and physical contact between the two organelles occurs in immature leaves (Wellburn, 1984). The observation that light activation of ChlMe2 expression occurs in 1-week-old (presenescent) but not 5-week-old (senescent) dark-grown cotyledons is consistent with a role in chloroplast biogenesis or biosynthesis. A similar study in developing leaves of peach (a C3 plant) showed that the activity of NADP-ME is high in young leaves and decreases steadily as the leaves mature (Merlo et al., 1993). Moreover, the strong expression of both ChlMe1 and ChlMe2 in the vascular tissues of stems and roots suggests that both genes play a role in the uptake and transport of ions, functions, which are influenced by the malate pool (Martinoia and Rentsch, 1994).
The Malate Valve and C4 Evolution
Several mechanisms exist to prevent photobleaching, whereby high-light intensities lead to a reduced status in chloroplasts with potential damage to the photosystems. One such mechanism is the malate valve system in which chloroplastic NADP-dependent malate dehydrogenase is induced to convert OAA and excess NADPH into malate for transport to the cytoplasm (Backhausen et al., 1994). The counter-exchange of cytosolic OAA and chloroplastic malate is mediated by a specific malate-OAA translocator present in the inner envelope membrane of the chloroplasts. It is plausible that the OAA for the valve system is maintained by PEPCase-mediated carboxylation of PEP, which is in turn maintained by pyruvate phosphate dikinase (PPdK)-mediated phosphorylation of pyruvate. Under high-light conditions, the presence of a malate-degrading (NADP-ME) activity in the chloroplasts would counteract this relief valve action, and might therefore be down-regulated. Therefore, the Mdh, ChlMe, PEPCase, and PPdK genes might exist as a light- and redox-regulated ensemble in non-C4 plants, explaining their recruitment as a group in numerous independent evolutions of the C4 pathway of the NADP-ME subtype.
Might enzymes coordinated for operation of a malate valve constitute a precursor of a C4 pathway? Signals arising from over-reduction of the electron transport components have been shown to provoke changes in nuclear and chloroplast gene expression both locally and systemically (Foyer and Noctor, 1999; Karpinski et al., 1999; Pfannschmidt et al., 1999). Direct evidence is currently lacking for redox regulation of the genes proposed above for a Flaveria spp. malate valve. However, it was reported that the injection of malate induced the activity of NADP-ME by 3-fold in mutant pea leaves lacking PSII (Karpilov et al., 1977). Moreover, we noted that high-light intensities suppress the expression of ChlMe genes (ChlMe1 and ChlMe2 in toto) in F. pringlei leaves (L. Lai and T. Nelson, unpublished data). Based on the profile of mRNA levels in developing leaves, ChlMe1 is the more likely of these to be responsive to chloroplastic reducing potential. In Figure 3A, the expression of ChlMe1 declined rapidly as the expression of ChlMe2 peaked. The down-regulation of ChlMe1 might be due to the elevation of NADPH resulting from CHLME2 activity. The testing of this hypothesis would require expression studies of all four of the proposed genes in C3 species under varying illumination and redox conditions. An intriguing possibility is that the redox up- and down-regulation of these genes for a malate valve system might provide a basis for the eventual compartmentalization of their expression in the different environments of BS and M cells in C4 species.
MATERIALS AND METHODS
Plant Material
Multiple isolates of Flaveria spp. were provided by Dr. Scott Holaday (Texas Tech University, Lubbock), Dr. Harold Brown (University of Georgia, Athens), Drs. Maurice S. B. Ku and Gerald Edward (Washington State University, Pullman), and Dr. Peter Westhoff (Heinrich-Heine-Universitat, Dusseldorf). Flaveria trinervia and Flaveria bidentis were grown from seeds, whereas all other Flaveria spp. were propagated from vegetative cuttings. Plants were maintained in a controlled growth room at 28°C with a 16-h light/8-h dark cycle at 700 to 900 μmol m−2 s−1 supplied by a combination of white fluorescent and incandescent lights. Where necessary, transgenic F. bidentis were grown to maturity in a greenhouse with an average temperature of 28°C during summer and 25°C during winter and supplemented lighting to maintain a 16-h light/8-h dark cycle in the range of 700 to 1,000 μmol m−2 s−1.
Oligonucleotides
The following oligonucleotides were used in the various procedures below. The FprME and FtrME primers were designed using the Flaveria pringlei ChlMe2 and the F. trinervia ChlMe1 cDNA sequences, respectively (Börsch and Westhoff, 1990; Lipka et al., 1994): FprME-5, 5′-CTTTACGGGGAATCTGATAC-3′; FprME-6, 5′-CTAATGACTTGTTTACTAAGTTG-3′; FprME-8, 5′-GTATCAGATTCCCCGTAAAG-3′; FtrME-1, 5′-GAGAGCTGCATGTACAGC-3′; FtrME-4, 5′-CATCCGCAAGATTTCTGCTC-3′; FtrME-6, 5′-GTTTAGCGGGAAAAAAAGACAG-3′; FtrME-7, 5′-CATTACAACATGCATCTAACAAC-3′; FtrME-8, 5′-GTATGTTGTTAGATGCATGTTG-3′; and FtrME-9, 5′-GTGAATAAACAAAAGTAGTATTAAG-3′.
Cloning of 3′ Regions of ChlMe1 and ChlMe2 Using TAIL-PCR
TAIL-PCR was carried out essentially as described by Liu et al. (1995) with the arbitrary degenerate primers reported therein as the reverse primers. Three sets of PCR were performed sequentially with three nested forward primers. The ChlMe-specific primers, FtrME-4 and FtrME-1 (downstream of FtrME-4), used in the primary and secondary PCR, respectively, were designed to anneal to 3′-coding regions that are identical in ChlMe1 and ChlMe2. The tertiary PCR was carried out (in separate reactions) with two gene-specific primers, ChlMe1-specific FtrME-6 and ChlMe2-specific FprME-5, which are downstream of the FtrME-1 region. These were designed from the cDNA sequences of F. trinervia ChlMe1 (Börsch and Westhoff, 1990; Rajeevan et al., 1991) and F. pringlei ChlMe2 (Lipka et al., 1994), respectively. The tertiary reactions assessed whether ChlMe1 and ChlMe2 products were successfully amplified and what sizes to expect of each product. Once it was confirmed that both ChlMe1 and ChlMe2 products were present, the products of the secondary reactions were cloned using TOPO TA cloning (Invitrogen, Carlsbad, CA). Therefore, only one ligation and one transformation step were performed, and both ChlMe1-3′ and ChlMe2-3′ clones were isolated based on the sizes determined by the tertiary reactions. After the sequences were obtained by dideoxy DNA sequencing, they were analyzed using GeneWorks (version 2.5.1, Intelligenetics, Inc., Mountain View, CA).
Genomic DNA-Blot Analysis
PCR was used to radiolabel probes (Mertz and Rashtchian, 1994) with 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. ChlMe1- and ChlMe2-specific probes were designed in the most divergent regions of these two genes, their 3′-UTRs. The ChlMe1-specific probe was obtained with the primers FtrME-8 and FtrME-9; using the F. trinervia ChlMe1 TAIL-PCR clone (accession no. AF288918) as template, these primers yielded a 103-bp radiolabeled product between regions II and III (Fig. 1A). This probe should detect ChlMe1 from all Flaveria spp., including the C3-C4 species of the 5-6 phyllary line. The ChlMe2-specific probe was obtained with the primers FprME-5 and FprME-6; using the F. trinervia ChlMe2 TAIL-PCR clone (accession no. AF288919) as template, these primers yielded a radiolabeled product containing the complete 3′-UTR.
For DNA-blot analysis, 10 to 15 μg of genomic DNA was digested with restriction enzymes, separated in a 1% (w/v) agarose gel and the DNA transferred to Nytran Plus membrane (Schleicher & Schuell, Keene, NH). Low-stringency hybridization was carried out in 25% (w/v) formamide, 5× SSC, 5× Denhardt's, 1% (w/v) SDS, 0.05% (w/v) tetrasodium pyrophosphate (Na4PPi), 250 μg mL−1 salmon sperm DNA, and 5% (w/v) dextran sulfate at 42°C. Two 20-min washes at 42°C were performed subsequently using 2× SSC, 0.1% (w/v) SDS and 1× SSC, 0.1% (w/v) SDS as the first and second wash solutions, respectively.
Measurement of RNA by Relative Quantitative RT-PCR
Using leaf length as an indicator of developmental stage, leaves ranging in length from 1 to 13 cm for F. pringlei and 1 to 8 cm for F. trinervia were collected and immediately frozen in liquid nitrogen. Senescing leaves were avoided by collecting leaves from only the top five nodes of plants with at least 10 nodes. Total RNA was isolated using the TRIzol reagent (Life Technologies, Grand Island, NY).
Leaf total RNAs (1 μg/reaction) were reverse transcribed with random decamers using the RETROscript kit (Ambion Inc., Austin, TX), and the resulting single-stranded cDNAs served as the templates for the subsequent relative quantitative PCR reactions. The latter procedure was carried out using the kit QuantumRNA 18S Internal Standards (Ambion Inc.). Both the ratio of 18S competimers to primers and the number of PCR cycles required to reach midexponential phase were empirically determined using RNA from fully expanded leaves. These optimized conditions were then applied to RNAs from all other leaf stages. The primers used for PCR amplification of ChlMe1 were FtrME-4 and FtrME-7 and of ChlMe2 were FtrME-4 and FprME-8. All PCR reactions were supplemented with a small amount of [α-32P]dCTP, and the products were separated on an 8% (w/v) polyacrylamide gel. The gel was then exposed to a phosphorimager plate and the signals quantitated using the Fujix BAS-2000 phosphorimager (Fuji, Tokyo).
Chimeric Constructs and Transformation of F. bidentis Plants
ChlMe1 Constructs
For construction of ChlMe1-nos, a 2-kb EcoRI fragment from the 5′ region of a F. trinervia ChlMe1 genomic clone (L. Lai and T. Nelson, unpublished data) was ligated in-frame to the 5′ end of the gusA gene in pBI101.2 (CLONTECH, Palo Alto, CA). This 5′ region included 1,758 bp upstream of the start codon plus the rest of exon 1 (30 bp), intron 1 (186 bp), and the first 89 bp of exon 2 (Fig. 4). The region including the ChlMe1 promoter, gusA, and the nopaline synthase (nos) terminator was cloned subsequently into the Agrobacterium sp. binary vector pPZP122 (Hajdukiewicz et al., 1994). The ChlMe1-3′ construct was generated by replacing the nos terminator with the 0.56-kb F. trinervia ChlMe1 3′ fragment obtained by TAIL-PCR (accession no. AF288918) before cloning into pPZP122. This 3′ region starts with the region coding for the last 14 amino acids of CHLME1 and contains 265 bp downstream from the poly(A) site (according to the cDNA sequence reported by Börsch and Westhoff [1990]).
ChlMe2 Constructs
For construction of ChlMe2-nos, a 1.9-kb BamHI-EcoRI fragment from the 5′ region of a F. pringlei ChlMe2 genomic clone (L. Lai and T. Nelson, unpublished data) was ligated in-frame to the gusA gene in pBI101.2. This 5′ region included 1,372 bp upstream of the start codon plus the rest of exon 1 (30 bp), intron 1 (393 bp), and the first 89 bp of exon 2 (Fig. 4). The region including the ChlMe2 promoter, gusA, and the nos terminator was cloned subsequently into the Agrobacterium binary vector pBIN19. The ChlMe2-3′ was constructed by substituting the nos terminator of ChlMe2-nos (in pBI101.2) with the 0.65-kb ChlMe2 3′ region (accession no. AF288908) obtained from the same genomic clone. This 3′ region starts with the region coding for the last 14 amino acids of CHLME2 and contains 405 bp downstream from the poly(A) site (according to the cDNA sequence reported by Lipka et al. [1994]). The region including the ChlMe2 promoter, gusA, and the 3′ region of ChlMe2 was cloned subsequently into pPZP122.
All four fusion constructs were used to transform Agrobacterium tumefaciens strain LBL4404. The subsequent transformation of F. bidentis was carried out as described by Chitty et al. (1994). Genomic DNA blots were used to determine the number of independent transformants with each construct: ChlMe1-nos (28), ChlMe1-3′ (11), ChlMe2-nos (6), and ChlMe2-3′ (11).
Histochemical Localization and Enzymatic Assay for GUS Activity
Histochemical localization was performed using only T1 seedlings and plants grown to maturity on germination medium (GM; Chitty et al., 1994). Seedlings were grown under selection (100 μg mL−1 gentamycin for ChlMe1-nos, ChlMe1-3′ and ChlMe2-3′; 50 μg mL−1 kanamycin for ChlMe2-nos) for 1 month before they were transferred to growth boxes (containing GM without antibiotics) to obtain mature plants. To avoid possible circadian-related discrepancies between experiments, growth was under continuous light (100–140 μmol m−2 s−1) in a growth chamber at 24°C. GUS localization was carried out by incubating plant tissues in 0.1 m sodium phosphate buffer (pH 7.0), 1% (w/v) Tween 20, 1 mm EDTA (pH 8.0), 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C overnight after a brief vacuum infiltration. For ChlMe1-nos and ChlMe1-3′ plants, the concentrations of both potassium ferricyanide and potassium ferrocyanide were increased to 5 mm to minimize diffusion of GUS product, because GUS activities in these lines were very high. Clearing was carried out in several changes of ethanol:acetic acid (3:1, v/v). Finally, several rinses with 70% (v/v) ethanol were carried out before microscopic examination. For thin-section analysis, histochemically stained material was fixed in propylene oxide before embedding in Spurr's resin according to the manufacturer's instruction (Electron Microscopy Sciences, Ft. Washington, PA).
GUS assays were performed on leaf extracts from plants grown to maturity in a greenhouse. As each leaf was collected, it was measured from tip to end of petiole and immediately frozen in liquid nitrogen. Crude extract was obtained by pulverizing the leaf using a polytron homogenizer in extraction buffer and a fluorometric assay for GUS activity carried out essentially as described by Jefferson (1987). For ChlMe1-nos leaves, time points were taken at 0, 5, and 10 min after incubation at 37°C; for ChlMe2-nos leaves, time points were taken at 0, 1, and 2 h after incubation at 37°C.
Growth under Different Light Conditions
Seedlings grown in the dark and different light qualities were sown on GM as described above. After 3 to 5 d of vernalization at 4°C, germination was induced by exposing seeds to white light for a period of 5 to 12 h before shifting to the different growth conditions at 22°C for 1 week. Growth conditions in darkness, blue, red, and far-red light were as described by McNellis et al. (1994). Whole seedlings were collected under the same light conditions used for growth. For dark-grown seedlings, green overhead light of very low intensity (approximately 0.01 μmol m−2 s−1) was used. To prevent white light-induced expression, these seedlings were kept in the dark throughout the whole staining and incubation period. Illumination was carried out by shifting the 1-week-old dark-grown seedlings into the growth chamber used for light-grown plants.
ACKNOWLEDGMENTS
We thank Dr. Brian McGonigle for providing the cDNA clone of the 5′ region of ChlMe1 from F. trinervia, which was used as a probe to isolate the ChlMe1 and ChlMe2 genomic clones. We are grateful to Drs. Vivian Irish, S. Lori Tausta, and Jane Langdale for numerous helpful discussions and for critical reading of the manuscript.
Footnotes
This work was supported by the Department of Energy (grant no. DE–FG02–91ER20038).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010448.
LITERATURE CITED
- Ali S, Taylor WC. Quantitative regulation of the Flaveria Me1 gene is controlled by the 3′-untranslated region and sequences near the amino terminus. Plant Mol Biol. 2001;46:251–261. doi: 10.1023/a:1010684509008. [DOI] [PubMed] [Google Scholar]
- Backhausen JE, Kitzmann C, Scheibe R. Competition between electron acceptors in photosynthesis: regulation of the malate valve during CO2 fixation and nitrite reduction. Photosynth Res. 1994;42:75–86. doi: 10.1007/BF00019060. [DOI] [PubMed] [Google Scholar]
- Bayraktaroglu L. The evolution and developmental expression of C4 photosynthesis in Flaveria. PhD thesis. CT: Yale University, New Haven; 1993. [Google Scholar]
- Berry JO, Niklou BJ, Carr JP, Klessig DF. Transcriptional and post-transcriptional regulation of ribulose-1,5-bisphosphate carboxylase gene expression in light and dark grown Amaranth cotyledons. Mol Cell Biol. 1985;5:2238–2246. doi: 10.1128/mcb.5.9.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börsch D, Westhoff P. Primary structure of NADP-dependent malic enzyme in the dicotyledonous C4 plant Flaveria trinervia. FEBS Lett. 1990;273:111–115. doi: 10.1016/0014-5793(90)81063-t. [DOI] [PubMed] [Google Scholar]
- Casati P, Drincovich MF, Andreo CS, Donahue R, Edwards GE. UV-B, red and far-red light regulate induction of the C4 isoform of NADP-malic enzyme in etiolated maize seedlings. Aust J Plant Physiol. 1998;25:701–708. [Google Scholar]
- Casati P, Fresco AG, Andreo CS, Drincovich MF. An intermediate form of NADP-malic enzyme from the C3-C4 intermediate species Flaveria floridana. Plant Sci. 1999;147:101–109. [Google Scholar]
- Chitty JA, Furbank RT, Marshall JS, Chen Z, Taylor WC. Genetic transformation of the C4 plant, Flaveria bidentis. Plant J. 1994;6:949–956. [Google Scholar]
- Dengler NG, Nelson T. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, editors. C4 Plant Biology. San Diego: Academic Press; 1999. pp. 133–172. [Google Scholar]
- Drincovich MF, Casati P, Andreo CS. NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 2001;490:1–6. doi: 10.1016/s0014-5793(00)02331-0. [DOI] [PubMed] [Google Scholar]
- Drincovich MF, Casati P, Andreo CS, Chessin SJ, Franceschi VR, Edwards GE, Ku MSB. Evolution of C4 photosynthesis in Flaveria species: isoforms of NADP-malic enzyme. Plant Physiol. 1998;117:733–744. doi: 10.1104/pp.117.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GE, Andreo CS. NADP-malic enzyme from plants. Phytochemistry. 1992;31:1845–1857. doi: 10.1016/0031-9422(92)80322-6. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Leaves in the dark see the light. Science. 1999;284:599–601. doi: 10.1126/science.284.5414.599. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Taylor WC. Regulation of photosynthesis in C3 and C4 plants: a molecular approach. Plant Cell. 1995;7:797–807. doi: 10.1105/tpc.7.7.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Kanai R, Edwards GE. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, editors. C4 Plant Biology. San Diego: Academic Press; 1999. pp. 49–88. [Google Scholar]
- Karpilov IS, Karpova RN, Oparina LA. Malic acid induction of decarboxylating NADP-malate dehydrogenase synthesis in C3 plant leaves. Biokhimiia. 1977;42:860–863. [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Bauwe H. H-protein of glycine decarboxylase is encoded by multigene families in Flaveria pringlei and F. conquistii (Asteraceae) Mol Gen Genet. 1995;249:111–116. doi: 10.1007/BF00290242. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Chu C-C, Bauwe H. Molecular phylogeny of Flaveria as deduced from the analysis of nucleotide sequences encoding the H-protein of the glycine cleavage system. Plant Cell Environ. 1996;19:1028–1036. [Google Scholar]
- Ku MSB, Kano-Murakami Y, Matsuoka M. Evolution and expression of C4 photosynthesis genes. Plant Physiol. 1996;111:949–957. doi: 10.1104/pp.111.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku MSB, Wu J, Dai Z, Scott RA, Chu C, Edwards GE. Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiol. 1991;96:518–528. doi: 10.1104/pp.96.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Zelitch I, Miller E, Nelson T. Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 1988;7:3643–3651. doi: 10.1002/j.1460-2075.1988.tb03245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, Walker RP. Regulation of the C4 pathway. In: Sage RF, Monson RK, editors. C4 Plant Biology. San Diego: Academic Press; 1999. pp. 89–132. [Google Scholar]
- Lipka B, Steinmueller K, Rosche E, Boersch D, Westhoff P. The C3 plant Flaveria pringlei contains a plastidic NADP-malic enzyme which is orthologous to the C4 isoform of the C4 plant F. trinervia. Plant Mol Biol. 1994;26:1775–1783. doi: 10.1007/BF00019491. [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Chitty JA, Surin B, Taylor WC. Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. Plant Cell. 1997;9:1515–1525. doi: 10.1105/tpc.9.9.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Taylor WC. Two genes encode highly similar chloroplastic NADP-malic enzymes in Flaveria. Plant Physiol. 1996;111:1251–1261. doi: 10.1104/pp.111.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Rentsch D. Malate compartmentation: responses to a complex metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:447–467. [Google Scholar]
- McNellis TW, von Arnim AG, Deng X-W. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell. 1994;6:1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo L, Ferritti M, Ghisi R, Passera C. Developmental changes of enzymes of malate metabolism in relation to respiration, photosynthesis and nitrate assimilation in peach leaves. Physiol Plant. 1993;89:71–76. [Google Scholar]
- Mertz LM, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- Monson RK. The origins of C4 genes and evolutionary pattern in the C4 metabolic phenotype. In: Sage RF, Monson RK, editors. C4 Plant Biology. San Diego: Academic Press; 1999. pp. 377–410. [Google Scholar]
- Moore BD, Cheng S-H, Edwards GE. The influence of leaf development on the expression of C4 metabolism in Flaveria trinervia, a C4 dicot. Plant Cell Physiol. 1986;27:1159–1167. [Google Scholar]
- Moore BD, Edwards GE. The influence of leaf age on C4 photosynthesis and the accumulation of inorganic carbon in Flaveria trinervia, a C4 dicot. Plant Physiol. 1988;88:125–130. doi: 10.1104/pp.88.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Powell AM. Systematics of Flaveria (Flaveriinae-Asteraceae) Ann MO Bot Gard. 1978;65:590–636. [Google Scholar]
- Rajeevan MS, Bassett CL, Hughes DW. Isolation and characterization of cDNA clones for NADP-malic enzyme from leaves of Flaveria: Transcript abundance distinguishes C3, C3-C4 and C4 photosynthetic types. Plant Mol Biol. 1991;13:371–384. doi: 10.1007/BF00040632. [DOI] [PubMed] [Google Scholar]
- Sheen J. C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:187–217. doi: 10.1146/annurev.arplant.50.1.187. [DOI] [PubMed] [Google Scholar]
- Stockhaus J, Schlue U, Koczor M, Chitty JA, Taylor WC, Westhoff P. The promoter of the gene encoding the C4 form of phosphoenolpyruvate carboxylase directs mesophyll-specific expression in transgenic C4 Flaveria spp. Plant Cell. 1997;9:479–489. doi: 10.1105/tpc.9.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-L, Long JJ, Hotchkiss T, Berry JO. C4 photosynthetic gene expression in light- and dark-grown amaranth cotyledons. Plant Physiol. 1993;102:1085–1093. doi: 10.1104/pp.102.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. Ultrastructural, respiratory and metabolic changes associated with chloroplast development. In: Baker NR, Barber J, editors. Topics in Photosynthesis: Chloroplast Biogenesis. Amsterdam: Elsevier Science Publishers; 1984. pp. 253–304. [Google Scholar]
- Wessler SR. Transposable elements associated with normal plant genes. Physiol Plant. 1998;103:581–586. [Google Scholar]