Abstract
A survey of the already characterized and potential two-component protein sequences that exist in the nine complete and seven partially annotated cyanobacterial genome sequences available (as of May 2005) showed that the cyanobacteria possess a much larger repertoire of such proteins than most other bacteria. By analysis of the domain structure of the 1,171 potential histidine kinases, response regulators, and hybrid kinases, many various arrangements of about thirty different modules could be distinguished. The number of two-component proteins is related in part to genome size but also to the variety of physiological properties and ecophysiologies of the different strains. Groups of orthologues were defined, only a few of which have representatives with known physiological functions. Based on comparisons with the proposed phylogenetic relationships between the strains, the orthology groups show that (i) a few genes, some of them clustered on the genome, have been conserved by all species, suggesting their very ancient origin and an essential role for the corresponding proteins, and (ii) duplications, fusions, gene losses, insertions, and deletions, as well as domain shuffling, occurred during evolution, leading to the extant repertoire. These mechanisms are put in perspective with the different genetic properties that cyanobacteria have to achieve genome plasticity. This review is designed to serve as a basis for orienting further research aimed at defining the most ancient regulatory mechanisms and understanding how evolution worked to select and keep the most appropriate systems for cyanobacteria to develop in the quite different environments that they have successfully colonized.
INTRODUCTION
The cyanobacteria constitute a very large and morphologically diverse group of oxygen-evolving photosynthetic prokaryotes. They can be found in most terrestrial, freshwater, and marine habitats (28). Like most bacteria, cyanobacteria use two-component regulatory systems proteins to regulate cell behavior and gene expression in response to changes in the external environment (3, 24, 54, 56, 80, 132, 136, 140). Two-component systems typically consist of two types of proteins, histidine kinases (HK) and response regulators (RR), which may sometimes be carried by a single polypeptide to form the hybrid kinases (HY). They are characterized by the presence of specific signatures: the HisKA (dimerization and phosphoacceptor) and HATPase (histidine kinase ATPase) domains, which make a histidine kinase, an aspartate-containing receiver domain for the response regulators. The so-called hybrid sensors have all three domains. Upon detection of a stimulus, the HisKA and HATPase domains function to autophosphorylate a histidine residue. The phosphate group is then transferred to an aspartate residue of the receiver cognate response regulator or hybrid sensor. As a result, a change in the activity of the protein that carries the receiver domain occurs, such that it modifies some aspect of cell behavior (such as taxis) or gene expression, or the phosphate group is further transferred in so-called phosphorelays (32, 54, 68, 132). The deduced sequences of two-component protein genes have been found to contain a number of other “sensory” domains, but precise functions for a large number of them still await definition (40). Surveys performed on complete annotated genomes of prokaryotes revealed that the differences in the total number of genes and the complexity of the ecophysiology of the bacterium and of its environment have an effect on the number of response regulators (11, 38). The total number of all signal transduction proteins increases for most bacteria as a square of the genome size (38).
Genome sequences of the Cyanobacteria have revealed that they likely make extensive use of a variety of two-component proteins to regulate responses to the environment (34, 89, 102, 118). In May 2005, 8 completely annotated cyanobacterial sequences were available in Cyanobase (http://www.kazusa.or.jp/cyano/), and a total of 16 sequences, not all completely annotated, were available from the U.S. Department of Energy Joint Genome Institute (the Integrated Microbial Genomes [IMG] system [http://img.jgi.doe.gov/cgi-bin/pub/main.cgi]). The possibility of performing an extensive comparative analysis of the repertoire of two-component genes in each organism was thus opened up. Available genomes come from unicellular and filamentous freshwater or terrestrial strains (including one thermophile) and from marine environments. Five of the 16 strains are capable of nitrogen fixation. The strains belong to three of the five subsections defined within the BX phylum (Cyanobacteria) of bacterial taxonomy (28). There is no representative of subsection II (species that reproduce by the formation of baeocytes, i.e., subsequent multiple fission of a cell that yields motile baeocytes) or of subsection V (branching filamentous heterocystous cyanobacteria [a heterocyst is a differentiated cell specialized in nitrogen fixation]). For subsection III (i.e., filamentous nonheterocystous cyanobacteria that divide in only one plane), the single sequenced genome is not really representative of the group because it can reduce molecular dinitrogen; it is the only strain known to be able to do so. In any case, it is worth noting that no single strain can truly be representative, because the subsection is polyphyletic and must now be considered an artificial grouping.
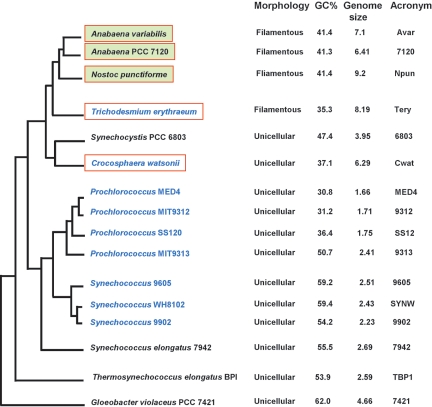
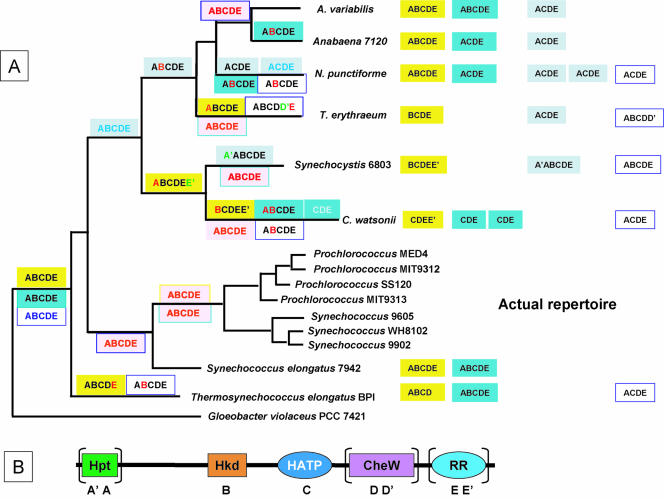
This survey presents a detailed analysis of the cyanobacterial two-component system repertoire. The species names of the 16 cyanobacterial strains and their morphologies, main features, and habitats, as well as the acronyms used at the beginning of gene names, are shown in Fig. 1 and in Table S1s in the supplemental material. The organization of the sensor, receiver, transmitter, and response domains is discussed in terms of the significance for the function of each family of two-component proteins and how the repertoire of such proteins found in each species of cyanobacteria relates to its requirement for regulation of its internal cellular activity. The 1,171 proteins found have been classified according to structural domain organization and orthology relationships. Whenever known, the function of the two-component proteins is mentioned and its occurrence within an orthology group is discussed in relation with the physiological properties and ecological niche of the strains that share it. Phylogenetic studies were performed to estimate the relative contributions of gene fusion, duplication, insertion, deletion, and shuffling during evolution. Finally, a generic gene name is proposed for each orthology group, even if at present the groups are composed of a single representative and the corresponding proteins do not yet have an assigned function, to aid in identification and future research. Corresponding names were attributed to the putative proteins: Chk for the histidine kinases, Crr for the response regulators, and Chy for the hybrid kinases (Table 1; see also Table S3s in the supplemental material). If within a group both HK and HY genes coexist, they have been named with chk and chy acronyms, respectively, with the same number attached, e.g., chk15 and chy15. To avoid, as much as possible, confusion within the literature, since numbers had already been assigned to almost all of the Synechocystis sp. strain PCC 6803 two-component proteins (described as HikX and RreY), we have kept the same numbers and used them for naming of the orthologues; numbering was continued from there for the new groups. Other sensing systems and regulators (S/T kinases, AC/GC, and one-component proteins) also contribute to the rather sophisticated regulatory pathways evolved by cyanobacteria, but they have not been considered in this review except when they are fused to two-component system protein domains.
FIG. 1.
Phylogenetic tree of the cyanobacterial strains whose genome sequences are available. The tree is adapted from previously published trees based on 16S rRNA sequences (55, 75, 99, 114, 116, 118; J. Elhai, personal communication). Names of the marine strains are in blue. Strains able to fix dinitrogen are boxed in red, and a yellow-green motif inside a box indicates that diazotrophy is linked to heterocyst differentiation.
TABLE 1.
Two-component proteins from the 16 complete cyanobacterial genomes available as of May 2005, listed according to orthology relationshipsa
| Group | Gene locus tag or GOIb | Classification | Functional or temporary gene namec |
|---|---|---|---|
| Histidine kinase | 7120alr1680, Avar_400189020, NpunNpF1895 | HKI | chk63 |
| orthologue | 7120all3359, NpunNpF1016, Avar_ 400209600 | HKI | chk56(†) |
| 6803sll0094 | HKI | chk37, hik37 | |
| Cwat_400871830 | HKII+GAF | ||
| 7421gll4098, NpunNpR1110 | HKI | chk174 | |
| 6803sll1590, 7421gll0572, NpunNpF0303, Avar_400173260 | HKI [N-ter-(TM)3] | chk20, hik20 | |
| 7120alr3155, Avar_400216080 | HKI [N-ter-(TM)3] | chk47(†) | |
| 6803slr0640, TBP1tll2351, 7120all3587, 7421gll1686, Tery _403243260, Avar_400209110, NpunNpF5193, 7942_ 403105770, Cwat_400868570 | HKI [N-ter-TM-150-TM] | chk27, hik27, manS | |
| 6803slr1147, TBP1tlr0195, 7120alr3982, Avar_400194500, NpunNpR4028, Tery_403263390, 7942_403094230, Cwat_400859120 | HKII+GAF | chk2, hik2, caeS(*) | |
| 7421glr1587, SS12Pro0301, 9313PMT1860, 9605_403118700, MED4PMM0269, 9312_ 402281550, SYNW0246, 9902_403146450 | HKI | ||
| 7120alr3037, 7421glr2258, Avar_400185970, NpunNpF1330 | HKI | chk48 | |
| 7120alr3547, Avar_400212820, NpunNpF3765 | HKI | chk49 | |
| 7120all3167, Avar_400216230, 6803sll0798 | HKI [N-ter-TM-150-TM] | chk30nrsS, rppB, hik30 | |
| 6803slr1414, 7120alr1665, Avar_400188750, Cwat_400884460, NpunNPR2485 | HKI | chk11, hik11 | |
| 6803sll1353 | HKI | chk15, hik15(*) | |
| Cwat_400846700 | HYII+(PAS)1-4-(PAC)1-6 | chy15 | |
| 7120alr4716, Avar_400196850 | HKI [N-ter-(TM)3] | chk50(*) (†) | |
| NpunNpR4835 | HKV+HNOBA | ||
| 7120alr4882, Avar_400198860, NpunNpF3835 | HKI | chk51(†) | |
| 7120all4502, Avar_400211220, NpunNpF4215, 7421glr0418 | HKI [TM] | chk7, hik7, sphS, phoR(*) | |
| TBP1tll0925 | HKII+[TM]-PAS | ||
| 6803sll0337, Cwat_400885230, 7942_403099950, SYNW0948, MED4PMM0706, 9605_403132780, 9312_402286050 | HKII+PAS | ||
| 7120alr0117, Avar_400192160, NpunNpR3854, Tery_403227130, Cwat_400846460, 7942_403112370 | HKI [TM for 7120 and Ava] | chk52, hepX | |
| 7120all2772, Avar_400177390 | HKI [N-ter-(TM)3] | chk53 | |
| 7120alr4905, Avar_400199100, NpunNpF3280 | HKI [N-ter-TM-150-TM] | chk54 | |
| TBP1tll1909, Avar_400218440, NpunNpR6203, 7421glr0282, 7120alr1171 | HKI [TM] | chk55 | |
| 6803slr6041, 7120all7583, NpunNpR1778 | HKI [N-ter-TM-125-TM] | chk46 | |
| 7421glr0347, NpunpNPBF145 | HKI (HisKA_3) | chk57 | |
| 7120all4636, Avar_400197610, NpunNpR2421 | HKI (HisKA_3) | chk58 | |
| 7120all2956 | HKI | chk59(*) | |
| 7421gll1662 | HKV (cNMP-HK) | ||
| Cwat_400862090 | HYI | chy59(*) | |
| 6803sll1888, 7120alr1308, Avar_400207800, NpunNpR1550, Cwat_400841670, 7942_403094500 | HKII+GAF | chk5, hik5 | |
| 6803slr0484, TPB1tll1659, 7120all0330, Tery_403264440, Avar_400224920, NpunNpR5149, 7942_403112800, Cwat_400889030 | HKII+GAF | chk26, hik26 | |
| 7120all1191, Avar_400183300, NpunNpR1432 | HKII+GAF | chk60, kinA | |
| 7120all0825, Avar_400221990, NpunNPR4745 | HKII+GAF | chk61 | |
| 7120all1088, Avar_400214770 | HKII+GAF | chk62 | |
| 7120all5074, Avar_400200660, NpunNpR0454 | HKII+GAF | chk44 | |
| 6803sll1334 | GAF-HisKA | ||
| 7120all1688, Avar_400188950, NpunNpF1000, TBP1tll0899 | HKII+GAF (Phyt_2) | chk24, hik24, cikA(*) | |
| Cwat_400887290, 6803slr1969, Tery_403229140, 7942_403096220 | HYII+GAF | chy24(*) | |
| 7120all0853, Avar_400222250, NpunNpF2781 | HKII+2 GAF | chk64 | |
| 7120all2699, Avar_400220270, NpunNpR4776 | HKII+(GAF)3 | chk65, aphC | |
| 7120alr5272, Avar_400202640, NpunNpR6125 | HKII+PAS/PAC-GAF(Phyt_2)-GAF | chk66 | |
| 7120all4261, Avar_400189370, NpunNpR1597 | HKII+GAF(Phyt_2)4 | chk67 | |
| Cwat_400877940, 6803slr1393 | HKII+GAF-GAF(Phyt_2)2-PAS | chk1, hik1 | |
| Avar_400182390, NpunNpF2854 | HKII+GAF[Phyt_2]4 | chk68 | |
| 7120all1145, Avar_400221670, NpunNpF1203 | HKII+PAS/PAC-GAF | chk69 | |
| 7120all1280, Avar_400207510, NpunNpF6001 | HKII+GAF-(PAS/PAC)2-PAS-GAF-HKA_2 | chk45, ETR1(*) | |
| 6803slr1212 | (TM)3-PAC-(PAS/PAC)2-GAF | ||
| 7120alr1966, Avar_400221110, 7421glr3432, NpunNpF6362, 7942_403103440 | HKII+GAF-PAS-PAS/PAC-GAF[Phyt_2] | chk71 | |
| 6803slr0210 | HKI | chk9, hik9(†) | |
| Cwat_400860390 | HKII + GAF | ||
| 7120all0542, Avar_400206910 | HKII + GAF-PAS/PAC | chk72(*) | |
| NpunNpR4748 | HYII | chy72 | |
| 6803slr0473, 7120alr3157, Avar_400216100, NpunNpF0020, Cwat_400857300-400857310 | HKII+phytochrome | chk35, hik35, cph1, aphA | |
| 7120all2899, Avar_400187280, NpunNpF1183 | HKII+phytochrome | chk73, aphB | |
| Avar_400212970, NpunNpR6148, 7120all3563 | HKII+(TM)5-PAS/PAC | chk74 | |
| 7120all3564, Avar_400212980 | HKII+PAS | chk16, hik16(*) | |
| NpunNpR6149, 6803slr1805, Cwat_400864000, TBP1tlr1215 | HKII+GAF-PAS/PAC (N-ter MASE1 for slr1805, Cwat_400864000, NpR6149) | ||
| 7120all5327 | HKII-PAS/PAC | chk75(*) | |
| NpunNpR3083 | HY (TM)4-PAS/PAC | chy75(*) | |
| Avar_400226560, 7120alr3225 | HKII+(PAS/PAC)5 | chk76 | |
| 7120all3767, Avar_400192890 | HKII+(PAS/PAC)2 | chk77 | |
| 7120all 0707, Tery_403245990, NpunNpF3837 | HKII+PAS | chk78 | |
| 7120alr0428, NpunNpF1277 | HKII+(PAS)2PAS/PAC | chk79 | |
| Avar_400181300, 7120alr2481, NpunNpR3263 | HKII+(PAS/PAC)3 | chk80 | |
| 7120alr1229, Avar_400183650 | HKII+TM-(PAS/PAC)4-GAF-(PAS/PAC)5 | chk32, hik32(*) | |
| NpunNpF3797, 6803sll1473-75 | TM-GAF[Phyt_2]-(PAS/PAC)2 | ||
| 7421glr3562 | HKV+CheR/B-(PAS)2 | ||
| Tery_403240680 | HKII+CBS-GAF | chk81(*) | |
| TBP1tll1831 | HKV+FHA | ||
| 7120alr0546, Avar_400180710 | HKII+(PAS)1 or 3 | chk83 | |
| 7421gll4161, 7942_403108630 | HKII+(PAS)1 or 3 | chk84 | |
| 6803slr0533, 7120all4726, Avar_400196760, NpunNpR6227, Cwat_400855750, 7421glr0861 | HKIII+HAMP [N-ter-TM-50-TM] | chk10, hik10 | |
| 6803sll1871, Tery_403261040 | HKIII+HAMP | chk6, hik6(*) | |
| 7942_403108070 | HYIII+HAMP | chy6(*) | |
| 7120alr4586, Avar_400202050, NpuNpR0565 | HKIII+HAMP [N-ter-TM-∼200-TM] | chk85 | |
| 7120all4496, Avar_400211160, NpunNpR1012, Tery_403248920 | HKIII+HAMP [N-ter-TM-175-TM] | chk86, hepK | |
| 7120alr5189, Avar_400201780, 7421gll0023 | HKIII+HAMP [N-ter-TM-150-TM] | chk87 | |
| 7120alr1192, Avar_400183310, NpunNpF1439, Cwat_400885690, 7942_403108630, 9902_403159810, 9313PMT0265 | HKIII+HAMP [N-ter-TM-140-TM] | chk88 | |
| 7120alr4105, Avar_400185210, Tery_403255460, NpunNpR1236 | HKIII+HAMP [TM] | chk89 | |
| 7120alr2739, Avar_400220660, NpunNpR5253 | HKIII+HAMP [N-ter-TM-150-TM] | chk90 | |
| 9313PMT0804, SYNW0807, 9605_403135040 | HKIII+HAMP [N-ter-TM-120-TM] | chk91 | |
| Avar_400212080, NpunNpR5315 | HKIII+HAMP [N-ter-TM-160-TM] | chk92 | |
| Avar_400185110, NpunNpR3052 | HKIII+HAMP [TM] | chk82 | |
| 7120alr1551, NpunNpR3816 | HKIII+HAMP | chk94 | |
| 6803sll0698, 7120alr3511, TBP1tlr0437, 7421gll0055, SS12Pro1422, MED4PMM1341, 9313PMT1417, SYNW0551, Tery_403227530, NpunNpR3716, Avar_400209790, 7942_403099080, Cwat_400840950, 9605_403137900, 9902_403149340, 9312_402293260 | HKIII+HAMP-PAS [N-ter-TM-150-TM] | chk33, nblS, ycf26, dspA, hik33 | |
| 7120alr0642, NpunNpR6504 | HKIII+HAMP-Cache [N-ter-TM-310-TM] | chk95 | |
| 6803slr0311 | HKIII+PAS/PAC (HisKA_3) | chk29, hik29(*) | |
| 7421glr3986, 7120alr2137 | HKII+PAS/PAC-GAF-(HKA_3) | ||
| NpunNpR2408 | Cache-HAMP-PAS/PAC-PAS-HK | ||
| 7120all0323, Avar_400224850, NpunNpR4085 | HKIV S/Tkin-GAF | chk96 | |
| 7120alr0354, Avar_400205470 | HKIV S/Tkin-GAF | chk97 | |
| 7120all4687, Avar_400197110, NpunNpF3830 | HKIV S/Tkin-GAF | chk98 | |
| 7120alr2258, Avar_400177980, NpunNpF2369 | HKIV S/Tkin-GAF | chk99, hstK | |
| 7120all3557, Avar_400212910, NpunNpF2670, Tery_403252070 | HKIV S/Tkin-GAF | chk100 | |
| 7120alr0900, Avar_400222720 | HKIV S/Tkin-GAF | chk101 | |
| 7120all0886, Avar_400222570 | HKIV S/Tkin-GAF | chk102 | |
| 7120alr2682, NpunNpF3778 | HKIV S/Tkin-GAF-PAS/PAC | chk103 | |
| 7120all3691, Avar_400213520 | HKIV+S/Tkin-(GAF)2 | chk104 | |
| NpunNpF3565, Avar_400175850 | HKIV+S/Tkin-(GAF)2 | chk105 | |
| Avar_400205900 | HKIV+S/Tkin-GAF-PAC | chk106(*) | |
| NpunNpF1766 | HKIV+S/Tkin-GAF-(PAS/PAC)2 | ||
| 7120alr0710, Avar_400217530, NpunNpR4071 | HKIV+S/Tkin-GAF | chk107 | |
| 7120all2095, Avar_400220860 | HKV (CBS)2-(PAS/PAC)5-PAC-(PAS/PAC)4-HKA_2 | chk108(*) | |
| 7421glr1767 | glr1767 has no CBS | ||
| 6803sll0750, 7120all3600, TBP1tlr0029, Tery_403262320, Avar_400209230, NpunNpR5764, Cwat_400864220, 7942_403111070, SS12Pro1121, MED4PMM1077, 9313PMT1099, 9312_402289750, SYNW0753, 9202_403151380, 9605_403135790 | HKV-KaiB | chk8, hik8, sasA | |
| 7421glr2212, NpunNpR0588 | HKV+cNMP | chk110 | |
| 7120alr4273, Avar_400189500, TBP1 tlr1447 | HATPase+(GAF)3-4 | chk111 | |
| 6803slr1285, TBP1tlr1565, 7120alr2572, 9313PMT1709, Tery_403232890, Avar_400182180, NpunNpR0564, 7942_403105070, Cwat_400872720, 9605_403120370, 9902_403163440, SYNW2043, SS12Pro1734, 9312_402295580, MED4PMM1579 | HisKA | chk34, hik34* | |
| 7421glr1586 | HKI | ||
| 7120all7218, Avar_400171460, NpunpNPBR204 | HATPase | chk112 | |
| Response regulator | Cwat_400853290 (+), NpunNpF2162 | RRI CheY | crr103 |
| orthologue | 6803sll1292, TBP1tll1024, Cwat_400882490 | RRI CheY | crr11, rre11 |
| 7120all2955, Cwat_400862100 | RRI CheY | crr79 | |
| 7120alr2049, Avar_400208360 | RRI CheY | crr45 | |
| 7120all2898, Avar_400187290, NpunNpF1184 | RRI CheY | crr46 | |
| 7120alr2240, Avar_400177800 | RRI CheY | crr47 | |
| 9313PMT1356, 9902_403161720, 9605_403122420 | RRI CheY | crr48 | |
| NpunNpR2902, 7942_403103430 | RRI CheY | crr49 | |
| NpunNpR3053, Avar_400185100 | RRI CheY | crr50 | |
| 6803slr1042, TBP1tlr0346, 7120all0929, Tery_403243600, Avar_400182250, NpunNpF5961, Cwat_400835560 | RRI CheY | crr7, rre7, pilH | |
| 6803slr2024, Cwat_400876810, Tery_403224660, NpunNpF3829, Avar_400207520, TBP1tlr2203, 7942_403100940, 7120all1281 | RRI CheY | crr13, rre13 | |
| 6803slr1982, Tery_403262690, 7942_403108080, 7421glr1684 | RRI CheY | crr21, rre21 | |
| 6803slr0474, 7120alr3158, Avar_400216110, NpunNpF0021, Cwat_400857320 | RRI CheY | crr27, rre27, rcp1 | |
| TBP1tll0571, 7120all2164, Avar_400177740, NpunNpF5637, 7942_403098370 | RRI CheY | crr51 | |
| 7120all0823, Avar_400221970, NpunNpR4743, 6803slr1037 | RRI CheY | crr10, rre10 | |
| 7120all3239, Avar_400226670, NpunNpF4460 | RRI CheY | crr52 | |
| 7120alr1967, 7421glr3433, Avar_400221100, NpunNpF6363, Tery_403260750 | RRI CheY | crr53 | |
| 7120all1071, Avar_400215320, NpunNpR6014, 6803sll0039 | RRI CheY | crr35, rre35, pixH, pisH | |
| 6803slr2041, 7120alr0442, Avar_400206000, NpunNpF6378, Cwat_400879540, 7942_403106840, Tery_403258240 | RRI CheY | crr42, rre42, devR, divK | |
| 7120all3766, Avar_400192900 | RRI CheY | crr54 | |
| 7120alr5328, NpunNpF3084 | RRI CheY | crr55 | |
| 7120alr2429, Avar_400179550, NpunNpR4768, 7421glr4213 | RRI CheY | crr56 | |
| 7120alr3594, Avar_400209180, NpunNpF5759 | RRI CheY | crr57 | |
| 7120alr2726, Avar_400220540, NpunNpR3959 | RRI CheY | crr58 | |
| 7120all5172, Avar_400201580 | RRI CheY | crr59 | |
| 7120alr3386, Avar_400211570 | RRI CheY | crr60 | |
| 7120alr0264, Avar_400204950, NpunNpF1447 | RRI CheY | crr61 | |
| 7120alr0774, Avar_400224390 | RRI CheY | crr62 | |
| Avar_400176300, NpunNpF2972 | RRI CheY | crr63 | |
| 7120all2165, Avar_400177730, NpunNpF5636 | RRI PatA | crr64 | |
| 7120all0521, Avar_400206700, NpunNpF5682 | RRI PatA | crr65, patA | |
| 7120all0930, Avar_400182260, NpunNpF5960, Tery_403243610, 6803slr1041, TBP1tlr0345, 7421gll0222 | RRI PatA | crr6, rre6 | |
| 7120all2821, Avar_400188280, NpunNpF1084, TBP1tll0572, 7942_403098360 | RRI PatA | crr66(*) | |
| Cwat_400866340 | RRIV-GGDEF | ||
| 7120all1072, Avar_400215310, NpunNpR6015, 6803sll0038 | RRI PatA | crr36, rre36, pixG, pisG, taxP1 | |
| Cwat_400853280, Tery_403234330, NpuNpF2161 | RRI PatA | crr67 | |
| 6803sll1291, TBP1tll1025 | RRI-PatA | crr12, rre12 | |
| TBP1tll1665, NpunNpR5896 | RRI other | crr68 | |
| 7120alr5150, Avar_400201400 | RRI other | crr69 | |
| 7120alr0960, Tery_403234380, NpunNpR5015, Avar_400182560, 6803sll1879, Cwat_400867290, TBP1tlr2323, 7942_403107930, SYNW0775, 9902_403151660, 9605_403135380, 9313PMT0522, SS12Pro0871, 9312_402286900, MED4PMM0795 | RRI-other (>200) | crr23, rre23, ycf55 | |
| 7120alr0356, Avar_400205490, NpunNpR1633 | RRI-other (>200) | crr70(*) | |
| 7421glr0744 | RRII-NarL | ||
| 6803sll0396, 7120alr1194, Avar_400183330, NpunNpR1688, 7942_403108640, Cwat_400882680, 9313PMT1357, SYNW1875, 9902_403161750, 9605_403122410, Tery_403242920, 7421glr2798 | RRII OmpR | crr28, rre28 | |
| 6803sll0649, 7120all4727, Avar_400196750, Cwat_400851810, NpunNpR6228, 7421glr0860 | RRII OmpR | crr3, rre3 | |
| 6803slr1584, Avar_400218430, NpunNpR3793, TBP1tll1910 | RRII OmpR | crr38, rre38 | |
| 6803slr1837, 7120all1964, Tery_403254660, Avar_400221130, NpunNpR6360, 7942_403103920, Cwat_400875050, TBP1tll2099, 7421glr1811 | RRII OmpR | crr16, rre16, manR | |
| 6803slr0947, TBP1tll2364, 7120all3822, 7421glr2669, SS12Pro0156, MED4PMM0134, 9313PMT1994, SYNW2246, Tery_403232990, Avar_400196090, NpunNpF5788, 7942_403104420, Cwat_400884040, 9605_403140590, 9902_403146840, 9312_402280200 | RRII OmpR | crr26, rre26, rpaB, ycf27 | |
| SS12PRO1780, MED4PMM1619, 9313PMT0146, 9312_402295990, SYNW0126, 9902_403145270, 9605_403117370, 7942_403114170 | RRII OmpR | crr71 | |
| 6803slr0115, TBP1tlr2423, 7120all0129, 7421gll4162, SS12Pro0150, MED4PMM0128, 9313PMT1988, SYNW2236, Tery_403229360, Avar_400192280, NpunNpF3659, 7942_403090590, Cwat_400890120, 9605_403140530, 9902_403146900, 9312_402280140 | RRII OmpR | crr31, rre31, rpaA | |
| 6803slr0081, TBP1tlr0589, 7120all4503, 7421glr4128, SYNW0947, Avar_400211230, NpunNpF4214, 7942_403099960, Cwat_400857010, 9605_403132790, MED4PMM0705, 9313PMT0994, 9312_402286040, Tery_403222900 | RRII OmpR | crr29, rre29, phoB, sphR | |
| 6803sll1330, 7120all4312, Tery_403234950, Avar_400189900, NpunNpR3907, Cwat_400846440, TBP1tlr1330, 7942_403114690, 7421glr2274, SS12Pro1083, MED4PMM1113, 9313PMT1097, 9312_402290110, SYNW0904, 9605_403133240, 9902_403158130 | RRII OmpR | crr37, rre37 | |
| 9313PMT0805, SYNW0808, 9605_403135030 | RRII OmpR | crr72 | |
| 7120all4750, TBP1tlr0261, Tery_403242920, Avar_400196530, NpunNpR4160, 7942_403113030, Cwat_400886040 | RRII OmpR | crr73, nblR | |
| 7120alr0072, Avar_400191760 | RRII OmpR | crr74 | |
| 7120alr3260, Avar_400226850, NpunNpR4101, 7942_403095260, Cwat_400849160 | RRII OmpR | crr75 | |
| 7120all3788, Avar_400196480, NpunNpR4165 | RRII OmpR | crr76 | |
| Avar_400212090, NpunNpR5316 | RRII OmpR | crr77 | |
| 7120alr5188, 7421gll0022, Avar_400201790 | RRII OmpR | crr78 | |
| 6803slr6040, 7120all7584, NpunNpR1779 | RRII OmpR | crr44 | |
| 7120alr8535, Avar_400218540 | RRII NarL | crr80 | |
| 7120alr3768, Avar_400192880, NpunNpF1453 | RRII NarL | crr81, orrA | |
| 6803sll0921, 7421glr4404, 7120all1704, Avar_400183790, NpunNpR5136 | RRII NarL CheY-LuxR | crr25, rre25 | |
| 6803slr1909, TBP1tll1048, 7120all3660, Tery_403237370, Avar_400213840, NpunNpF0832 | RRII- NarL | crr9, rre9(*) | |
| Cwat_400859330 | RRI-CheY | ||
| 6803sll1592, 7120alr3156, NpunNpF0304, 7421gll0571, Avar_400173270 | RRII NarL | crr19, rre19 | |
| 7421glr0348, NpunpNPBF146 | RRII NarL | crr82 | |
| 6803slr0312, 7120alr2138, 7421gll1683, Avar_400175840, NpunNpR2407 | RRII NarL | crr32, rre32 | |
| 6803sll1708, TBP1tlr0724, 7120alr0913, Avar_400222860, NpunNpF0484, Cwat_400864300, 7421gll0813 | RRII NarL | crr17, rre17 | |
| 6803slr1783, TBP1tlr1263, 7120all1736, 7421glr1588, SS12Pro0194, MED4PMM0169, 9313PMT2043, SYNW2289, Tery_403238910, Avar_400180170, NpunNpF1516, 7942_403108510, Cwat_400851460, 9312_402280550, 9902_403165290, 9605_403141020 | RRII NarL | crr1, rre1, ycf29 | |
| 7120all5069, Avar_400200620, NpunNpR0450, Cwat_400880970, Tery_403262490, TBP1tll2446 | RRII NarL | crr83 | |
| 7120all4635, Avar_400197620, NpunNpR2420 | RRII NarL | crr84 | |
| NpunNpR3308, Avar_400208190, 7421glr3742 | RRII NarL | crr85 | |
| 9902_403160010, SYNW1385, 9605_403131970 | RRII NarL | crr86 | |
| Avar_400176620, 7120alr7652 | RRII NarL | crr87 | |
| 7120alr7219, NpunpNPBF205, Avar_400171470 | RRII NarL | crr88 | |
| 7421gll4097, NpunNpR1109 | RRII NarL | crr89 | |
| Cwat_400878390, 6803sll0485 | RRII NarL | crr30, rre30 | |
| 7120all3232, Avar_400226600, NpunNpF4909 | RRII AraC | crr90 | |
| 7120all5174 | RRIII+Hpt-GGDEF | crr91(*) | |
| Avar_400201600 | RRIII+Hpt-RR-GGDEF | ||
| Avar_400176310, NpunNpF2968 | RRIII+Hpt | crr92 | |
| 7421glr4211 (+), Avar_400193590 | RRIII+Hpt-(RR)2 | crr93(*) | |
| NpunNpF5034, 7942_403091170 | RRIII+Hpt-(RR)2-GGDEF | ||
| TBP1tll1049 | RRIII+(RR)2-GGDEF | ||
| 7120all5323, NpunNpR3078 | RRIII+Hpt | crr97 | |
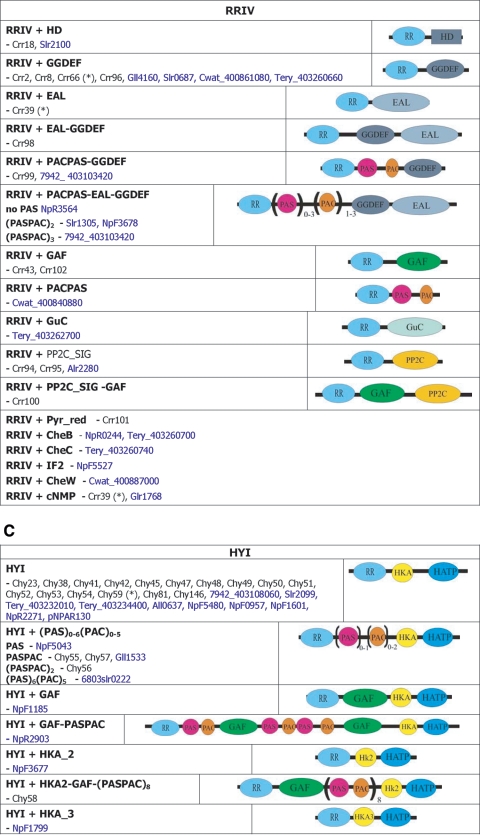
| 7120all3759, Avar_400192960, NpunNpR1683 | RRIV PP2C_SIG | crr94 | |
| 7120alr1086, Tery_403233220, NpunNpR3733), Avar_400214790, TBP1tlr1153 | RRIV PP2C_SIG | crr95 | |
| 7120alr3920, Avar_400195080, NpunNpF0906, 6803sll1624, Cwat_400848680 | RRIV+HD | crr18, rre18 | |
| 7120all2874, Avar_400187560, NpunNpF2364, 6803slr1760 | RRIV+GGDEF | crr8, rre8 | |
| 6803sll1673, Cwat_400862110 | RRIV+GGDEF | crr2, rre2 | |
| 7120alr3599, Avar_400209220, NpunNpF5763 | RRIV+GGDEF (+HTH-FIS except Npun) | crr96 | |
| 7120alr1230, Avar_400183660, NpunNpR4503, 6803slr1588 | RRIV+CheY-EAL | crr39, rre39(*) | |
| 7421glr3563 | RR+cNMP-HTH_CRP_2 | ||
| 7120alr2306, Avar_400178380, NpunNpF5361, Cwat_400880830 | RRIV+GGDEF-EAL | crr98 | |
| 7120all1012, Avar_400214250, NpunNpF3972 | RRIV+PACPAS-GGDEF | crr99 | |
| 7120all1703, Tery_403240160, Avar_400183800, NpunNpR5135 | RRIV+GAF-PP2C_SIG | crr100 | |
| 7421glr0480, NpunNpR0589 | RRIV+Pyr_red | crr101 | |
| 7120alr5251, Avar_400191500, NpunNpR4435, Tery_403262910 | RRIV+GAF | crr102 | |
| 6803sll5059, Cwat_400866740 | RRIV+GAF | crr43 | |
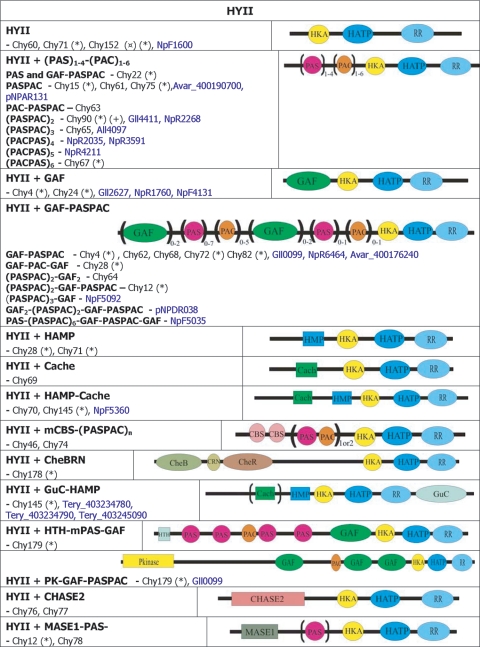
| Hybrid kinase | 7120alr2307, Avar_400178390, NpunNpF5362 | HYI | chy81 |
| orthologue | 7120all1388, Avar_400217340, NpunNpR6346 | HYI | chy45 |
| 7120all5308, Avar_400203040, NpunNpF1212, Tery_403261340 | HYI | chy146 | |
| 7120alr4879, Avar_400198830 | HYI | chy47 | |
| 6803sll1229, Cwat_400850270 | HYI | chy41, hik41 | |
| 7120alr3159, Avar_400216120, NpunNpF0022, Cwat_400892220, 6803slr1324 | HYI | chy23, hik23 | |
| 7120alr2241, Avar_400177810 | HYI | chy48 | |
| 7120alr1231, Avar_400183670, 6803sll1555 | HYI | chy42, hik42 | |
| 7120alr3121, Avar_400224710 | HYI | chy49 | |
| 7120alr4880, Avar_400198840, NpunNpF3833 | HYI | chy50 | |
| 7120all3764, Avar_400192920, NpunNpR1448 | HYI | chy51 | |
| 6803slr1400, 7120all4096 | HYI | chy38, hik38 | |
| 7120all1177, Avar_400218940 | HYI | chy52 | |
| 7120alr3671, Avar_400213720, NpunNpF0839, 7421gll3122 | HYI | chy53 | |
| 7120all3765, Avar_400192910, NpunNpR1449 | HYI | chy54 | |
| 7120all1279, Avar_400207500, NpunNpF6002 | HYI+PAS/PAC | chy55 | |
| 7120all2897, Avar_400187300 | HYI+(PAS/PAC)2 | chy56 | |
| 7120all2094, Avar_400220870 | HYI+PAC/PAS | chy57 | |
| 7120all2239, Avar_400177790 | HYI+HKA2-GAF-(PAS/PAC)8 | chy58 | |
| 7120all2883, Avar_400187470, NpunNpF2889, Tery_403264330, Cwat_400840100 | HYII | chy152(*) (†) | |
| TBP1tll0480 | HKI | chk152(*) | |
| 7120alr1285, Avar_400207560, NpunNpR3825 | HYII | chy60 | |
| 7421glr0033 | HYII+PAS | chy22, hik22(*) | |
| 6803slr2104 | HYII+GAF-PAS/PAC-Hpt) | ||
| 7120all0182, NpunNpR1868, Avar_400204180 | HYII+(TM)3-PAS/PAC | chy61 | |
| Avar_400215270, NpunNpR3784 | HYII+GAF-PAS/PAC | chy62 | |
| 7120all3985, Avar_400194470 | HYII+PAS/PAC | chy63 | |
| 6803sll1228 | HYII+GAF | chy4, hik4(*) | |
| NpunNpR5897 | HYII+GAF-(PAS/PAC)n | ||
| NpunNpR3562, 7120all5210, Avar_400205150 | HYII+(PAS/PAC)2-GAF | chy64 | |
| NpunNpF6350, 7120alr3442, Avar_400212230 | HYII | chy65 | |
| 7120all0824, NpunNpR4744 (1 more PAS N-ter), Avar_400221980 | HYII (PAS/PAC)3-PAS-HK-CheY | chy66 | |
| 7120alr3092, Avar_400215770 | HYII (TM)3-(PAS/PAC)n | chy67(*) | |
| 7421glr0376 | HKV+HTH_4-PAS-PAS/PAC | chk67(*) | |
| Avar_400225770, 7120all1804, NpunNpR3548 | HYII+(TM)9-GAF-PAS/PAC (Npun no GAF) | chy68 | |
| 7120all1389, Avar_400217330, NpunNpR6347 | HYII+Cache | chy69 | |
| TBP1tll1367, NpunNpR1798 | HYII+HAMP-GAF | chy70 | |
| 7120all0638 | HYII | chy71(*) | |
| Tery_403261350 | HYII+HAMP | ||
| NpunNpF1211 | HYIII+HAMP | ||
| 7120alr4878, Avar_400198820, NpunNpF5479 | HYII+Cache-HAMP | chy145(*) | |
| Tery_403250580 | HYII+GuC-HAMP | ||
| 7120all2379, Avar_400179140, NpunNpF0354, 6803slr6001 | HYII+mCBS-(PAS/PAC)n | chy46 | |
| 7120all1914, Avar_400207900, NpunNpF4948 | HYII+mCBS-(PAS/PAC)n | chy74 | |
| 7120alr3120, Avar_400224700 | HYII+(CBS)4-GAF-PAC-GAF | chy28, hik28(*) | |
| 6803sll0474 | HYII+TM-160-TM-HAMP-PAC | ||
| 7421gll1854 | HKV+CheR/B | chk178(*) | |
| 7120all1716, Avar_400180300 | HYII+CheR/B | chy178 | |
| 7120all5309, Avar_400203050 | HYII+CHASE2 | chy76 | |
| 7120all1178, Avar_400218930 | HYII+CHASE2 | chy77 | |
| 7120all3275, Avar_400226990 | HYII+MASE1-PAS | chy78 | |
| 7120all0978, Avar_400180780 | HYII+HTH_4-(PAS/PAC)n-GAF | chy179(*) | |
| NpunNpF5679 | HYII PK-GAF-PAC-(GAF)2 | ||
| 7421gll3736 | HKV+HTH_4 | chk179 | |
| 7120all5173, Avar_400201590 | HYIII+m(PAS/PAC) | chy80 | |
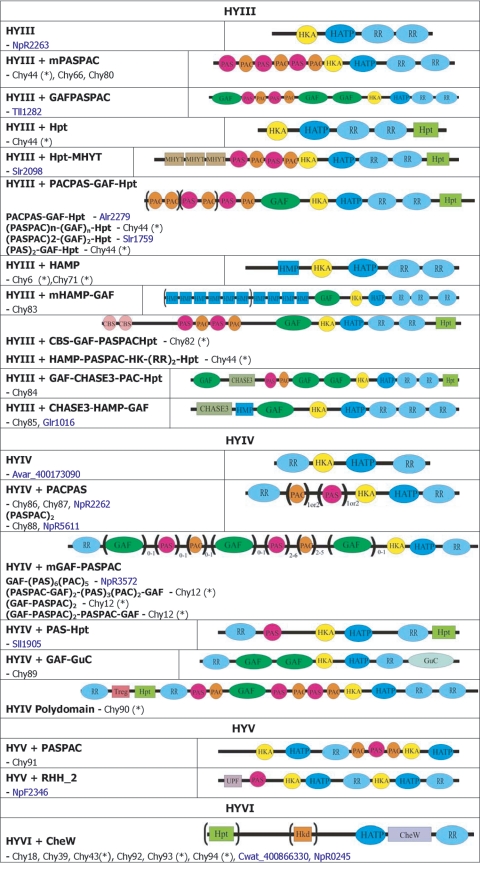
| TBP1tll0876 | HYIII+PAS/PAC | chy44(*)d | |
| Cwat_400866730 | HYIII+Hpt | ||
| NpunNpF2363, 7120all2875, Avar_400187550 | HYIII+PAS/PAC-GAF-Hpt | ||
| 6803sll5060 | HYIII+(PAS)2-GAF-Hpt | ||
| Tery_403233060 | HYIII+HAMP-PAS/PAC-GAF-Hpt | ||
| 7421glr0718 | HYII PAS/PAC-GAF | chy82(*) | |
| Tery_403238490 | HYIII+CBS-(PAS/PAC)2-GAF-Hpt | ||
| 7120alr1121, Avar_400224650, NpunNpF2686, 7421gll0635 | HYIII+(HAMP)n-GAF | chy83 | |
| 7120alr3761, Avar_400192940, NpunNpR1685 | HYIII+GAF-CHASE3-PAC-Hpt | chy84 | |
| 7120alr1883, Avar_400225480, NpunNpR2375 | HYIII+CHASE3-HAMP-GAF | chy85 | |
| 7120all1846, NpunNpF3541, Avar_400225520, 7421gll0634 | HYIV+PAS/PAC | chy86 | |
| 7120all1640-all1639 (+), NpunNpR2901, Avar_400225220 | HYIV+PAS/PAC | chy87 | |
| 7120alr1968, Avar_400221090, NpunNpF6364, 7421glr3434 | HYIV+(PAS/PAC)2 (7421 only 1 PAS/PAC) | chy88 | |
| 6803sll1672* | HYII MASE1 | chy12, hik12(*) | |
| Cwat_400850260 | HYII+(PAS/PAC)2-GAF-PAS/PAC | ||
| Avar_400223990, 7120all0729 | HYIV+GAF-(PAS/PAC)3-(GAF)2 | ||
| NpunNpR3691 | HYIV+(PAS/PAC-GAF)3 | ||
| 7120all4963, Tery_403242830, Avar_400199740, NpunNpR0896 | HYIV+(GAF)2-GuC | chy89, cyaC | |
| 7120alr2428, Avar_400179540, NpunNpR4769 | HYIV polydomain (Npun no GAF) | chy90(*) | |
| 7421glr4212 (+) | HYII+(PAS/PAC)2 | ||
| 7421glr2749, NpunNpF2908, Avar_400188120 | HYV+PAS/PAC | chy91 | |
| 7120all1068, Avar_400215350, NpunNpR6010 | HYVI+Hpt-CheW | chy92 | |
| 6803sll0043, Tery_403235320 | HYI (Hpt)n-CheW | chy18, hik18, pixL, taxAY1 | |
| Cwat_400853350 | HKV+Hpt | chk93(*) | |
| NpunNpF2165 | HYVI+Hpt-CheW | chy93 | |
| 7120all0926, Avar_400182220, NpunNpF5964, TBP1tlr0349, 7942_403099980 | HYVI+Hpt-CheW | chy43, hik43, taxAY3(*) | |
| 6803slr0322 | (6803 Hkd and no Hpt) | ||
| Cwat_400838100, Tery_403243570 | (Tery and Cwat, no Hpt) | ||
| 7120all2161, Avar_400177770, NpunNpF5640, 7942_403098400, TBPtll0568, Cwat_400850380 | HYVI+Hpt-CheW (7942, TBP1+Hpt-Hkd-CheW) | chy94(*) | |
| 6803sll1296, Cwat_400882520, TBP1tll1021 | HYVI+Hpt-CheW | chy39, hik39, taxAY2 | |
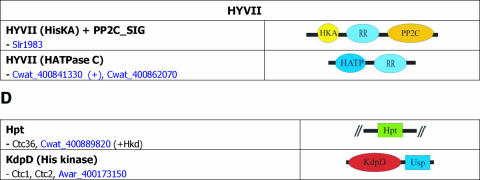
| Other orthologue groups for two- | 6803slr0073, Tery_403253950, 7942_403101090, Cwat_400883640 | Hpt | ctc36, hik36, pilN |
| component | 6803slr1731, 7421gll0570 (2KdpDUsp), Avar_400173080, | KdpD Avar (plasmid B) | ctc1, kdpD(*) |
| related ORFs | 7942_403107200 | ||
| 7120all4242, NpunNpR0299, Avar_400189180 | KdpD | ctc2, kdpD | |
| Avar_400173150 | KdpD (plasmid B) | ctc3, kdpD |
The Anabaena sp. strain PCC 7120 and E. coli K-12 sequences used in the searches to retrieve the putative proteins containing HK and RR domains were as follows: RR-CheY, 7120alr0442 and E. coli NP_416396.1; RR-OmpR, 7120all3822 (aa 7 to 124) and E. coli NP_417864.1 (aa 5 to 124); T_reg, 7120all3822 (aa 151 to 233) and E. coli NP_417864.1 (aa 156 to 232); HisKA-HATPase, 7120all0330 (aa 429 to 656) and E. coli NP_417863.1 (aa 234 to 439); Hpt, 7120all1068 (aa 8 to 111) and E. coli NP_415513.1 (aa 815 to 896). Analyses were performed at the Integrated Microbial Genomes website (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) and at the European Bioinformatics Institute website (http://www.ebi.ac.uk/InterProScan/); domain structure assignments were made at http://www.cbs.dtu.dk/services/TMHMM/.
Boldface type indicates that orthologues exist in the 16 genomes. (+), putative sequencing error.
Whenever a single gene or a member of a group has been assigned a function in the literature and/or an annotation in databases, the gene name is mentioned. (*), proteins with different domain structures belong to this group of orthologues; (†), proteins have no bacterial orthologues.
All seven gene loci for this gene have an E value of 0.0 e+00.
BIOINFORMATIC GENOME ANALYSIS
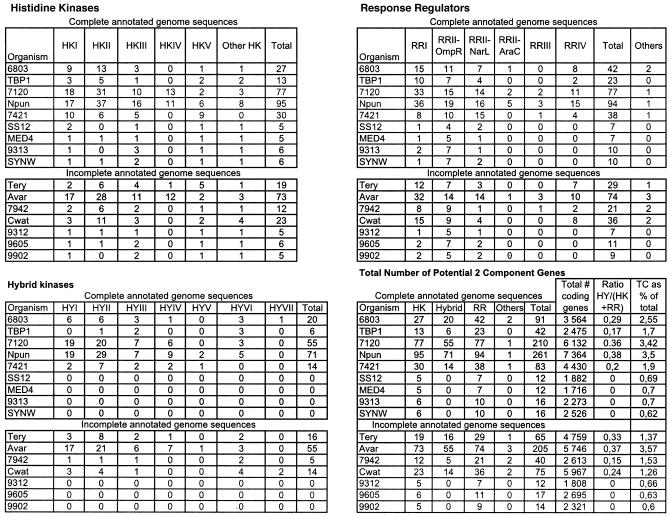
The cyanobacterial genome sequences were accessed (as of May 2005) at Cyanobase (http://www.kazusa.or.jp/cyano/) and IMG (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). Synechococcus sp. strain PCC 6301 is very closely related to PCC 7942 in terms of genome size and physical map (44, 116). Since its sequence is not yet fully released, it was not considered in our comparisons, but most of what is said for Synechococcus elongatus PCC 7942 applies to PCC 6301 as well. The identities of potential two-component genes were derived from the published assignments (http://www.kazusa.or.jp/cyano/ and http://img.jgi.doe.gov/cgi-bin/pub/main.cgi [89, 102]) and analyses performed at Interpro (http://www.ebi.ac.uk/InterProScan/) and supplemented with PBLAST searches of each genome with a battery of cyanobacterial and Escherichia coli K-12 two-component domains (domains used include receivers from CheY and OmpR, Lux_R, HisKA/HATPase, and Hpt [see Table S2s in the supplemental material]). A few protein sequences have been kept even if they presently no longer carry the key residues required to form a canonical HK or RR domain, when they clearly are part of an orthology group gathering sequences from almost all of the 16 cyanobacterial genomes (see below). Though some sequence and/or assembly errors may have occurred, they do not seem to be numerous (95), and they permit the in silico survey presented here. In the course of this study, we noticed a few probable sequencing errors; these are mentioned in the legend to Fig. 2. For clarity, each gene name begins with a four-character acronym, referenced to the organism (Fig. 1), followed by the locus tag that comes from Cyanobase (e.g., 6803slr0396) and the locus tag or gene object identifier (GOI) (gene_OID) from the IMG site (NpunNpR0564, Tery_403232890, or Avar_400182180, etc.). All sequences can thus be easily downloaded, with both locus tags and GOIs remaining available at IMG for searches even after completion of the genome annotations.
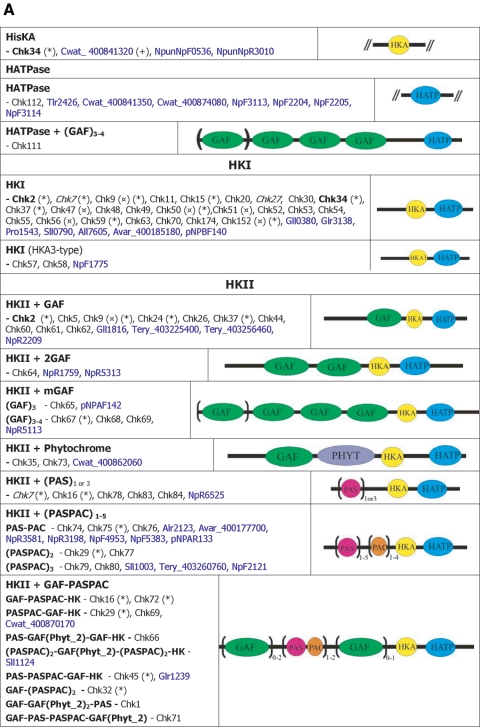
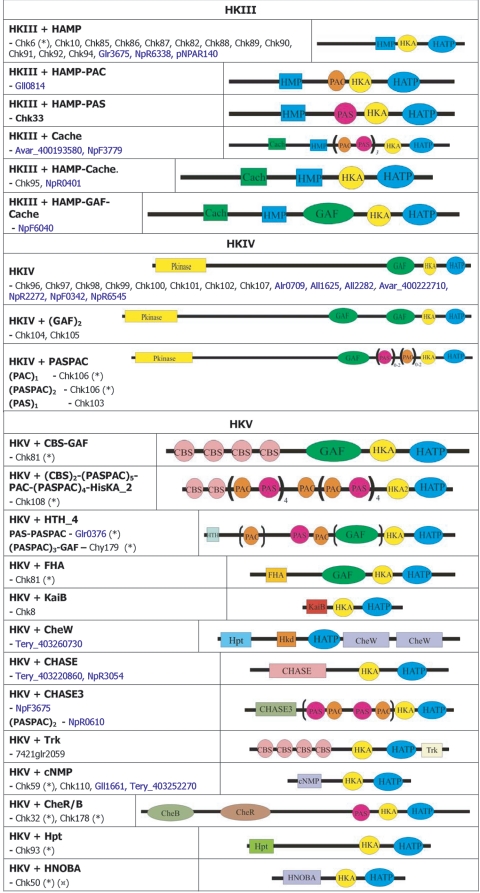
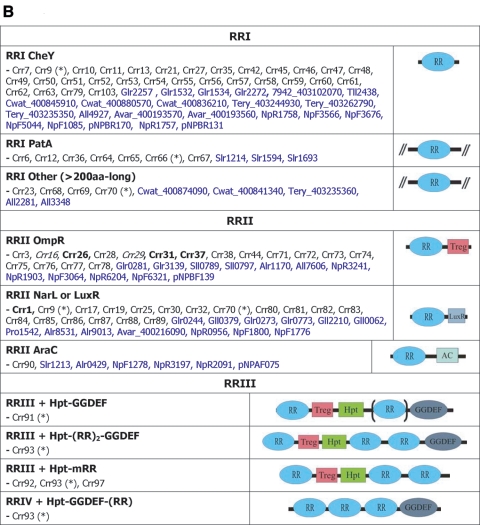
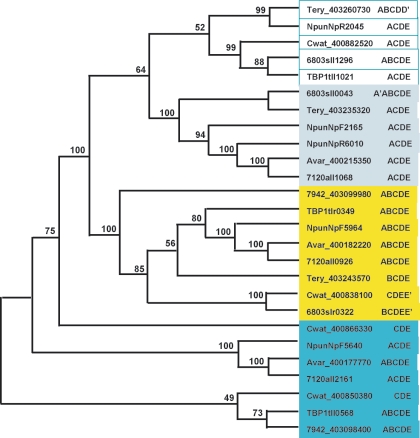
FIG.2.
Cyanobacterial two-component ORF repertoire. (A) HKs. (B) RRs. (C) HYs. (D) Others. Subscript numbers following parentheses are the numbers of similar domains that may be found in the proteins listed in a given subclass. Boldface type indicates that orthologues exist in the 16 genomes, and the corresponding phylogenetic trees are shown in Fig. 5. *, proteins with different domain structures belong to this group of orthologues; ( ), protein that has no bacterial orthologue; (+), putative sequencing error (see below). Putative proteins that have no cyanobacterial orthologue are shown in blue and are identified by their locus tag or GOI. Putative sequencing errors: 7120all1640-1639 (7A instead of 8A would create a frameshift, giving a protein with 95% identity to Avar_400225220); the A of the 7421glr4211 stop codon can serve in the ATG initiation codon for 7421glr4212; Cwat_400841320 could form an HY with Cwat_400841330; and the HATP-RR Cwat_400841330 could form an HYI with Cwat_400841320.
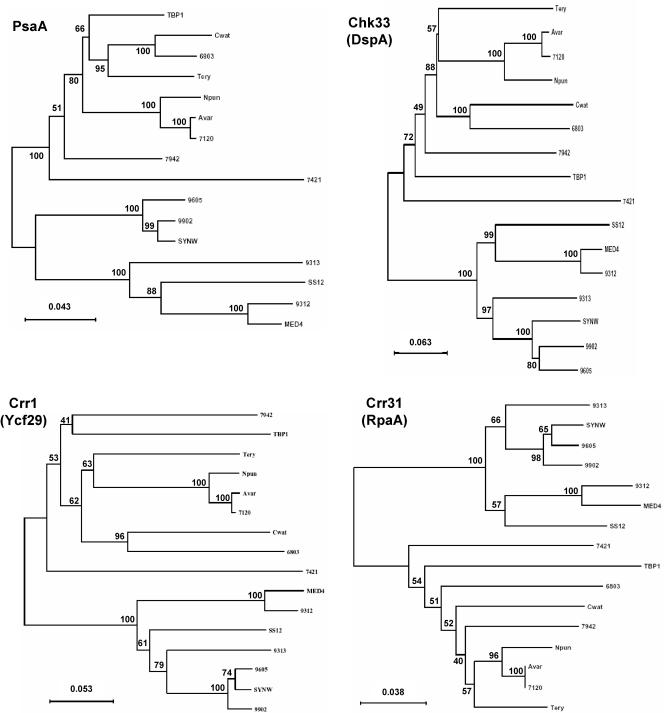
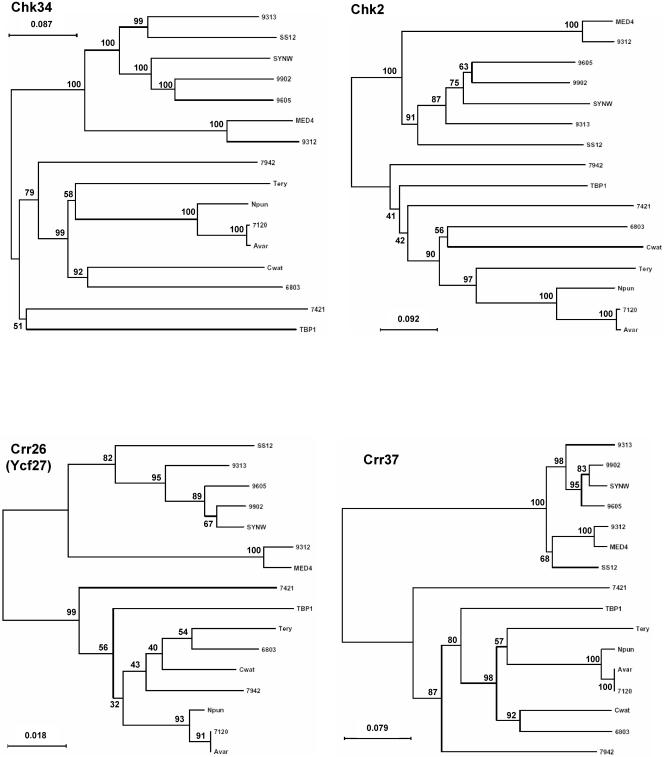
Domains were initially assigned by Pfam batch analysis at http://www.sanger.ac.uk/Software/Pfam/ (18). Domains were recorded for each two-component gene only if they were scored as “Pfam's trusted match thresholds.” Domain assignments were checked and modified by using the more extensive domain assignments at InterPro (http://www.ebi.ac.uk/interpro/).The data have been manually checked to avoid false-negative and false-positive hits, which may arise from automated analyses. The results from the 16 different cyanobacterial species were used to classify the putative two-component proteins by domain organization with a nomenclature adapted from Ohmori et al. (102), with each group subdivided by the organization of the identified signaling domains. The cartoon-style diagrams presented in Fig. 2 were constructed from these data, with the sizes of the domains roughly in proportion. The putative open reading frames (ORFs) that exhibit similar domain structures have been gathered into groups and listed within each according to their orthology relationship (Table 1; see also Table S3s in the supplemental material). Parentheses followed by figures indicate the number of similar domains that may be found in the proteins listed in a given subclass. Orthology information, based on the bidirectional best hits from BLAST searches of each organism against other organism polypeptides, is accessible at the IMG site. It was ascertained by performing CLUSTALW alignments for each subclass and by making phylogenetic trees with the PHYLO_WIN program (41, 138). The domain structures of some orthologues may vary slightly as a result of gene fusion, shuffling, and/or insertions-deletions. Paralogues are the homologues present within a given organism. These definitions are not fully accurate but can be considered a useful approximation inasmuch as we cannot always be sure of whether the polypeptides arose from a single gene present in the last common ancestor (orthologues) or from gene duplication within a genome (paralogues) (128).
CYANOBACTERIAL TWO-COMPONENT ORF REPERTOIRE: STRUCTURE AND FUNCTION
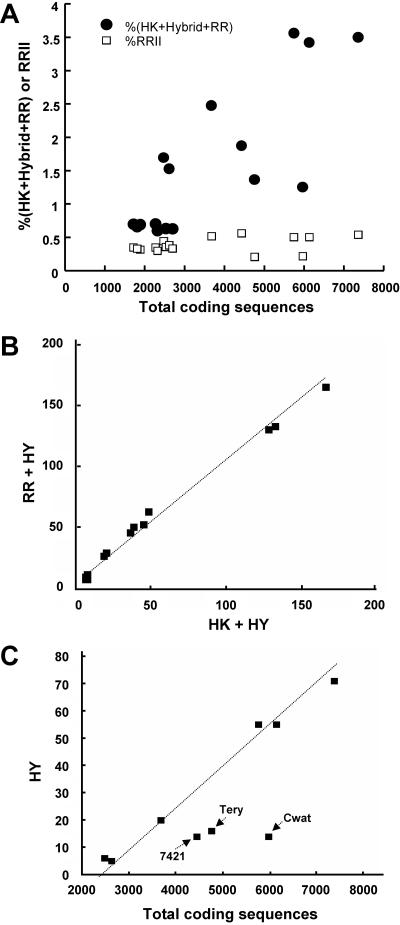
For each organism, a preliminary list of all histidine kinases, hybrid kinases, and response regulators was constructed from the functional annotations (http://genome.jgi-psf.org/mic_home.html[89, 102]). Table 1, Fig. 2, and Table S3s (in the supplemental material) present all of the putative two-component proteins that could be retrieved. The number of two-component systems (Fig. 3) agrees well with published data (11, 34, 38, 93, 94, 102, 106, 118). The small differences result, in particular, from the integration of previously unrecognized, “atypical” HisKA domains, HisKA_2 (HWE) and HisKA_3 in a few proteins, and of ORFs detected on later sequenced plasmid genomes.
FIG. 3.
Cyanobacterial genes encoding two-component system proteins (see Fig. 1 and 2 for acronyms and abbreviations).
Structural Domains Found in Cyanobacterial Two-Component Proteins
Many of the putative polypeptides that were found, whether kinases or response regulators, are multidomain proteins. A list of all of the domains and their acronyms that have been identified in cyanobacterial two-component proteins is given in Table S2s, in the supplemental material, together with short definitions of their functions. The most common ones are GAF, PAS, and PAC for the kinases in particular and DNA-binding domains (Treg and NarL or LuxR) for the response regulators. Some are only seldomly found: CBS, CheB, CheR, CheW, CHASE, cNMP, FHA, GuC, MASE, MHYT, Pkinase, PP2C, Trk, and UPF. With the exception of GerR and LytTR, all of the different types that compose the bacterial domain repertoire can be found in cyanobacterial proteins (141).
A HisKA domain is about 60 amino acids (aa) long and constitutes the dimerization and phosphoacceptor domain of the HKs. HisKA_2 (HWE) and HisKA_3 are alternative dimerization and phosphoacceptor domains. Hkd is the homodimer interface of the signal transducing histidine kinase family, which often overlaps the HisKA domain. To form a histidine kinase (HIS_KIN), a HATPase_c domain (HATP), which is usually adjacent to and downstream of HisKA or homologues, is required. Such domains are found in many ATP-binding proteins and are necessary for kinase activity.
A canonical basic response regulator domain (RR or CheY) can be schematically described as two aspartate (D) residues and a lysyl (K) residue appropriately spaced within an ∼120-aa sequence. They usually are N terminal to output domains, some of which, the transcriptional regulators, have the property of binding to specific DNA sequences. Depending on sequence similarities, response regulator proteins are often classified in subfamilies named from the best-studied CheY, OmpR, NarL, or LuxR proteins, for example.
As the acronym indicates, GAF domains (for “cGMP phosphodiesterase, adenylyl cyclases, and the bacterial transcription factor FhlA,” about 130 aa long) have been linked to small-molecule binding, in particular the cyclic nucleotides cyclic AMP (cAMP) and cGMP, which are common second messengers in signal transduction (5, 9, 23, 53). They are found in different proteins related to the cNMPs, cyclases, and phosphodiesterases, as well as to light-signaling phytochromes (9, 85). The GAF family is among the largest of all classes of signaling domains. GAF (Phyt_2) is a member of this large family. PHYT is a light wavelength sensor domain to which a linear tetrapyrrole is bound through a thioether linkage via a Cys residue (113). It permits the reversible photochemical conversion of a protein between two forms.
PAS and PAC domains are often found to be associated. PAS derives from the names of three proteins that the domain occurs in: Per, period circadian protein; Arnt, Ah receptor nuclear translocator protein; Sim, single-minded protein. The acronym PAC is derived from “PAS-associated, C-terminal,” such sequences contributing to the PAS domain fold. The division between the PAS and PAC domains is caused by major differences in sequences in the region connecting these two motifs. A subset of PAS domains, the best-characterized members of this family, binds cofactors such as heme and flavin adenine dinucleotide. Sensing of light, oxygen, or redox potential requires cofactors, while signals such as voltage, xenobiotics, and nitrogen availability do not (16, 21, 43, 110). PAC domains can be found without an associated PAS domain. GAF and PAS domains exhibit striking similarity in their structures, and proteins carrying such domains are clearly linked in their evolution (53). The common theme among both classes of proteins with such domains is the binding, either covalent or not, of a remarkably diverse set of small regulatory molecules that often remain unidentified (5). The two domains are presumed to be functionally similar.
Histidine Kinases
Incomplete HKs.
For 18 putative proteins, which are about 450 aa long, only a HisKA domain could be recognized. Most of them form an orthology group with one sequence from each genome, except for that of Gloeobacter violaceus. The orthologous protein from this strain, Glr1586, is a complete HK, having both a HisKA and a HATPase domain (see below). One of these proteins, Slr1285 (Hik34, Chk34), has just been shown to be involved in salt sensing and hyperosmotic stress response in Synechocystis sp. strain PCC 6803 and to pair with the response regulator Slr1783 (Rre1, Crr1) (105, 126). The genes reported to be under the control of the Hik34-Rre1 pair following hyperosmotic or salt stress are rather general stress response genes; this may explain why the authors could not identify the sensor partner upstream of Hik34 in the regulatory pathway. In agreement with their previous data, the same group recently showed that Hik34 is required for thermotolerance (probably by regulating the expression of some heat shock genes) and that the purified protein could autophosphorylate in vitro (134). There do not appear to be any proteins orthologous to this Chk34 group in any of the other 110 completed bacterial genome sequences.
Thirteen putative proteins, originating from only five species (all diazotrophs except Thermosynechococcus elongatus) only have a HATP domain. Half of them are from Nostoc punctiforme, with NpF3113 (128 residues) being 100% identical at the amino acid level to NpF2204 and the adjacent NpF2205 (75 residues) being 100% identical to NpF3114. These may represent recent gene duplications. The fifth N. punctiforme representative, NpunpNPBR204, is carried by a plasmid and has orthologues in Anabaena sp. strain PCC 7120 and Anabaena variabilis, all being about 250 residues. Three of these putative proteins have additional N-terminal GAF domains and form a group of orthologues. Three additional proteins, listed as HYVII, consist of either a HisKA or a HATPase N-terminal domain linked to a response regulator (RR) downstream. Finally, a class composed of 26 proteins formed by different combinations of five basic domains, of which one is HATPase-c, appears in Fig. 2 as HYVI and HKV+CheW for the Trichodesmium erythraeum representative. It is discussed in more detail below. Whether such polypeptides act by forming complexes with specific HisKA proteins is a hypothesis that must be tested.
HKI.
HK class I kinases (HKIs), having only HisKA and HATP domains, can be considered basic proteins, i.e., serving as building blocks for the more sophisticated domain arrangements that also exist in cyanobacteria. However, it is likely that many of these ORFs have signaling domains that have not yet been identified. This possibility is highlighted by a number of HKIs that have one or more transmembrane (TM) domains that could flank putative signaling domains (Table 1; also see Table S3s in the supplemental material). None were found in the marine unicellular non-N2-fixing strains, with the exception of Pro1543 in Prochlorococcus sp. strain SS120. Depending on strains, they may represent one-fifth to one-twentieth of the whole histidine kinase repertoire, being more abundant in the unicellular species. There are 36 groups of orthologues; six of them are proteins found only in the three filamentous heterocystous strains, and three are made up of Anabaena sp. strain PCC 7120 and A. variabilis proteins only. In nine instances, an HKI may have orthologues which have a more complex structure (highlighted with asterisks in Table 1 and Fig. 2). As an example, Anabaena sp. strain PCC 7120 All2956 has an HYI orthologue in C. watsonii (Cwat_400862090) and an HKIV (cNMP-HK) (Gll1662) orthologue in G. violaceus. Chk7s constitute another example, as in most of the orthologues a PAS domain is also present (see the discussion of SphS, below). Six HKIs do not have any bacterial orthologue, 7120all7605 and NpunpNPBF140 being plasmid encoded. It is interesting that a G. violaceus protein (Gll0380) has a single orthologue in all of the sequenced bacterial genomes found in Archaeoglobus fulgidus, a hyperthermophilic marine sulfate reducer isolated from a hydrothermal environment. Phylogenetic analyses place G. violaceus close to the root of the cyanobacterial lineage, and the Archaeoglobales are the only archaebacteria that can grow by sulfate reduction, a property restricted to relatively few groups of eubacteria.
The largest group of orthologues (Chk27) contains a representative from every species except the marine unicellular non-N2-fixing strains. Synechocystis sp. strain PCC 6803 ManS, a protein involved in manganese homeostasis (100), is one of them. On the other hand, the Anabaena sp. strain PCC 7120 HepX (Alr0117, Chk52) has been reported to be involved in heterocyst development (heterocyst envelope polysaccharide [97]). It has orthologues not only in the other two heterocystous strains but also in the other two N2-fixing strains, as well as, surprisingly, the unicellular freshwater S. elongatus 7942.
Synechocystis sp. strain PCC 6803 Sll0798 (Chk30; termed RppB or NrsS) has been shown to control the Ni2+-dependent induction of the nrsBACD operon and to be involved in Ni2+ sensing (76). Such a member of the bacterial binding protein-dependent transport systems would also be present in A. variabilis and Anabaena sp. strain PCC 7120. On the other hand, the inactivation of both sll0790 (hik31, chk31) and slr6041 (chk46), two HKI paralogs sharing 97.5% identity, leads to the conclusion that the gene products are involved in the regulatory mechanisms that allow Synechocystis sp. strain PCC 6803 to adapt from photoautotrophic to photomixotrophic growth (62). This HKI would be required for the expression of icfG (encoding glucokinase) and the modulation of the glucose-6-phosphate dehydrogenase, thus having a dual role.
HKII.
HK class II (HKII) groups the putative proteins that have HK linked only to either one or more GAF and/or PAS or PAC domains. These domains are encountered in quite large numbers in bacteria and euryarchaeota (40), with PAS domains being more common than GAF, except in Synechocystis sp. strain PCC 6803. Compared to most other bacteria, the large number of GAF domains correlates and underlines the role of light in the regulation of gene expression and metabolic activities for photosynthetic organisms (40). None of the seven marine unicellular non-N2-fixing strains have any HKII+GAF, and only four of them have HKII+(PAS)1-3. Within this group there are 68 ORFs with only GAF sensor domains, 50 with PAS/PAC domains, and 28 with both. The analysis by Narikawa et al. (95) gives 17 PAS-containing ORFs in Synechocystis sp. strain PCC 6803, 61 in Anabaena sp. strain PCC 7120, and 84 in N. punctiforme (compared to 9 for E. coli and 10 for B. subtilis).
Only one orthology group, Chk2 (HKII+GAF), has one protein from each of the 16 species, but for G. violaceus and the marine Synechococcus and Prochlorococcus spp. the proteins are shorter and lack a detectable GAF domain; they are thus classified as HKI. Functional data have been reported for one of its members, Synechocystis sp. strain PCC 6803 Slr1147 (Hik2), which would interact with the response regulator Rre1, as does Slr1285 (Hik34, which has no detectable HATPase_c domain; see above). In this strain it would regulate the expression of sigB and four other genes in response to hyperosmotic stress (105, 126).
Within HKII, the subclass HKII-phytochrome is one in which proteins of well-known function occur. Synechocystis sp. strain PCC 6803 Slr0473 (Cph1, Chk35), for example, has been characterized as a photoreceptor (35, 36, 149). Light-induced conformational change of the chromophore in Cph1 results in inhibition of the histidine kinase activity (35). Two paralogues, aphA and aphB, exist in Anabaena sp. strain PCC 7120 as well as in the two other heterocystous strains (101). Besides these four strains, only C. watsonii has a Cph1-like protein, as well as a paralogue that does not group with AphB. Other marine species do not have any. It is worth noting that orthologues are not as widely distributed as could have been expected from the study conducted on the chromophore-binding (PHYT) domain of these proteins (49). Most of the cyanobacteria examined there were indeed shown to share a rather well-conserved chromophore binding sequence. The other HKs with multiple GAF domains are essentially from the filamentous heterocystous strains. Following the observation that red light decreased whereas far-red light increased cellular cAMP content in Anabaena sp. strain PCC 7120, Ohmori and coworkers disrupted 10 ORFs having putative chromophore-binding GAF domains. The all2699 (chk65, aphC) mutant failed to respond to far-red light. They concluded that the far-red light signal could be received by AphC and then transferred to the N-terminal RR domain of the CyaC adenylyl cyclase, stimulating its catalytic activity. The increased cAMP concentration would then drive the subsequent signal transduction cascade (104).
About half of the HKII+(PAS)1-3 subclass corresponds to one orthology group (Chk7). This group is constituted of proteins from 12 species that do not all possess a PAS domain. Those which do not possess a PAS domain have an N-terminal TM domain instead, and the T. elongatus orthologue (tll0925) has both. One member, 7942_403099950, has been identified as SphS, a sensor whose cognate response regulator is SphR (Crr29), by complementation of an E. coli phoR creC mutant for the expression of alkaline phosphatase (1). The genes are adjacent to the RR upstream from the HK. The S. elongatus 7942 mutant that lacks these genes is defective in the ability to produce alkaline phosphatase and some inducible proteins in response to phosphate limitation. This was one of the very first cyanobacterial two-component systems to be characterized. The Synechocystis sp. strain PCC 6803 Hik7 and Rre29 orthologues have since been shown to be the dominant sensory system that controls gene expression in response to phosphate limitation (51, 136). Murata and coworkers (136) suggested that a two-component system homologous to SphS-SphR is likely conserved in all cyanobacterial species. However, no direct orthologue could be detected in T. erythraeum, Synechococcus sp. strain 9902, or P. marinus SS120 and MIT9313. T. erythraeum (Chk78) has a HKII+(PAS)1-3 that is orthologous to those of Anabaena sp. strain PCC 7120 and N. punctiforme paralogues of Chk7.
Putative histidine kinases with either PAS or PAS/PAC domains that occur either in single or multiple copies were essentially found in the filamentous heterocystous strains. There is one in T. erythraeum and two in Synechocystis sp. strain PCC 6803. Ten of the 24 putative proteins do not have any orthologues, and one of them (NpunpNPAR133) is plasmid encoded.
Another HKII subclass is made up of proteins with both PAS and/or PAC and GAF domains. Such proteins are totally absent in marine strains, except C. watsonii. The Ssl1473-75 acronym (Chk32) is used in Table 1 and Fig. 2 because it corresponds to the Synechocystis sp. strain PCC 6803 wild-type sequence, which is interrupted by an insertion (IS) element in the “Kazusa” strain that was sequenced (103). This fusion protein is about 40% identical to the Fremyella diplosiphon (Tolypothrix PCC 7601) RcaE protein (GAF-PAS-PAC-HK), which has been shown to be a photoreceptor involved in complementary chromatic adaptation (137). From the microarray data obtained with a Synechocystis sp. strain PCC 6803 chk16 mutant, the Chk16 protein could be directly involved in sensing NaCl concentration (80). Under hyperosmotic conditions, it would be part of a phosphorelay cascade involving Synechocystis sp. strain PCC 6803 Chk41 (Hik41) and Crr17 (Rre17) (105, 126). Interestingly, the Synechocystis sp. strain PCC 6803 and C. watsonii Chk16s, which possess an N-terminal MASE1 domain (the function of which is currently unknown [96]) in front of a GAF (Phyt_2) domain, have orthologues in Anabaena sp. strain PCC 7120 and A. variabilis, with only a PAS domain. The N. punctiforme Chk16 orthologue has a GAF (Phyt) domain between the PAS/PAC and HK domains. Notably, for each of the three heterocystous strains, the closest paralogues of Chk16 (i) have rather similar structures, (ii) are orthologues (Chk74), and (iii) are located immediately downstream of the chk16 genes. Gene duplication thus probably occurred in an ancestor common to these three strains before their divergence, and the two genes, chk74 and chk16, have subsequently evolved differently.
HKIII.
Kinases of HK class III (HKIII) possess, in addition to the HisKA and HATPase, a HAMP (or “linker”) domain. The latter is typically found downstream from the last TM segment of a protein, and it has been shown that two symmetrical HAMP domains dimerize and cooperate to transfer the signal across the membrane via a linker to the histidine kinase (155). The presence of a HAMP domain suggests that the corresponding putative ORFs likely function as a dimer. In many cases, it is linked to transmitting signals across a membrane from periplasmic ligand-binding domains (6, 7, 10). The HAMP domain localizes upstream from HisKA. One protein has a PAC (Chk159, 7421gll0814), 16 have a PAS (Chk33), and 6 have an additional Cache (a signaling domain common to calcium-channel subunits and chemotaxis receptors [4]) upstream from the HAMP, of which one has a GAF (Chk161, NpunNpF6040) in between the two domains and two PAS/PAC domains (Chk155 and Chk177). Cache is a signaling domain that is found in animal calcium channel subunits and a certain class of prokaryotic chemotaxis receptors. It is thought to form an extracellular or periplasmic ligand sensor (4). All of these proteins originate from filamentous N2-fixing strains, and four were found in the endosymbiosis-forming species N. punctiforme.
Synechocystis sp. strain PCC 6803 Chk10 (Hik10) has been reported to be involved in the response to hyperosmotic stress, forming a pair with the response regulator Crr3 (Rre3) (105). No function has been described for its orthologues. Another HKIII (Chk33), which possesses a PAS domain, would also be involved in this stress response. Remarkably, orthologues of this Hik33 protein exist in all of the 16 genomes, and they are the only examples of cyanobacterial proteins with such architecture. Other bacterial orthologues (without any function yet defined) are at present restricted to the Firmicutes (gram-positive bacteria). This protein (termed DspA or Hik33 in Synechocystis sp. strain PCC 6803 and NblS in S. elongatus) has been reported to sense many environmental cues: cold, osmotic changes, high light, and nutrient limitations (56, 80, 87, 90). Since it is present even in the strains that have only a small number of two-component systems, it likely plays a key role in cyanobacteria by integrating cellular metabolism with environmental parameters. It also has homologues in the plastid genomes of the red algae Porphyra purpurea, Gracilaria tenuistipitata, and Cyanidium caldarium and was termed Ycf26. The cyanobacterial sequences, as well as those from G. tenuistipitata and P. purpurea, have a unique putative periplasmic signaling domain that has not been detected in any other protein (90).
HKIV.
The HK class IV (HKIV) polypeptides have an N-terminal S/T kinase domain and a C-terminal histidine kinase domain, with GAF domains in between. They are restricted to species belonging to the Nostocales family, i.e., filamentous heterocystous N2-fixing strains (11 to 13 each), with the exception of T. erythraeum, which has one. These proteins are quite interesting, as they are able to directly couple Ser/Thr kinase activities and transduction pathways involving two-component systems. One of them, HstK (Alr2258, Chk99) from Anabaena sp. strain PCC 7120, has been characterized; its expression depends on the type of nitrogen source that is available (109). Anabaena sp. strain PCC 7120 Alr0709 (Chk162) and Alr0710 (Chk107) are very large proteins (1,799 and 1,796 aa, respectively) which have the same modular organization and are adjacent on the chromosome; they align all along their length, with only one gap (10 aa) in the middle. They are the closest paralogues, with 63% identity and 74% similarity. The same physical organization exists for Avar_400222710 (Chk165) and Avar_400222720 (Chk101), and the two proteins are 61% identical. Only Chk101 however, has an orthologue in Anabaena sp. strain PCC 7120, which is neither Chk165 nor Chk107. Gene duplications thus probably occurred rather recently, i.e., after their divergence. Four HKIVs have a second GAF domain, and one protein from Anabaena sp. strain PCC 7120, one from A. variabilis, and two from N. punctiforme have PAS and/or PAC domains in between the GAF and the HK. They all are about 2,000 residues or more. The physiological functions of these proteins should be looked at closely to determine the role of each kinase and whether they act independently or synergistically, or if these proteins are nodes receiving signals from two different transduction pathways to achieve a single output function.
HKV.
In the last class, HK class V (HKV), there are 37 multidomain proteins, corresponding to the combination of different types of domains linked to a histidine kinase. One group of orthologous proteins (Chk8) has a representative in all genomes but G. violaceus. The S. elongatus 7942 (SasA) and Synechocystis sp. strain PCC 6803 orthologues have been characterized. They are clock-associated histidine kinases, necessary for the robustness of the circadian rhythm of gene expression, and have been implicated in clock output (57, 61) as well as in heterotrophic carbohydrate metabolism when cells are grown in light-dark cycles (127). The protein has been crystallized from Synechocystis sp. strain PCC 6803, and its structure has been determined to 1.9-Å resolution. It forms an open tetramer (52). Its cognate response regulator, tentatively named SasR, awaits identification. Another group (Chk178, Chy178) contains a protein from G. violaceus that associates with CheB, CheR, PAS, and HK domains, the Anabaena sp. strain PCC 7120 and A. variabilis orthologues being hybrid kinases (HYII) with an additional C-terminal RR. Within this subclass, which contains proteins with cNMP-binding domains, the Chk110 group gathers orthologues originating from quite distant strains: G. violaceus, presumed to be at the root of the cyanobacterial lineage, and N. punctiforme, which is the cyanobacterium with the largest genome (among the characterized ones) and the more complex ecophysiology.
Response Regulators
RRI.
The RR class I (RRI)-CheY class groups proteins with an RR domain within a polypeptide less than 200 aa long; 121 cyanobacterial ORFs that do not have any additional recognizable domains have been found. There are 30 groups containing orthologous genes, of which only 10 have more than three ORFs. Among the unicellular marine strains, Prochlorococcus sp. strain MIT9313 and Synechococcus sp. strains 9605 and 9902 are the only species to have such a protein, the three being orthologues (Crr48).
A few of these proteins have known functions. PilH (Crr7, Rre7, taxAY3) is required for motility in Synechocystis sp. strain PCC 6803 (151) and is also found in T. elongatus and the five N2-fixing species. Another RRI-CheY, PisH or PixH (Crr35, Rre35), is required for positive phototactic movement (152). Orthologues exist only in the three filamentous heterocystous strains. Rcp1 (Crr27, Rre27) is the cognate response regulator for the phytochrome Cph1 (Chk35, Hik35 [150]). Orthologues are found only in the strains that possess such an HKII phytochrome-like protein (Chk35), and they are always adjacent to and downstream from the corresponding gene. Anabaena sp. strain PCC 7120 DevR (Alr0442, Crr42) makes with HepK (All4496, Chk86) the first two-component system identified that regulates the biosynthesis of a polysaccharide as part of a patterned differentiation process (154). Orthologues can be found not only in the other N2-fixing strains but also in S. elongatus 7942 and Synechocystis sp. strain PCC 6803. In the latter, the Crr42 orthologue is annotated as DivK, a cell division response regulator, but on bases which have not been explicated; it is 66% identical and 79% similar to DevR. All of the Crr42 orthologues are adjacent to and divergently transcribed from genes which also are orthologues and potentially encode subunit A of DNA gyrase/topoisomerase IV. Since heterocysts do not divide, it may be that the phenotype observed for the devR mutant results from global regulation involving chromosome structure.
About 80% of the small RRI-CheY domains are less than 150 aa long. The absence of any identifiable output domain raises the question of their mode of action. Each of these probably interacts with not more than one partner besides its cognate kinase. A phosphorylated (P-RR) and a nonphosphorylated (RR) form would be in equilibrium, probably differing by their conformation. Under specific conditions, the cognate kinase will provide a phosphate (P) to form P-RR that could then establish specific interactions with a partner of which it regulates the activity, either positively or negatively. In E. coli after autophosphorylation of the CheA histidine kinase, the phosphoryl group is transferred to the CheY, an RR which then interacts with flagellar motor proteins (22, 145). Rhizobium meliloti, which does not possess CheZ, has two cognate CheYs (∼120 aa long) that interact with CheA: phospho-CheY2 (CheY2-P) is the chief regulator of flagellar rotation, its action being modulated by CheY1, which functions as a phosphatase of CheY2-P and becomes a sink for phosphate (129). A similar process may occur in Rhodobacter sphaeroides, which has two classic and two atypical CheA proteins and eight associated response regulators (six CheY proteins and two CheB proteins [111, 112]), as well as in cyanobacteria, which also do not have any CheZ homologues but possess a large number of “CheY”-like proteins. It will be of interest to determine whether the expression levels of the cyanobacterial genes and/or protein levels change upon alterations in the environment, as well as to look for a specific intracellular location of the gene products, if any.
The same basic RR-CheY domain also occurs in ORFs of more than 200 residues, usually about 400 aa long, with no characteristic associated domains. They have been classified as RRI PatA, because one such protein from Anabaena sp. strain PCC 7120, All0521 (Crr65), was the first of this group to have been characterized. Its name comes from the phenotype of the corresponding mutant, which is impaired in the pattern formation of the heterocysts (73). Another protein belonging to that class has been studied, Sll0038 (Rre36, Crr36), which is part of the pathway for perception and transduction of low temperature signals and might specifically regulate the expression of the desB gene in Synechocystis sp. strain PCC 6803 (135). Crr36 orthologues exist in the three filamentous heterocystous strains.
Another subclass, RRI-other (RRVI in Ohmori's nomenclature [102]), also contains a single RR domain in a polypeptide more than 200 aa long, with no other (as yet) identifiable domain but low overall sequence identity with PatA-type ORFs. This subclass mostly consists of a group of orthologues, Crr23 (Rre23, previously named Ycf55). Orthologues exist in all strains but G. violaceus; the marine unicellular non-N2-fixing strains, as well as T. elongatus and S. elongatus, however, do not presently have a canonical RR domain. They no longer exhibit in their N-terminal sequences the critical D and K residues which make recognizable RRs. They have nevertheless been kept in Fig. 2 and 3 because PBLAST searches performed with this domain, although less conserved than the C-terminal part, still pick the RR domains of the other orthologues. No function has yet been assigned to this probably very ancient and well conserved protein, present only in photosynthetic organisms.
RRII.
RR class II (RRII) proteins contain the more “classical” RRs in that they correspond to the structure of the first described response regulators, all being two-component DNA-binding response regulators. They have an N-terminal RR domain fused to an output DNA-binding domain, either a T_reg (for OmpR type [81]), HTH_LuxR (or Ger_E for LuxR/NarL type), or AraC. Thus, they probably function as transcriptional regulators. Examples of these RRs are found in all species of cyanobacteria, with the number of OmpR types (4 to 19, depending on the species) outnumbering (141 versus 89) the NarL types (1 to 16). Almost all of the RR repertoire found in unicellular non-N2-fixing strains belongs to this class, the rest (at most two proteins) being RRIs.
(i) OmpR-type subclass (T_reg output domain).
Within the OmpR-type subclass there are three groups of 16 orthologues and one in NarL. Two of them, Synechocystis sp. strain PCC 6803 RpaA (Crr31, Rre31) and RpaB (Crr26, Rre26, Ycf27), have been linked to long-term regulation of energy distribution by phycobilisomes (12). RpaA would also be a partner of Hik33 (also termed DspA or Chk33), and Ycf26 orthologues are present in all strains (see above). Synechocystis sp. strain PCC 6803 Sll0649 (Crr3 or Rre3), which has five orthologues, would pair with Hik10 (Slr0533 or Chk10), which also has orthologues in the same five strains (see Table S4s in the supplemental material). These two pairs are involved in the response of Synechocystis sp. strain PCC 6803 to hyperosmotic stress (105, 126). Interestingly, the Chk10 HKIII is adjacent to and downstream of Crr3 in all strains but Synechocystis sp. strain PCC 6803. In contrast, the Crr31 response regulators and Chk33 kinases are never adjacent in any of the species. For the third group of 16 proteins (Crr37), none of the RRs is adjacent to a histidine kinase and all of the corresponding genes except G. violaceus glr2274 are monocistronic transcriptional units, the adjacent genes being divergently transcribed on both sides. Expression of the Anabaena sp. strain PCC 7120 representative (all4312) is directly controlled by the global nitrogen regulator NtcA, suggesting that Crr37 might be related to cellular responses to nitrogen deprivation. The fourth one is Crr1 (Ycf29), which also has orthologues in algal plastid genomes (see below).
SphR/PhoB (Crr29) is the partner of the histidine kinase SphS (Chk7), which regulates the pho regulon in the signaling pathway of phosphate limitation (see above) (1, 136). Orthologues are distributed as for SphS, but they do not form an operon with the Chk7 proteins. Another group (Crr28) is made up of 12 sequences, no representative existing in T. elongatus and the Prochlorococcus spp. except MIT9313. No function is known for any of these, the only information being that in Synechocystis sp. strain PCC 6803, a Kdp kinase (Slr1731, Ctc1) might transfer a phosphate to 6803sll0396 (Crr28).
ManR (Crr16, Rre16) regulates manganese homeostasis in Synechocystis sp. strain PCC 6803 together with the HKI ManS (Chk27, Hik27) (100, 148). ManR orthologues exist in all of the strains that possess ManS, but they are never adjacent to their putative cognate kinases. NblR (7942_403113030, Crr73) has been described as an NblS partner that regulates expression of NblA, a protein required for the degradation of phycobilisomes under stress conditions in S. elongatus, but its precise cognate kinase awaits identification (125). Crr73 orthologues with more than 60% identity are found only in the N2-fixing species and in T. elongatus. Another group consists of seven sequences, Crr71, that originate from each of the unicellular marine non-N2-fixing strains plus S. elongatus 7942. RppA (Sll0797, NrsR, Crr33) is the RppB (Sll0798, NrsS, Chk30) partner and is located upstream from it on the Synechocystis sp. strain PCC 6803 genome. This pair was first found to be involved with redox control of photosynthesis and pigment-related genes (71) and more recently in nickel sensing (76). No orthologue was found, though Chk30 proteins seem to also exist in Anabaena sp. strain PCC 7120 and A. variabilis. For these two strains, however, no RR is adjacent.
(ii) NarL subclass (LuxR output domain).
Relatively few of the NarL-type RRs (14) have assigned functions. Ycf29 (Crr1) is the only one found in all 16 sequences (Fig. 2 and 3). As mentioned above, the Slr1783 protein (Rre1) would be the partner of Hik2 and Hik34 in the response of Synechocystis sp. strain PCC 6803 to hyperosmotic stress (105, 126). In this strain, crr1 may be an essential gene, as no group has reported segregated interposon mutants (V. Zinchenko, CyanoMutants, at http://www.kazusa.or.jp/cyano/; N. Burnett, personal communication). Copies of this gene are also found on the plastid genomes of the red algae Guillardia theta, Porphyra purpurea, Cyanophora paradoxa, Cyanidioschyzon merolae, Gracilaria tenuistipitata, and Cyanidium caldarium.
In Anabaena sp. strain PCC 7120, the RRII-NarL OrrA (Alr3768, Crr81) has been found to be involved with the response to osmotic stress (124). It is not an orthologue of either of the two proteins, Crr3 and Crr31, identified for similar stress responses in Synechocystis sp. strain PCC 6803, but it has orthologues in the other two filamentous heterocystous species.
(iii) AraC subclass (AraC output domain).
The last RRII group has an RR domain fused to HTH-AraC domains which, as a pair, form the DNA-binding domain of the AraC family of response regulators (139). In general, AraC transcriptional regulators are classified as having any receiver domain fused to the HTH_AraC domains. Only nine cyanobacterial sequences were found to have an RR fused to AraC. As usually observed for the sequences belonging to this family, the HTH motif is situated toward the C terminus. The three-dimensional structure of such a protein, E. coli MarA, has been solved. It showed that the two HTH_AraC subdomains are separated by 27 Å, which causes the cognate DNA to bend. There is a single such gene in Synechocystis sp. strain PCC 6803 and A. variabilis, two in Anabaena sp. strain PCC 7120, five in N. punctiforme (one plasmid encoded), and only one group (Crr90) with orthologues in the three filamentous heterocystous strains.
RRIII.
Some cyanobacterial response regulators have two or even three RR domains, together with Treg and Hpt (for “histidine phosphotransfer”), and in one group GGDEF domains. The ORFs that have one RR upstream and two downstream of the T_reg-Hpt domains are from the heterocystous N2 fixers, with one from G. violaceus (Crr93). They presumably function as conditional transcriptional regulators via phosphotransfer relays. Hpt domains are known to interact with more than one RR domain and are thus particularly well suited for cross-talks. The recently demonstrated coordination of synthesis and proteolysis of RpoS in E. coli by the two-component phosphotransfer network that involves ArcB, ArcA, and RssB is a good example (86). RcaC from F. displosiphon has a domain organization similar to that of Crr93. This protein has been described as involved in complementary chromatic adaptation (30). Both the N-terminal RR and Hpt domains were found to be important for the light-regulated control of phycocyanin gene expression, whereas the C-terminal RR only had a minor role (72).
RRIV.
The vast majority of the proteins in RR class IV (RRIV) do not have any DNA-binding domains, but a number have output domains with putative catalytic activities. More than 40% of these polypeptides possess a GGDEF domain, also named DUF1. This domain was first recognized in Caulobacter crescentus PleD, a response regulator controlling cell differentiation, before being found in proteins involved in cellulose biosynthesis, cell adhesion, or aggregation (119). It is highly “promiscuous,” as it is found associated as a module with a multitude of different domains. It has recently been demonstrated that PleD possesses catalytic guanylate cyclase activity (107). Expression of recombinant GGDEF domains from ORFs found in six very different bacteria (including the Synechocystis sp. strain PCC 6803 Slr1143, a GAF-GGDEF protein) demonstrated that (i) they all possess diguanylate cyclase activity and (ii) for Borrelia burgdorferi Rrp1 (a RR-GGDEF protein), phosphorylation of the RR is required for activity of the GGDEF domain (120). Thus, the GGDEF domains will represent the output of complex bacterial signal transduction networks, which convert different signals into the production of a secondary messenger, cyclic diguanylic acid (c-di-GMP). The cyclase activity correlates well with the correspondence between GGDEF and the catalytic domain of adenylate cyclases (40, 108). GGDEF domains can be found associated with an EAL domain (also known as DUF2), which is a good candidate for a diguanylate phosphodiesterase function (40). The corresponding proteins would then have opposing cyclase and hydrolase activities (107). Cyclic diguanylate-specific phosphodiesterase activity has recently been demonstrated from an overexpressed E. coli ORF containing an EAL domain (122). Some cyanobacterial RRs exhibit this kind of association, eventually with additional PAS and/or PAC domains, but most of them have only one of these two domains. It is worth mentioning, however, that although Synechocystis sp. strain PCC 6803 has one such protein (Crr41), the strain possesses very little, if any, c-di-GMP, at least under standard conditions (J. Houmard, unpublished data). Synechocystis sp. strain PCC 6803 Crr4 has both a GAF (Phyt_2) and GGDEF domain fused to an RR. RRs with a GGDEF domain are found in all cyanobacterial species except the open-ocean non-N2 fixers. In two instances, multiple N-terminal RRs are associated with a GGDEF domain, one (7942_403091170) also having a DNA-binding Treg domain.
There are six examples of RRs with an HD (for “phosphohydrolase activity”) output domain. The latter is found in enzymes such as cyclic nucleotide phosphodiesterase, 2′-nucleotidase, and phosphatase (8, 147). A knockout of the Synechocystis sp. strain PCC 6803 slr2100 gene (Crr20) indeed results in changes in the intracellular cyclic nucleotide (cGMP) concentrations and in an increased sensitivity of the cells to UV-B radiation (24). This protein is thus involved in cGMP homeostasis and light signaling. The other five RR-HD proteins form an orthology group. A Synechocystis sp. strain PCC 6803 crr18 (sll1624) null mutant has also been constructed and did not exhibit a phenotype similar to the crr20 mutant (24). T. erythraeum is the only diazotroph which does not possess such a protein, but it is also the only one to have an RR-GuC, which thus probably has purine nucleotide cyclase activity (discussed below).
The protein phosphatase 2C-like domain (PP2C, also referred to as SpoIIE) is found in PP2C and adenylate cyclase and in SpoIIE, which is known for its role in sporulation in Bacillus subtilis (17). Some of these proteins may have a role in cell division or differentiation. A PP2C domain is found as a C-terminal fusion to an RR in all filamentous species and T. elongatus but not in C. watsonii. This distribution closely resembles that observed for the HKIVs, which have S/T kinase domains. For one orthology group (Crr100), there is an additional GAF domain associated. Finally, there are examples of N-terminal RRs fused to GAF, PAS, cNMP, CheC, CheW, CheB, Pyr_red, or IF2 domains. Many of these ORFs are found in only one species and may result from recent fusions of domains.
Hybrid Kinases
The important feature to notice for this group of complex multidomain proteins is their complete absence from the open-ocean non-N2-fixing species. The nomenclature of each subclass is based firstly on the position of the RR domain relative to the HisKA-HATPase domains. HY class I (HYI) groups have ORFs with a single RR N terminal to the HK, and HYII groups are those with a single RR C terminal to the HK. HYIIIs correspond to those ORFs with either two or three RRs C terminal to a single HisKA-HATPase. An HYIV-type protein has a single HK with at least one RR on each side. HYVs have two HKs with one RR in between; HYVI groups are the ORFs that have HATP (a domain found in several ATP-binding proteins), CheW, and RR domains with additional Hpt and/or Hkd domains; and HYVIIs are incomplete hybrids with either HisKA or HATPase domains that may be linked to additional modules.
HYI.
HYI-type proteins are totally absent from S. elongatus and T. elongatus. About half of the “orthology” groups consists of a single protein. Only one HYI has a known physiological function, 6803sll1229 (Hik41, Chy41). It has been found to respond to salt (NaCl) stress, together with Synechocystis sp. strain PCC 6803 Hik16 (Chk16) (80). Synechocystis sp. strain PCC 6803 has five “simple” HYIs, of which three (Chy38, Chy40, and Chy41) have in some strains another hybrid kinase immediately upstream and of which one (Chy23) has an HK-RR pair (AphA-Rcp1). Thus, they may belong to multiphosphorelay systems, although the colocalization of the genes involved in phosphorelays is not a prerequisite. Indeed, although Chy41 would be part of such a relay for the response of Synechocystis sp. strain PCC 6803 to hyperosmotic stress, its partners Chk16 and Crr17 are not encoded by adjacent genes (105, 126). Similarly, the Anabaena sp. strain PCC 7120 genes for AphC and CyaC, between which phosphotransfer has been evidenced (see below), are not closely localized. Some HYIs have a variable number of PAS and PAC domains in between the RR and the HK plus, for a few of them, one or two GAFs. No function has yet been assigned to any of these ORFs.
For some HYIs, an HWE (HisKA_2) histidine kinase domain substitutes for the HisKA. Members of this family differ from most other HKs by lacking a recognizable F box and the presence of uniquely conserved residues: a His in the N box and the sequence WE in the G1 (64). Though found in many different species, such proteins are not as widely distributed as HisKA. They are particularly abundant in the Rhizobiaceae family. HWE domains were previously not detected in cyanobacteria, but the present analysis shows that each of the heterocystous species has one. Anabaena sp. strain PCC 7120 and A. variabilis each have a very large HWE kinase (Chy58, ∼1,700 aa long), which also has GAF and PAS-PAC domains. One N. punctiforme HYI (Chy109, NpF1799) has an HKA_3 kinase domain, another HisKA alternative.
HYII.
Only 11 of the 97 HYIIs do not have additional domains. For the others, various associations involving 14 different structures exist, a large number of these ORFs having PAS, PAC, and/or GAF domains. About one-fifth of the HYII “orthology” groups have HK orthologues with similar structural organization but without the C-terminal RR. For example, 7120all1716 and Avar_400180300 (Chy178) are orthologues of HKIII-CheR/B 7421gll1854. 7120all0978, Avar_400180780, and NpunNpF5679 (Chy179) are orthologues of the fairly similar G. violaceus HKV-HTH_4 (Chk179, Gll3736).
S. elongatus HYII-GAF (Chy24) corresponds to CikA, a bacteriophytochrome that resets the circadian clock (123). No orthologue exists in G. violaceus or in the genomes of the marine non-N2 fixers, and the structure differs between the strains. For T. elongatus and the three filamentous heterocystous strains, it is HKII-GAF(PHYT_2) (Chk24) without any RR. A detailed characterization of S. elongatus 7942 CikA showed that (i) it can covalently bind bilin chromophores in vitro, even though it lacks the expected ligand residues (it may not serve, however, as a photoreceptor itself); (ii) deletion of the GAF domain or the N-terminal region adjacent to GAF dramatically reduced autophosphorylation of the HK domain, whereas elimination of the receiver domain increased activity by 10-fold; and (iii) the RR domain, which lacks the conserved aspartyl residue that serves as a phosphoryl acceptor in response regulators, would not work as bona fide receiver domain in a phosphorelay but could interact with an unknown protein partner to modulate the autokinase activity of CikA (92). In CikA, both the GAF and RR noncanonical modules would act as protein-proteininteraction domains that induce conformational changes in another domain to modulate its activity.
There is one subclass that contains only four sequences, all from T. erythraeum. All of these ORFs have a C-terminal GuC domain and thus likely possess a purine nucleotide cyclase activity. Though the presence of multiple nucleotide cyclases (AC/GC) has already been reported for cyanobacteria (see, for example, references 67 and 99), the different proteins were usually made of different domain arrangements. T. erythraeum has by far the highest number of such enzymes (13, compared to 5 or 6 for the heterocystous strains). Among the four HY-GuC proteins, three have the requirements for being adenylyl cyclases (Chy145, plus Chy129 and Chy130, which are adjacent on the chromosome), the fourth one (Chy131) having those for a guanylyl cyclase (99).
HYIII.
Thirty-one ORFs differ from the previous hybrid kinases by having at least two C-terminal RRs in tandem. N. punctiforme NpR2263 is the only one that does not posses any additional domains, almost all having either PAS and/or PAC or GAFs. One member of this subclass, Anabaena sp. strain PCC 7120 Alr2279 (Chy133), has an additional N-terminal HNOBA domain (not identified by Pfam). The HNOBA domain could potentially contain a PAS-like fold. A homologous domain is also found in the first 200 aa of the N. punctiforme NpR4835 (Chk50 [58]). The two other Chk50s do not have it. HNOBA domains functionally interact with HNOB (for “heme, no binding”) domains located on a second protein. The HNOB domain is predicted to function as a heme-dependent sensor for gaseous ligands (NO, CO, or possibly O2). Proteins carrying such domains (7120alr2278 and its orthologue NpunNpR4836) are encoded by the upstream genes in the two cyanobacterial examples. As stated by Iyer et al. (58), the co-occurrence of the HNOB and HNOBA domains in either the same protein or proteins encoded by the same operon suggests a strong functional interaction between them. The potential role, if any, of NO in cyanobacteria deserves further studies.
About one-third (13/31) of the “orthology” groups have only one representative, and another third have orthologues but with a different domain structure. Synechocystis sp. strain PCC 6803 Chy21 is of particular interest. It is, at present, the only cyanobacterial two-component protein that has an MHYT domain, a newly identified conserved protein domain with a likely signaling function (39). A model of the membrane topology of the MHYT domain indicates that its conserved residues could coordinate one or two copper ions, suggesting a role in sensing oxygen, CO, or NO. This protein is just upstream from and cotranscribed with the Chk40 HYI, which is followed by the RRIV-HD Crr20, a protein involved in cGMP homeostasis and UV-B response (see above) (24). This cluster, to which Chy22 (HYII-GAFPAC-Hpt) is very close, could thus form a large multiphosphorelay system sensing changes in the environmental parameters and involving cGMP as a second messenger (98). Cyclic nucleotide concentrations have already been shown to vary in some cyanobacteria upon oxic-anoxic transitions, for example (reviewed in reference 26).
HYIV.
The HYIV hybrid kinases have RRs on both sides of the kinase domain. All but one of the orthology groups have additional sensing domains: PAS (plus PAC for most of them) and/or GAF. One group, Chy90, shows sequences with a histidine kinase and four RRs and up to seven different types of associated domains. G. violaceus Chy90 is about 1,000 residues less than the three others and is annotated as a (PAS/PAC)2-HK-RR HYII. However, a 611-aa-long RR [Glr4211, RR-Treg-(RR)2] is immediately upstream, the A of its stop codon also being used as the first base for the ATG of Glr4212 (Chy90). A careful analysis of the sequence would be required to ascertain that there was no sequencing frameshift error. The four proteins have an adjacent RR downstream, which is also orthologous. The cluster organization would thus have been conserved through evolution from G. violaceus to the filamentous heterocystous strains.
Synechocystis sp. strain PCC 6803 Chy19 (Hik19) has been found to be an essential gene (based on no complete segregation of the mutation) involved in the transduction of low-temperature signals (135). It might function downstream from Chk33 (DspA), transducing the low-temperature signal by phosphorylating Crr36 (PixG), which in turn controls desB gene expression.
The HYIV+GAF-GuC Chy89 proteins, known as CyaC, are orthologues present in every filamentous strain, whether an N2 fixer or not, as it is also present in Spirulina platensis (66). An orthologue has also been found in Tolypothrix sp. strain PCC 7601, also known as Calothrix sp. strain PCC 7601 or Fremylla diplosiphon (L. Jia and J. Houmard, unpublished data). Kasahara and Ohmori (65), studying CyaC from S. platensis, demonstrated that the HK domain will autophosphorylate and will transfer the phosphate to the adjacent C-terminal RR domain, whereas the N-terminal RR domain, separated from the HK domains by two GAF domains, was not phosphorylated by it. Replacement of the conserved aspartate residue by alanine in the N-terminal RR did not affect the activation of cyclase activity in vitro. S. platensis CyaC has been crystallized, and the mechanism of bicarbonate activation has been studied (130). CyaC is one of the six AC/GC purine nucleotide cyclases found in Anabaena sp. strain PCC 7120 (101). Because a cyaC mutant has a very low cAMP level, it has been proposed to be responsible for the maintenance of the steady-state level of cellular cAMP. On the other hand, it has been demonstrated that in Anabaena sp. strain PCC 7120 the phytochrome-like AphC (Chk65) mediates the increase in cAMP concentration induced by far-red light. Okamoto et al. (104) have proposed a model in which far-red light illumination provokes the autophosphorylation of AphC, followed by a phosphotransfer to the N-terminal RR domain of CyaC (Chy89). The HK domain of CyaC will then autophosphorylate, the phosphate will be transferred to the downstream RR, and the catalytic activity domain will in turn be activated. The cAMP produced could then, through binding to CRP-like proteins, regulate different adaptation processes. This is one of the very first examples of a signal transduction mechanism involving a two-component system phosphorelay described for cyanobacteria.
HYV.
A few hybrid kinases possess two complete HK and one or two RR domains. None has any known or putative function yet. They also have either PAS or PAC domains. N. punctiforme Chy139 (NpF2346) has a UPF/RHH_2-type N-terminal domain, described for a few 80-aa-long hypothetical proteins, members of the MetJ/Arc repressor superfamily clan of unknown function.
HYVI.
Another group of 25 ORFs all have an additional CheW domain, between the HATP and the RR, as well as an Hkd and/or Hpt domain upstream of those. They would thus be involved in chemotaxis signaling mechanisms. The Synechocystis sp. strain PCC 6803 Chy18 (PixL, TaxAY1, Hik18) and Chy39 (TaxAY2, Hik39) proteins have been shown by analysis of the phenotype of the corresponding mutants to regulate phototaxis (20). Chy43 could represent the C-terminal part of a CheA-like protein which is required for motility, transformation competency, and the assembly of thick pili, the N-terminal Hpt domain of this CheA-like protein being separately encoded by pilN (Hik36 and Ctc36, an orphan Hpt protein). Though sharing a very similar organization and gene repertoire with the tax1 cluster (slr0038 to slr0043), the tax2 cluster (sll1291 to sll1296) would not be involved in motility (19). It is worth noting that each of the three Synechocystis sp. strain PCC 6803 TaxAY (Chy18, Chy39 and Chy43) proteins would be connected to two different (adjacent on the chromosome) RRs, an RRI-CheY and an RRI-PatA: Chy18 working with Crr36 (Sll0038) and Crr35 (Sll0039), Chy39 with Crr12 (Sll1291) and Crr11 (Sll1292), and Chy43 with Crr6 (Slr1041) and Crr7 (Slr1042).
HYVII.
The last class, HYVII, groups putative hybrid proteins with RRs but which only have either a HisKA or an HATPase domain. Synechocystis sp. strain PCC 6803 Chy180 (Rre22) consists of an N-terminal HisKA domain with an RR domain and a PP2C-like (PP2C_SIG) domain downstream. Although the gene name ppcE was assigned to Chy180, no description of its function could be retrieved. Such an acronym exists for probable peptidases. C. watsonii has two paralogous HATP-RRs, one of which could be a complete HYI if the putative sequencing error does exist. None has any known function.
Other Two-Component Related ORFs
There were only five examples of an orphan Hpt domain detected. The Hpt domain, more frequently found in histidine kinases and hybrid sensors, is known to act as a phosphoacceptor and phosphodonor in phosphorelays from one RR domain to another (132). As mentioned above, Hpt domains, because they interact with more than one RR domain, are especially well suited for cross-talks (86). The orphan Hpt YPD1 from Saccharomyces cerevisiae is known to greatly increase the half-life of its phosphorylated cognate response regulator (59). This could apply for the function of Ctc36 (PilN) in Synechocystis sp. strain PCC 6803 phototaxis (see above). For two Anabaena sp. strain PCC 7120 ORFs, mentioned by Ohmori et al. (102), Alr4086 and All8565, we could not find any identifiable Hpt domain by InterProScan, SMART, or PBLAST (searches performed at NCBI) (2).
KdpD proteins form a different family of histidine kinases. In E. coli, the KdpD domain senses turgor pressure and Usp forms the output domain (144, 153). It phosphorylates and interacts with its cognate (RRII-OmpR type) RR, KdpE (46). There are three examples in A. variabilis, two of which are encoded by plasmid B, and single examples in the other heterocystous strains, as well as in Synechocystis sp. strain PCC 6803, S. elongatus, and G. violaceus. The G. violaceus copy is interesting, as it has a tandem duplication of the two domains. Ballal et al. (15) have demonstrated an interaction between the N-terminal TM domains of Anabaena sp. strain L-31 with E. coli KdpD, which alters the phosphatase activity. KdpD (Ctc1) is also mentioned, probably on the basis of two-hybrid experiments, as the cognate phosphodonor for the RRII-OmpR Crr28 (Rre28) (http://www.genome.ad.jp/dbget-bin/show_pathway?syn02020+slr1731).
ORTHOLOGOUS GROUPS
Many examples of highly versatile permutations and combinations of a number of conserved modules have been found. Fusions to various domains increase the versatility of a protein family and allow its recruitment into various cellular regulatory pathways. It has been reported that multidomain proteins have significantly less functional conservation than single-domain ones, except when they share the exact same combination of domain folds. In addition, for two proteins containing the same combination of two structural superfamilies the probability of them sharing the same function increases to 80%, and even up to >90% in the case of complete coverage along the full length of both proteins (47). Sensory domains, which are highly represented in multidomain proteins, have also been shown to evolve faster than other domains (receiver, transmitter, and output domains in the case of two-component proteins [146]). Finally, domain insertion may occur without affecting the function of a protein.
“Orthology” groups were thus defined that are based on the bidirectional best hits from BLAST searches of each organism against each other organism, completed by phylogenetic analyses. A tentative gene nomenclature is also proposed in Table 1 and in Table S3s in the supplemental material. Direct comparisons between ORFs will be restricted to the orthologues, the time of divergence being assumed to be the same, i.e., that of speciation. Sixteen proteins, plus six groups of two or more, do not have any orthologue in any other sequenced bacterial genomes. In contrast, for the cyanobacterial Chy83 proteins (which are 1,000 to 2,000 residues long), orthologous proteins of more than 1,000 amino acids can be found in some 22 bacterial genomes with BLAST E values of 0.0.
HKs and RRs Common to All Genomes
All of the sequenced genomes would encode a Chk33 protein (nblS, dspA, or ycf26). As mentioned above, this multidomain protein probably acts as a “hub” connecting various environmental signals to their specific signal transduction pathways. A Chk2 protein would also be present in all cyanobacteria, though with a different modular organization. It is either a GAF-HK or a “basic” HK without GAF for the unicellular marine nondiazotrophic strains and G. violaceus. The Chk34 orthologues which might be involved in salt stress (see above) have only a HisKA domain in all species except the G. violaceus representative, in which it is a complete histidine kinase. For them, orthologues do not exist in any of the other fully sequenced bacterial genomes. Quite interestingly, this Chk34 protein was also shown to be essential for thermotolerance in Synechocystis sp. strain PCC 6803, possibly by negatively regulating the expression of certain heat shock genes (134). It is thus likely a quite important protein for cyanobacteria. S. elongatus 7942 Chk8 (SasA) has orthologues in all genomes but G. violaceus, but without cognate response regulators yet identified. This protein is implicated in the circadian rhythm clock output (57, 61). It will be of interest to check whether or not G. violaceus exhibits a circadian rhythm. Four classes of RRs are made of orthologues from all genomes: Crr1 (Ycf29), Crr26 (RpaB, Ycf27), Crr31 (RpaA), and Crr37. These proteins must perform key roles in regulating important processes in cyanobacteria, particularly as they are retained in the “streamlined genomes” of Prochlorococcus species. Crr23 (Ycf55) occurs in all species but G. violaceus.
As the acronyms indicate, Ycf26, Ycf27, Ycf29, and Ycf55 orthologues are also found in the plastid genomes of red algae and/or diatoms, suggesting that the corresponding genes are very ancient and should have already existed in the cyanobacterial ancestor who gave rise to the plastids (13, 45). Most two-component genes are not essential to the growth of cyanobacteria under standard laboratory conditions and can be inactivated. Fully segregated Synechocystis sp. strain PCC 6803 knockout mutants could not, however, be easily obtained for chk33 (ycf26), crr1 (ycf29), crr23 (ycf55), crr26 (ycf27), and crr37, further supporting the key roles of the corresponding gene products (12, 90, 135, 142; N. Burnett, unpublished data). A similar result was obtained for crr37 in Anabaena sp. strain PCC 7120 (91).
As mentioned above, in Synechocystis sp. strain PCC 6803, RpaA (Crr31) would be a partner of Chk33 (DspA/NblS) in the response of the cells to hyperosmotic stress (105, 126). The pair will thus have been highly conserved throughout evolution. RpaA (Crr31) and RpaB (Crr26) are the closest paralogues, and both of them are about 41% identical and 61% similar to B. subtilis YycF, an RR which modulates the expression of the ftsAZ operon (36a). On the other hand, NblS (Chk33) has homology (27% identity and 48% similarity) with the last two-thirds of YycG, the cognate B. subtilis HK of YycF. Based on the observed phenotypes of the corresponding Synechocystis sp. strain PCC 6803 mutants, a role in the long-term regulation of energy distribution by phycobilisomes has been proposed for RpaA and RpaB (12). Because of their homology with B. subtilis YycF, and of the presence of orthologues in Prochlorococcus spp. that do not have phycobilisomes, the primary function of RpaA and RpaB might be more directly related to thylakoid biogenesis and/or composition, the phenotype observed by Ashby and Mullineaux (12) resulting from rearrangements of the photosynthetic complexes within the membrane. In Prochlorococcus sp. strain MED4, the expression of these two genes greatly increases upon a shift to high light (82). Their main function could thus be to tune the transfer of excitation energy to the reaction centers with the metabolic capacity or state of the cells.
A few other HKs have wide distribution: SphS (Chk7, PhoR) orthologues are present in all but T. erythraeum, Synechococcus sp. strain 9902 and the two low-light-adapted Prochlorococcus spp., but the cognate regulator SphR (Crr29) has a wider distribution, being also present in T. erythraeum and Prochlorococcus sp. strain MIT913. ManS (Chk27) is in all but the seven marine non-N2-fixing unicellular strains. The same distribution was observed for the cognate ManR (Crr16). These protein pairs are involved in phosphate and manganese homeostasis, respectively. Their absence in some strains could mean that the corresponding organisms use different regulatory systems to handle phosphate and manganese or do not experience any starvation for these oligoelements in their usual ecological niches.
HY Subclasses
Slightly more than a fifth of the HYs do not have any cyanobacterial orthologues. Twenty-one groups contain representatives from the three filamentous heterocystous strains, with no orthologues from other cyanobacterial species. Twelve groups are specific for Anabaena sp. strain PCC 7120 and A. variabilis proteins and two for A. variabilis and N. punctiforme. The three filamentous strains share a number of properties, among which are (i) the differentiation of 5 to 10% of the vegetative cells into intercalary heterocysts in the absence of combined nitrogen and (ii) the possibility of entering into a developmental cycle involving the formation of hormogonia that usually exhibit transient gliding motility (116). N. punctiforme shares with A. variabilis only the ability to differentiate into akinetes (see Table S1s in the supplemental material). The orthologues that are found only in the A. variabilis and N. punctiforme genomes might thus be involved in the control of akinete differentiation. At least some of the genes specific to N. punctiforme may be important for symbiosis and/or complementary chromatic adaptation, and those specific to A. variabilis may be related to the expression and/or functioning of the Fe and/or V nitrogenases, the alternative nitrogen-fixing systems. Indeed, each of these physiological properties is restricted to a single cyanobacterium within the species for which complete genome sequences are available.
DISTRIBUTION OF TWO-COMPONENT ORFs
To regulate their cellular functions in response to changes in environmental cues, prokaryotes use both one- and two-component systems, and it has been proposed that the former dominate (141). Cyanobacteria differ, however, as their repertoire of two-component proteins is either similar to or much larger than those of the one-component regulators, in particular for the filamentous heterocystous species (210/148 and 258/146 for Anabaena sp. strain PCC 7120 and N. punctiforme, respectively). Many paralogues that still exhibit a high degree of sequence identity may sometimes be recognized within a given species. Such proteins may have closely related functions, and their co-occurrence is probably meaningful. Each of them must, however, have a slightly different role, as recently shown for NarX and NarQ, for example (131).
The number of each subclass and class and the total number of potential two-component ORFs for each species are shown in Fig. 3. Also presented is the percentage of the protein-coding capacity of the genome that is represented by two-component ORFs, as well as the ratio of the number of hybrid kinases to histidine kinases plus response regulators. For the nine complete annotated genomes, it is apparent that the total number of two-component genes is a function of the genome size, ecophysiology, and physiological properties of the organism, as has been noted in general for the Bacteria (11, 38). Thus, about 3.5% of the coding capacity is taken up by two-component genes for the filamentous heterocystous strains. Though A. variabilis has the highest percentage (3.57%), the maximum number of two-component system proteins is found in N. punctiforme. They both are able to differentiate into functional hormogonia and akinetes, but, in addition, N. punctiforme is a type II complementary chromatic adapter and can form symbioses with plants. The seven oceanic unicellular non-N2-fixing species have a much smaller percentage of total coding capacity dedicated to two-component genes (0.6 to 0.7%) and do not have any hybrid kinases. Compared to the terrestrial strains, the marine diazotrophs also have fewer two-component genes (1.2 to 1.3%). T. elongatus and S. elongatus 7942 have similar genome sizes and devote about the same percentage of their total coding capacity to the two-component systems. G. violaceus exhibits a slightly higher percentage (1.9%). In contrast to the total number (HKs, RRs, and HYs), the number of DNA-binding RRs is rather constant whatever the strain (Fig. 4A). Thus, the differences in the physiological and adaptation properties, as well as the fine tuning of the regulatory processes, seem not to depend too much on the transcription factors.
FIG. 4.
Relative abundance of the two-component system proteins in cyanobacterial genomes. (A) Percentage of total putative two-component system proteins (HK+HY+RR) and transcriptional factors (RRII) as a function of the total coding capacity. (B) Total number of response regulators (RR+HY) versus that of histidine kinases (HK+HY). (C) Number of hybrid kinases as a function of the total coding capacity. For each genome, the total coding capacity is given in Fig. 3.
Synechocystis sp. strain PCC 6803 has, however, a somewhat anomalously large number of two-component proteins for its total coding capacity (2.55%). It also possesses an impressive repertoire of IS sequences, of which certain ones are still active. Indeed, at least three different genotypes are presently used around the world as wild-type Synechocystis sp. strain PCC 6803: the Pasteur culture collection (PCC) strain differs from the Kasuza strain (used for the complete sequencing of the genome) by the presence of a 1,174-bp IS sequence (ISY203g) between sll1473 and sll1475 (Chk32) and another one between sll1574 and sll1575 (103); the third strain (GT), a spontaneous mutant isolated as tolerant to glucose, from which the Kasuza strain derives, also has its spkA (a Ser/Thr-type protein kinase) as a split gene comprising sll1574 and sll1575, while the PCC strain does not. On the other hand, examination of the Synechocystis sp. strain PCC 6803 genome showed a group of three very similar clusters, with RRs preceding HKs, and a group of two with HYs followed by RRs. The first one is composed of Crr34-Chk31 (Sll0789 and Sll0790); the Crr33-Chk30 (Sll0797 and Sll0798) pair, located nine genes downstream; and Crr44-Chk46 (Slr6040 and Slr6041), which is encoded on the plasmid pSYSX. Crr34 is 91% identical to Crr44, and Chk31 is 87% identical to Chk46, each couple being 53% and 45% identical to Crr33 and Chk30, respectively. Interestingly, there is a putative transposase (Slr0799) situated two genes upstream from Sll0789. Both ancient and more recent transposition events might be responsible for the larger repertoire observed in this strain. Despite having nearly 800 more protein-coding genes than Synechocystis sp. strain PCC 6803, G. violaceus has slightly fewer two-component genes. Phylogenetic analyses using multiple criteria strongly suggest that G. violaceus, which was isolated from a calcareous rock, is a member of an early branching lineage. This strain (i) lacks thylakoids and has rod-shaped instead of fan-like phycobilisomes directly attached to the cytoplasmic membrane, (ii) is an obligate photoautotroph, (iii) does not exhibit gliding motility, and (iv) is not halotolerant.
Plotting the total number of kinases (HKs plus HYs) versus that of receivers (RRs plus HYs) for the 16 cyanobacteria, and considering whole polypeptides (even if some have more than one HK or RR domain), gives a linear relationship with a slope of 0.97 ± 0.02 (Fig. 4B). A quite similar relationship is obtained when the total number of HK domains is plotted against that of RRs. These data suggest that most often a specific RR pairs with each kinase. The ratio of hybrid kinases to histidine kinases plus response regulators (HY/[HK+RR]) ranges from 0.15 to 0.24, for the unicellular species, to around 0.37, for the filamentous heterocystous species, and is comparatively high for Synechocystis sp. strain PCC 6803 (0.29). It probably reflects the sophistication and flexibility of the regulatory pathways built up during evolution. Indeed, the presence of both functions (HK and RR) on a single polypeptide allows intramolecular phosphotransfer, as demonstrated for CyaC (104; also see above), and might permit the large multimodular structures to serve in more than one regulatory pathway as part of regulatory networks. Chk33 could serve as a paradigm in cyanobacteria, since it has been shown to be involved in the transduction of different signals: nutrient limitation, cold, and high light stresses (see above). Signal transduction might thus occur more easily and integrated structures could lead to more secure and finer tuning of the regulatory processes. As the physiological potentialities and the complexity of the regulatory circuits increase within a cell, the controls that are to be set up must be tighter so as to avoid unwanted cross-talks. This may in part be facilitated by the increasing proportion of hybrid kinases that are observed. Interestingly, there is again a linear relationship between the number of HYs and the total coding capacity, except for G. violaceus and the marine strains that reduce molecular dinitrogen (Fig. 4C). None of the marine strains that do not fix nitrogen have any HYs, and the two N2 fixers possess only about half or a third of the number expected from their coding capacity. Although G. violaceus has only 14 HYs, 8 of them have cyanobacterial orthologues. Eleven of the 14 HYs in C. watsonii, and 11 of the 16 in T. erythraeum, have cyanobacterial orthologues. For Anabaena sp. strain PCC 7120, the proportion is 51 of 55. This suggests that such bifunctional proteins may have evolved rather early during evolution.
The marine non-N2-fixing strains have a very limited repertoire of two-component proteins: 5 to 6 potential HKs and 7 to 11 potential RRs. As mentioned above, this correlates well with their thriving in oligotrophic marine environments that are more stable than terrestrial or freshwater environments. They also have a very compact genome, close to the theoretical lower limit for an oxyphototrophic organism (33, 34). Each pair of phylogenetically close strains has the same number of HKs and RRs, but not all fall in the same class of orthologues. Prochlorococcus sp. strain MIT9313, as well as Synechococcus sp. strains WH8102 and 9605, have, in particular, a pair of genes adjacent on the chromosome (Crr72-Chk91) that are orthologues and that are absent from the other Prochlorococcus spp. and Synechococcus sp. strain 9902. In contrast, Pro1543 (Chk129) does not have any homology with the others, or with any cyanobacterial protein, which could suggest that the gene may have arisen via a bacteriophage or horizontal gene transfer. It must be stressed, however, that a TBLASTN search performed with this sequence against all sequences (GenBank, EMBL, DDBJ, and PDB) did not permit retrieval of any similarity between its N-terminal sequence (∼250 aa) and that of another organism. The genome of the source organism, if any, has thus not yet been sequenced.
LOCALIZATION AND PHYSICAL ORGANIZATION OF TWO-COMPONENT GENES
Unlike in most bacterial species, not many of the histidine kinases and response regulators are encoded by adjacent genes in cyanobacteria, even when they form an identified cognate pair. In E. coli, B. subtilis, and Pseudomonas aeruginosa, 80% or more of the two-component system proteins are encoded by adjacent genes (29). As an example, the sphS and sphR gene are distant by some 500 kilobases on the genome of Synechocystis sp. strain PCC 6803. The orthologous genes are also not adjacent in C. watsonii, T. erythraeum, and G. violaceus, while they are adjacent in the other strains with sphR upstream from sphS.
Synechocystis sp. strain PCC 6803 has 16 clusters of two-component genes totaling 36 (39.6%) out of the 91 two-component genes, with up to four genes in close vicinity (slr2098 to slr2104) (98). Of these 16 sets, two are pairs located on plasmids, one (slr6040-slr6041, Chk46-Crr44) on the 106 kbp-long pSYSX and one (sll5059-sll5060; Chy44-Crr43) on the ∼120 kbp-long pSYSM. The Slr6001 HY is also encoded by pSYSX. No two-component proteins were found on any of the five other plasmids, but three of them are only ∼5 kbp long or less. The heterocystous strains also contain endogenous plasmids. There are probably four in A. variabilis, presently identified as separate contigs: two small ones of about 36 and 37 kbp, the ∼366 kbp-long plasmid B, and the ∼301 kbp-long plasmid C. Three pairs of adjacent two-component proteins occur on plasmid B (Chy135-Ctc1, Chk20-Crr19, and Chk112-Crr88). On plasmid C, there is a pair of response regulators (Crr63-Crr92) and a single one (Crr87). N. punctiforme has five plasmids (A to E, which are ∼355, ∼255, ∼123, ∼66, and ∼26 kbp long, respectively). Three carry two-component proteins: one pair of adjacent genes and two single ones on plasmid A (encoding pNAPF130-pNAPF131, HYI-HYII+PAS/PAC; pNPAF142, HKII+mGAF; and pNPAF075, RRII+AraC), two pairs (chk123-crr138 and chk112-crr88, the latter two being on opposite strands) and three single RR genes (crr82, an RRII-NarL, and crr120 and crr121, two RRI-CheY genes) on plasmid B, and a single HY gene on plasmid D (chy126, pNPDR038). Anabaena sp. strain PCC 7120 has six plasmids: alpha (∼408 kbp), beta (∼187 kbp), gamma (∼102 kbp), delta (∼55 kbp), epsilon (∼40 kbp), and zeta (∼5.5 kbp). Four harbor two-component proteins: a gene pair, chk112 (All7219) and crr87 (an RRII-NarL), on plasmid alpha; two pairs, chk46-crr44 and chk126-crr130 (two HKI-RRII-OmpR proteins), on plasmid beta; and two single RRII-NarL genes, crr80 and crr143, on plasmids delta and epsilon, respectively. Notably, the plasmid HATPase and RRII-NarL genes of the pairs found in the heterocystous strains, though adjacent, are encoded by opposite strands of the DNA, and they are in a conserved cluster of 19 orthologous genes with exactly the same physical organization on the three plasmids, large divergences being observed on both sides of it. The occurrence of orthologous pairs of proteins on plasmids in different strains could indicate that these plasmids had a common ancestor.
In a few instances, orthologues can be found on the chromosome for some strains and on a plasmid for others. The A. variabilis Chk20-Crr19 (HKI-RRII-NarL) pair is located on plasmid B, while its orthologues in N. punctiforme, Synechocystis sp. strain PCC 6803, and G. violaceus are on the chromosome. An additional cluster of two orthologous genes (α and β subunits of a K+-transporting ATPase) is adjacent but divergently transcribed. It is present on the four genomes, but the similarities in the physical map stop there. Similarly, the orthologue of the above-mentioned plasmid Synechocystis sp. strain PCC 6803 and Anabaena sp. strain PCC 7120 Chk46-Crr44 (HKI-RRII-OmpR) pairs is on the chromosome for N. punctiforme, the gene environment differing immediately on both sides in the three strains. Another example concerns the plasmid-encoded 6803slr6001 (Chy46), an 861-residue-long PAS-(PAS/PAC)2-HK-RR protein, for which orthologues are longer. They have CBS and two or three more PAS/PAC domains at the N terminus and are located on the chromosome in A. variabilis, Anabaena sp. strain PCC 7120, and N. punctiforme. For each of these genes, the environment completely differs. All these genes and/or clusters appear to form conserved islands, the list of such occurrences being not exhaustive.
For the other complete annotated genome sequences, Anabaena sp. strain PCC 7120 has 38 and N. punctiforme has 51 clusters of two-component genes, G. violaceus has 18, T. elongatus has 5, and the open-ocean species have 1 or 2 (see Table S4s in the supplemental material). Large rearrangements of genetic clusters have occurred between phylogenetically related strains such as Prochlorococcus species and marineSynechococcus species (33, 50). The observable degree of synteny likely is a reflection of the time of divergence between the various strains. Large rearrangements seem to have happened in the heterocystous strains as well. More species-specific events have also occurred. In Synechococcus sp. strain 9902, for example, there has been a specific deletion of the sphR-sphS pair, while the physical map is perfectly conserved for the adjacent genes compared to Synechococcus sp. strains 9605 and WH8102. For none of the Chk and Crr classes for which orthologues exist in the 16 species are the corresponding genes in a similar environment on the genome. There is no case of gene-clustering conservation. The most-conserved couple of adjacent genes is observed for the “hub” histidine kinase DspA (NblS), the corresponding chk33 gene being downstream from purD in all the strains except N. punctiforme, Synechocystis sp. strain PCC 6803, C. watsonii, and T. elongatus. The purD gene encodes the phosphoribosylamine-glycine ligase, an enzyme involved in purine metabolism, connecting the pentose phosphate pathway and glutamine metabolism to thiamine metabolism. In the five strains that have a Cph1 (AphA, Chk35) phytochrome orthologue, the cognate response regulator (Rcp1, Crr27) is downstream, but similarities in the physical map are restricted to this gene pair. A chemotaxis-related cluster of five genes starting with the two response regulators Crr36 and Crr35 also exists for the five strains that have orthologues, but again the neighborhoods differ.
EVOLUTION AND PHYLOGENY
The large number of orthologue subclasses indicates that there has been a large amount of gene duplication, even in the ancestor, because we were able to find representatives of each cyanobacterial genome in different orthologue subclasses. We can also see there a reflection of the bias in evolution rates that exists for protein-forming pairs, such as the two-component partners that must maintain protein-protein recognition and/or coevolve (60). Beside duplication and evolution, new proteins could have been generated by shuffling of domains and/or sets of domains, as well as gene fusion (95). Proteins or their domains that do not interact with each other may have different evolutionary rates, and domain shuffling may occur without interfering with the function encoded by the protein. However, proteins that establish multiple interactions with different partners probably have low evolution rates; the high conservation of Chk33 may be an illustration of this. As stated by Deeds et al. (31), new structural domains are likely to evolve much more slowly than new sequences (in the case of sequence comparisons between widely conserved orthologues such as the 16S rRNA) and more slowly than new genes (given that new genes may be discovered through novel permutations of existing structural domains). The longer time scales characterizing domain evolution thus hold great promise for illuminating the “deeper” branches of prokaryotic phylogeny.
Strain Phylogeny
From the phylogenetic relationships based on 16S rRNAs, it is generally accepted that G. violaceus, which exhibits many peculiarities, is at the bottom of the trees constructed from 16S rRNA sequences, whether the maximum-likelihood (FastDNAml) or neighbor-joining methods (37, 55, 75, 114, 116, 118) are used. A schematic tree combining the available data are presented in Fig. 1. This tree is similar to that obtained by Ochoa de Alda et al. (99) and J. Elhai (personal communication).
How do trees constructed from orthologous proteins present in all 16 genomes relate to those made from 16S rRNAs? Such trees were constructed by the neighbor-joining method (PHYLO_WIN[41]) for the orthology groups that contain 15 or 16 proteins—Chk2, Chk33 (DspA), Chk34, Chk8 (SasA), Crr1 (Ycf29), Crr23 (Ycf55), Crr26 (RpaB, Ycf27), Crr31 (RpaA), and Crr37—with PsaA (for “photosystem I core protein A1”) used for comparison. The psaA gene is always present in a single copy, and PsaA is essential for the photoautotrophic growth of cyanobacteria. Identities between the various PsaA sequences and the Synechocystis sp. strain PCC 6803 protein (651 residues long) range from 63 to 86%. Similar tree topologies (Fig. 5) that compare very well with the consensus 16S rRNA tree were obtained. The seven marine unicellular non-N2-fixing strains always cluster with 98 to 100 bootstrap values. The three marine Synechococcus spp. form a clade distinct from the Prochlorococcus clade, in which high-light-adapted strains (MIT9312 and MED4) always group with bootstrap values of 100, MIT9313 being the more distantly related. The three heterocystous strains form a distinct group, with Anabaena sp. strain PCC 7120 and A. variabilis being closer to each other than to N. punctiforme. T. erythraeum sequences most often are close to the other filamentous N2-fixing strains, but bootstrap values may be only in the range of 50. Synechocystis sp. strain PCC 6803 and C. watsonii also usually cluster. The G. violaceus sequences appear at the root of the trees and separate the group of non-N2-fixing marine unicellular strains. The positions in the trees of the freshwater unicellular S. elongatus and the thermophilic T. elongatus relative to the others are more variable. Altogether, there is good congruence of the protein and 16S rRNA trees, which could support the hypothesis that, for each orthology group, a similar gene was already present in the cyanobacterial ancestor.
FIG.5.
Unrooted trees for orthologous proteins present in the 16 cyanobacterial genomes constructed from the amino acid sequences by the neighbor-joining method with the Phylo_Win program (41). The numbers of sites kept by the program for analysis, after global gap removal, are given in parentheses: PsaA (730), Chk33 (580), Crr1 (212), Crr31 (238), Chk34 (362), Chk2 (187), Crr26 (191), and Crr37 (219). Figures at the nodes correspond to the values produced by bootstrap analysis (1,000 replicates). Abbreviations for species and gene names are given in Fig. 1 and in Table S1s in the supplemental material.
Trees were also constructed for two pairs of HK-RR for which it has been shown in at least one species that they are partner proteins: SphS (PhoR or Chk7)-SphR (PhoB or Crr29) and ManS (Chk27)-ManR (Crr16). The clustering of the three heterocystous strains is always kept, as is the marine grouping, and the architectures of the trees are rather similar. It must be stressed, however, that for the sphS-sphR couple there has been a differential loss of genes: Prochlorococcus sp. strain SS120 and Synechococcus sp. strain 9902 have lost both, while MIT9313 and T. erythraeum have lost only sphS.
Gene Origin
Horizontal gene transfers have been reported, in particular between the marine nondiazotrophic unicellular strains and other bacteria during adaptation to specific ecological niches (50, 106, 121). The extent of its importance in the currently observable repertoire of two-component proteins cannot be easily appreciated. There does not exist any protein orthologous to Chk34, a key regulator for cyanobacteria (see above), in any of the 110 finished bacterial genome sequences. In contrast, for DspA/NblS (Chk33), which exists in all of the 16 genomes and has a unique architecture among the cyanobacterial two-component proteins, bacterial orthologues can be found but they are at present restricted to the Firmicutes, a gram-positive species.
If a domain architecture occurs only in cyanobacteria, there is a good probability that the corresponding polypeptide evolved by gene duplication, followed by fusion, early on during evolution, especially if it is already present in G. violaceus. Gene duplications followed by mutations and gain of new functions have certainly been important phenomena (see below). Examples of probably recent gene duplication may also be detected. One concerns the class of the orphan HATPases. Half of them come from N. punctiforme, with NpF3113 (128 residues) being 100% identical at the amino acid level to NpF2204 and its adjacent NpF3114 (75 residues) being 100% identical to NpunNpF2205. The fact that ORFs NpF2201 to NpF2205 are all 100% identical to the similarly arranged corresponding NpF3110 to NpF3114 proteins underlines this conclusion that the duplication is recent. Noticeably, the fifth N. punctiforme HATPase, pNPBR204 (Chk112), is carried by a plasmid and has orthologues in Anabaena sp. strain PCC 7120 and A. variabilis. Anabaena sp. strain PCC 7120 Alr0709 (Chk162) and Alr0710 (Chk107, an HKIV) constitute a second example. They are very large proteins (1,799 and 1,796 aa, respectively), have the same modular organization, and are adjacent on the chromosome; they align all along their length with a single 10-residue-long gap in the middle. They are the closest paralogues, with 63% identity and 74% similarity. The same physical organization exists for A. variabilis Chk165 and Chk101, and the two proteins are 61% identical. None, however, is an orthologue of the Anabaena sp. strain PCC 7120 proteins. Another example was found in Synechocystis sp. strain PCC 6803: the Sll0789-Sll0790 pair is a paralogue of the plasmid-encoded Slr6040-Slr6041, with more than 95% identity between the putative proteins (see above). Putative histidine kinases (HKII), with either PAS or PAC domains that occur either in single or multiple copies, are essentially found in the filamentous heterocystous strains. There is also one in G. violaceus, T. erythraeum, and C. watsonii and two in Synechocystis sp. strain PCC 6803, and many do not have any orthologue, the N. punctiforme pNPAR133 being plasmid encoded. All of these proteins have probably evolved rather recently.
Domain Shuffling, Fusion, and Gene Loss
Extensive repetitive DNA facilitates prokaryotic genome plasticity, and cyanobacteria can be rich in such sequences, called STRRs (for “short tandem repetitive sequence”), some being palindromic (HIP1, for “highly iterated palindrome”) (84, 117). Longer sequences (LTRRs, 32 to 37 bp long) have also been recognized in Anabaena sp. strain PCC 7120, and the presence of an LTRR-like DNA region in mitochondrial plasmids of Vicia faba indicates strong conservation of such structures during evolution (83). IS sequences are also numerous.
In Synechocystis sp. strain PCC 6803, there is on average one HIP every 1.25 kb and 99 ORFs derived from perhaps 77 IS-like elements (70). In Anabaena sp. strain PCC 7120, a total of 145 genes were identified as presumptively encoding transposases, with 86 in the chromosome and the remaining 59 in the plasmids (63), and frequent transposition of IS892 was observed during prolonged culture (25). On the other hand, the four ISs specific for the Synechocystis Kazusa strain probably result from recent transposition events (103).
STRR sequences mostly occur outside of the coding sequences, and the same repeats can be found on both sides of a gene either in the same or opposite direction (84). That STRR sequences are found in the genomes of many heterocyst-forming cyanobacteria would support the idea that they spread before diversification of this group of cyanobacteria. However, the presence of unrelated STRR sequences in the same gene for the sister strains Anabaena sp. strain PCC 7120 and A. variabilis would indicate a more recent and independent origin of the insertions. This is difficult to reconcile with parsimony criteria. One possible scenario is that the several families of STRR sequences invaded the genome of a common ancestor of heterocystous cyanobacteria but have spread and multiplied their copy numbers recently, different STRR sequences having been amplified in different strains in a random fashion. That would explain why strains that have a very recent common ancestor contain different kinds of STRR sequences, not only within the rnpB gene but also at other positions in the genome (143). To our knowledge, no data have yet been reported concerning the presence and abundance of repeated sequences in the other 10 available genomes.
An additional property of the cyanobacteria lies in the large number of genome copies that they carry in every cell, usually around 10 copies per cell. It is nevertheless rather easy to get mutants because of the high tendency of the cells to quickly homogenize their genomes so as to reach isogeny. It is only when a mutation affects an essential gene that wild-type and mutated versions of the chromosome coexist (see, for example, references 27 and 42).
Finally, bacteriophages are also probably important and could serve as a reservoir for gene insertion. Cyanobacterial phages do exist, and the recent discovery that a photosynthetic gene can be on such genomes opens a new way of thinking about genetic exchanges. The genes psbA and psbD, encoding the D1 and D2 core components of the photosynthetic reaction center photosystem II, key genes for photosynthesis, are present in the genome of the bacteriophage S-PM2, a cyanomyovirus (79, 88). Different arrangements of these genes, plus other photosynthesis-related genes (petE, petF, and hli), have been found in three cyanophages (podovirus P-SSP7 and myoviruses P-SSM2 and P-SSM4) that infect Prochlorococcus species (74). In addition, some cyanophages that infect Prochlorococcus spp. can also cross-infect Synechococcus spp. (133). All of these features thus offer many possibilities for genetic rearrangements: duplications, insertions, losses, fusions, and shuffling.
To illustrate this section, we have analyzed what could have happened for the proteins that carry the basic structure HATP-CheW-RR. All of them belong to the HYVI subclass except Tery_403260730 (Chk169), which belongs to the HKVI family. Figure 6 describes a possible scenario of what could have happened during evolution. This is a parsimonial trial for the description of gene loss (in red) or duplication (in blue) and domain deletion (in red) or duplication (in green). A letter has been assigned to each of the five domains from which all of the proteins are made. Not so many events are required to obtain the present situation. The phylogenetic tree constructed for these 26 sequences is presented in Fig. 7. On the other hand, many examples of duplication of domains and sets of domains within a conserved structure can be observed: GAFs and GAF-PAS-PAC in HKII, HKV, HYI, HYII, and HYIV; PAS or PAC, also in the HYI; and HYIV. Whether they arise from insertion or duplication will require a more thorough investigation.
FIG.6.
Parsimonial hypothesis for the origin of the extant cyanobacterial HKs possessing a CheW domain. (A) The consensus phylogenetic tree based on 16S rRNA sequences (Fig. 1) was used as a scaffold. Three gene copies, coded as yellow, dark blue, and white boxes, should have already existed in the ancestor common to the strains that possess such proteins. The actual repertoire is shown to the right, with the domain structure represented by the letters (A to E′) within the boxes and the color code reflecting the present similarities between the proteins (phylogenetic tree presented in Fig. 7). The letters correspond to the different domains present in the proteins, as shown in the diagram in panel B. Gene duplications that occurred during evolution are represented by blue letters within the boxes, and gene losses are indicated by red letters in dotted boxes. Gains of domains within a gene are shown as green letters, and losses are shown as red letters. Gene names corresponding to the boxes on the right are as follows for each strain, from left to right: Avar_400182220, Avar_400177770, and Avar_400215350; 7120all0926, 7120all2161, and 7120all1068; NpunNpF5964, NpunNpF5640, NpunNpF2165, NpunNpR6010, and NpunNpR0245; Tery_403243570, Tery_403235320, and Tery_403260730; 6803slr0322, 6803sll0043, and 6803sll1296; Cwat_400838100, Cwat_400850380, Cwat_400866330, and Cwat_400882520; TBP1tlr0349, TBP1tll0568, and TBP1tll1021; 7942_403099980 and 7942_403098400. (B) Diagram showing the correspondence between letters and domains (see the legend to Fig. 2 and Table S2s, in the supplemental material, for domain abbreviations).
FIG. 7.
Unrooted phylogenetic tree constructed from amino acid sequences by the neighbor-joining method with the Phylo_Win program (41). A total of 406 sites were kept by the program for analysis, after global gap removal. Figures at the nodes correspond to the values produced by bootstrap analysis (1,000 replicates). Abbreviations for species and gene names are given in Fig. 1 and in Table S1s in the supplemental material. The proteins belong to the HYVI+CheW subclass and to HKV+CheW for Tery_403260730. Color coding corresponds to that used in Fig. 6.
Another interesting case is the orthology group Chk110, which includes 7421glr2212 and NpunNpR0588 (E value of 0.0). These ORFs occur in most distant species with no close homologue in any of the other species, and the neighborhoods of these orthologues completely differ. The sequencing of more cyanobacterial genomes should permit precise determination of whether this results from horizontal gene transfer, gene loss of the ancestral gene in most species, or, less likely, convergent evolution. Determining whether each of these proteins assumes a similar function(s) is worth testing. This is quite in contrast to the chk71, crr53, and chy88 groups, in which only ORFs from G. violaceus, Anabaena sp. strain PCC 7120, A. variabilis, and N. punctiforme orthologues are found still clustered and in the same order on the four genomes, and for which an ancestral cluster may thus be predicted. A similar observation can be made for chk35 and crr27, for which orthologues are present in the same four strains plus Synechocystis sp. strain PCC 6803. For chk87 and crr78, orthologues with a conserved gene order could be detected in G. violaceus, Anabaena sp. strain PCC 7120, and A. variabilis but not in N. punctiforme, that strain having probably recently lost these genes. The many different genetic mechanisms ensuring genome plasticity would thus have operated in cyanobacteria.
As reported previously for other genera (29, 69), although some cyanobacterial two-component system proteins probably result from a recruitment mechanism as well as from domain shuffling and/or fusion, coevolution by duplication from ancestral pairs appears to be the predominant mechanism by which novel regulator-sensor couples arise. In a few instances, we found that “orthologous” sensor domains were fused to different output modules. As an example, the Chk108s (7120all2095, Avar_400220860, and 7421glr1767) have an “orthologue” in N. punctiforme (NpF5467) in which the HKA_2 output domain is replaced by a GGDEF-EAL domain, the E value being 0.0. Similarly, in Synechocystis sp. strain PCC 6803, the orthologue (Sll0779) has a GGDEF domain instead of the HK domain. Different response regulatory mechanisms thus probably operate upon sensing similar stimuli. This observation fits well with the observed differences found in acclimation processes within the cyanobacterial phylum. The Chy44 group is also worth being looked at. It contains HYIII ORFs with different structures: TM-(270 aa)-(PAS/PAC)3-PAS-PAS/PAC-GAF-HK-(RR)2-Hpt for Anabaena sp. strain PCC 7120 and A. variabilis (∼1,800 residues), TM-(235 aa)-HAMP-PAS/PAC-GAF-HK-(RR)2-Hpt for T. erythraeum (∼1,400 residues), (TM)2-(115 aa)-TM-(PAS/PAC)3-GAF-HK-(RR)2-Hpt for N. punctiforme (∼1,400 residues), PAS/PAC-HK-(RR)2 for T. elongatus (∼1,035 residues), (PAS)2-GAF-HK-(RR)2-Hpt for the plasmid-encoded Synechocystis sp. strain PCC 6803 protein (∼1,200 residues), and HK-(RR)2-Hpt (∼730 residues) for C. watsonii. All seven of these proteins have an RR downstream and have an E value of 0.0.
CONCLUDING REMARKS
It has been proposed that the number of signal transducers and/or their fraction in the total protein set represents a measure of the organism's ability to adapt to diverse conditions, the “bacterial IQ” (38) or “rudimentary form of intelligence” (48). In that sense, cyanobacteria can be considered “pretty clever” organisms, including the marine strains, which exhibit fewer two-component system proteins. Indeed, living in more stable environments, they need fewer of these proteins and, as judged by their success in populating oligotrophic areas, they have made an efficient compromise, avoiding unnecessary protein synthesis. Compared to other bacteria, which have about 60% of sensor proteins with transmembrane regions, cyanobacteria only have around 30% (38). This seems at first paradoxical, as, with the exception of G. violaceus, they have a greatly higher membrane content because of the presence of the thylakoids. The latter, however, may act as “compartmentalizers” within the cell, thus reducing the need to anchor proteins. Recent developments in imaging techniques should be very useful in tracing the probable compartmentalization of the different partners involved in a given signal transduction pathway.
Even though some functions must be common to all of the cyanobacteria, the regulatory networks seem to be achieved by different sets of proteins, or at least the domains which interact may belong to proteins that do not have the same multimodular arrangements. Although no physiological function has yet been assigned to the phytochrome and its associated RR (Rcp1), it does not seem to exist in all of the species. The nbl (for “nonbleaching”) set of genes is another good example. Not all of the cyanobacteria that bleach upon nitrogen starvation have the same gene repertoire, including the NblS-NblR pair. In addition, differences have been observed in the regulation of expression of the nbl genes (77, 78, 115, 125).
The table of orthologues (Table 1; see also Table S3s in the supplemental material) clearly shows that very few ORFs already have a precise gene name, meaning that no function has yet been assigned to these genes. The orthologous genes certainly deserve in-depth investigation, in particular those that are present in many strains, as well as the genes that have kept clustering and those that are still also present in red algal genomes.
Nowadays, genome availability is a very precious tool for all biologists but constitutes only the starting material. Most of the work for understanding life is still to come. It will be necessary to gather complementary studies performed in a number of different fields. In silico bioinformatics and modeling are interesting and useful because they represent global approaches. They rely, however, on the data and hypotheses used to build the algorithms. There must be a constant cross-feeding between bioinformaticians and experimentalists. Thanks to the work of the former, a more limited and focused number of experiments may need to be performed, particularly where orthologous groups have been identified, instead of “fishing blind.” The latter, whether geneticists, biochemists or physiologists, must validate the predictions and/or ask for changes in the algorithms.
Evolution has been driven by the constraints resulting from the conditions prevailing at a given time within an ecosystem. These conditions may have differed from those of the present, and they certainly do differ from laboratory conditions. Thus, finding a phenotype and deciphering regulatory pathways are real challenges for the experimentalists.
Supplementary Material
Acknowledgments
M.K.A. was supported by a new-initiative grant from the Mona campus of the University of the West Indies. Support by the Centre National de la Recherche Scientifique (CNRS) and grant IMPB012 to J.H. are acknowledged.
We are indebted to Jesús A. Gómez Ochoa de Alda for insightful help with data analysis and critical reading of the manuscript. We thank Kamel Jabbari and Hugues Roest-Crollius for valuable and helpful discussions.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Aiba, H., M. Nagaya, and T. Mizuno. 1993. Sensor and regulator proteins from the cyanobacterium Synechococcus sp. PCC7942 that belong to the bacterial signal-transduction protein families: implication in the adaptive response to phosphate limitation. Mol. Microbiol. 8:81-91. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman.1997. . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves, R., and M. A. Savageau. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25-51. [DOI] [PubMed] [Google Scholar]
- 4.Anantharaman, V., and L. Aravind. 2000. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535-537. [DOI] [PubMed] [Google Scholar]
- 5.Anantharaman, V., E. V. Koonin, and L. Aravind. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 307:1271-1292. [DOI] [PubMed] [Google Scholar]
- 6.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appleman, J. A., L. L. Chen, and V. Stewart.2003. . Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases.Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 9.Aravind, L., and C. P. Ponting. 1997. The GAF domain: and evolutionary link between diverse phototransducing proteins.Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 10.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 11.Ashby, M. K. 2004. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol. Lett. 231:277-281. [DOI] [PubMed] [Google Scholar]
- 12.Ashby, M. K., and C. W. Mullineaux. 1999. Cyanobacterial ycf27 genes regulate the coupling of phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181:253-260. [DOI] [PubMed] [Google Scholar]
- 13.Ashby, M. K., J. Houmard, and C. W. Mullineaux.2002. . The ycf27 genes from cyanobacteria and eukaryotic algae: distribution and implications for chloroplast evolution. FEMS Microbiol. Lett. 214:25-30. [DOI] [PubMed] [Google Scholar]
- 14.Baikalov, I., I. Schroder, M. Kaczor-Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL.Biochemistry 35:11053-11061. [DOI] [PubMed] [Google Scholar]
- 15.Ballal, A., M. Bramkamp, H. Rajaram, P. Zimmann, S. K. Apte, and K. Altendorf. 2005. An atypical KdpD homologue from the cyanobacterium Anabaena sp. strain L-31: cloning, in vivo expression, and interaction with Escherichia coli KdpD-CTD.J. Bacteriol. 187:4921-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balland, V., L. Bouzhir-Sima, L. Kiger, M. C. Marden, M. H. Vos, U. Liebl, and T. A. Mattioli. 2005. Role of arginine 220 in the oxygen sensor FixL from Bradyrhizobium japonicum. J. Biol. Chem. 280:15279-15288. [DOI] [PubMed] [Google Scholar]
- 17.Barák, I., J. Behari, G. Olmedo, P. Guzman, D. P. Brown, E. Castro, D. E. Walker, J. Westphelling, and P. Youngman.1996. . Structure and function of the Bacillus SpoIIE protein and its location to sites of sporulation septum assembly.Mol. Microbiol. 19:1047-1060. [DOI] [PubMed] [Google Scholar]
- 18.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database.Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhaya, D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53:745-754. [DOI] [PubMed] [Google Scholar]
- 20.Bhaya, D., A. Takahashi, and A. R. Grossman. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98:7540-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 23.Bruder, S., J. U. Linder, S. E. Martinez, N. Zheng, J. A. Beavo, and J. E. Schultz.2005. . The cyanobacterial tandem GAF domains from the cyaB2 adenylyl cyclase signal via both cAMP-binding sites. Proc. Natl. Acad. Sci. USA 102:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadoret, J. C., B. Rousseau, I. Perewoska, C. Sicora, O. Cheregi, I. Vass, and J. Houmard. 2005. Cyclic nucleotides, the photosynthetic apparatus and response to a UV-B stress in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 280:33935-33944. [DOI] [PubMed] [Google Scholar]
- 25.Cai, Y. 1991. Characterization of insertion sequence IS892 and related elements from the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cann, M. J. 2004. Signalling through cyclic nucleotide monophosphates in cyanobacteria. New Phytol. 159:289-293. [Google Scholar]
- 27.Capuano, V., J. C. Thomas, N. Tandeau de Marsac, and J. Houmard.1993. . An in vivo approach to define the role of the LCM, the key polypeptide of cyanobacterial phycobilisomes.J. Biol. Chem. 268:8277-8283. [PubMed] [Google Scholar]
- 28.Castenholz, R. W. (2001). The archaea and the deeply branching and phototropic bacteria, p. 473-599. In G. Garrity, D. R. Boone, and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 29.Chen, Y. T., H. Y. Chang, C. L. Lu, and H. L. Peng. 2004. Evolutionary analysis of the two-component systems in Pseudomonas aeruginosa PAO1.J. Mol. Evol. 59:725-737. [DOI] [PubMed] [Google Scholar]
- 30.Chiang, G. G., M. R. Schaefer, and A. R. Grossman. 1992. Complementation of a red-light-indifferent cyanobacterial mutant. Proc. Natl. Acad. Sci. USA 89:9415-9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeds, E. J., H. Hennessey, and E. I. Shakhnovich.2005. . Prokaryotic phylogenies inferred from protein structural domains. Genome Res. 15:393-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditty, J. L., S. B. Williams, and S. S. Golden. 2003. A cyanobacterial circadian timing mechanism. Annu. Rev. Genet. 37:513-543. [DOI] [PubMed] [Google Scholar]
- 33.Dufresne, A., L. Garczarek, and F. Partensky. 2005. Accelerated evolution associated with genome reduction in a free-living prokaryote.Genome Biol. 6:R14. [Online.] http://genomebiology.com/2005/6/2/R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weisenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanaobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 100:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteban, B., M. Carrascal, J. Abian, and T. Lamparter. 2005. Light-induced conformational changes of cyanobacterial phytochrome Cph1 probed by limited proteolysis and autophosphorylation.Biochemistry 44:450-461. [DOI] [PubMed] [Google Scholar]
- 36.Fischer, A. J., and J. C. Lagarias. 2004. Harnessing phytochrome's glowing potential. Proc. Natl. Acad. Sci. USA 101:17334-17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis.Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 37.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galperin, M. Y., T. A. Gaidenko, A. Y. Mulkidjanian, M. Nakano, and C. W. Price.2001. . MHYT, a new integral membrane sensor domain.FEMS Microbiol. Lett. 205:17-23. [DOI] [PubMed] [Google Scholar]
- 40.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 41.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Dominguez, M., J. C. Reyes, and F. J. Florencio.2000. . NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 35:1192-1201. [DOI] [PubMed] [Google Scholar]
- 43.Gilles-Gonzalez, M.-A., and G. Gonzalez. 2004. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96:774-783. [DOI] [PubMed] [Google Scholar]
- 44.Golden, S. S., M. S. Nalty, and D. S. Cho.1989. . Genetic relationship of two highly studied Synechococcus strains designated Anacystis nidulans.J. Bacteriol. 171:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallick, R. B., and A. Bairoch. 1994. Proposals for naming of chloroplast genes. III. Nomenclature for open reading frames encoded in chloroplast genomes. Plant Mol. Biol. Rep. 12(Suppl. 2):S29-S30. [Google Scholar]
- 46.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilises the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 51:51277-51284. [DOI] [PubMed] [Google Scholar]
- 47.Hegyi, H., and M. Gerstein. 2001. Annotation transfer for genomics: measuring functional divergence in multi-domain proteins.Genome Res. 11:1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellingwerf, K. J. 2005. Bacterial observations: a rudimentary form of intelligence? Trends Microbiol. 13:152-158. [DOI] [PubMed] [Google Scholar]
- 49.Herdman, M., T. Coursin, R. Rippka, J. Houmard, and N. Tandeau de Marsac.2000. . A new appraisal of the prokaryotic origin of eukaryotic phytochromes. J. Mol. Evol. 51:205-213. [DOI] [PubMed] [Google Scholar]
- 50.Hess, W. 2004. Genome analysis of marine photosynthetic microbes and their global role. Curr. Opin. Biotechnol. 15:191-198. [DOI] [PubMed] [Google Scholar]
- 51.Hirani, T. A., I. Suzuki, N. Murata, H. Hayashi, and J. J. Eaton-Rye. 2001. Characterization of a two-component signal transduction system involved in the induction of alkaline phosphatase under phosphate-limiting conditions in Synechocystis sp. PCC 6803. Plant Mol. Biol. 45:133-144. [DOI] [PubMed] [Google Scholar]
- 52.Hitomi, K., T. Oyama, S. Han, A. S. Arvai, and E. D. Getzoff. 2005. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.J. Biol. Chem. 280:19127-19135. [DOI] [PubMed] [Google Scholar]
- 53.Ho, Y.-S. J., L. M. Burden, and J. H. Hurley. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor.EMBO J. 19:5288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 55.Honda, D., A. Yokota, and J. Sugiyama. 1999. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 48:723-739. [DOI] [PubMed] [Google Scholar]
- 56.Hsiao, H.-Y., Q. He, L. G. van Waasbergen, and A. R. Grossman. 2004. Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: negative and positive regulation and interactions between signal transduction pathways. J. Bacteriol. 186:3882-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwasaki, H., S. B. Williams, Y. Kitayama, M. Ishiura, S. S. Golden, and T. Kondo. 2000. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101:223-233. [DOI] [PubMed] [Google Scholar]
- 58.Iyer, L. M., V. Anantharaman, and L. Aravind.2003. . Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janiak-Spens, F., D. P. Sparling, and A. H. West.2000. . Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J. Bacteriol. 182:6673-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jordan, I. K., Y. I. Wolf, and E. V. Koonin.2004. . Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kageyama, H., T. Kondo, and H. Iwasaki. 2003. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J. Biol. Chem. 278:2388-2395. [DOI] [PubMed] [Google Scholar]
- 62.Kahlon, S., K. Beeri, H. Ohkawa, Y. Hihara, O. Murik, I. Suzuki, T. Ogawa, and A. Kaplan. 2006. A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC 6803 to the presence of glucose. Microbiology 152:647-655. [DOI] [PubMed] [Google Scholar]
- 63.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213, 227-253. [DOI] [PubMed] [Google Scholar]
- 64.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling.J. Bacteriol. 186:267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kasahara, M., and M. Ohmori. 1999. Activation of a cyanobacterial adenylate cyclase, CyaC, by autophosphorylation and a subsequent phosphotransfer reaction. J. Biol. Chem. 274:15167-15172. [DOI] [PubMed] [Google Scholar]
- 66.Kasahara, M., K. Yashiro, T. Sakamoto, and M. Ohmori. 1997. The Spirulina platensis adenylate cyclase gene, cyaC, encodes a novel signal transduction protein. Plant Cell Physiol. 38:828-836. [DOI] [PubMed] [Google Scholar]
- 67.Katayama, M., and M. Ohmori. 1997. Isolation and characterization of multiple adenylate cyclase genes from the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:3588-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kehoe, D., and A. R. Grossman. 1997. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J. Bacteriol. 179:3914-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2000. Evolution of two-component signal transduction. Mol. Biol. Evol. 17:1956-1970. [DOI] [PubMed] [Google Scholar]
- 70.Kotani, H., and S. Tabata. 1998. Lessons from sequencing of the genome of a unicellular cyanobacterium, Synechocystis sp. PCC6803.Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:151-171. [DOI] [PubMed] [Google Scholar]
- 71.Li, H., and L. A. Sherman. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:4268-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li, L., and D. M. Kehoe. 2005. In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol. Microbiol. 55:1538-1552. [DOI] [PubMed] [Google Scholar]
- 73.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm.2004. . Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 101:11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Litvaitis, M. K. 2002. A molecular test of cyanobacterial phylogeny: inferences from constraint analyses.Hydrobiologia 468:135-145. [Google Scholar]
- 76.Lopez-Maury, L., M. Garcia-Dominguez, F. J. Florencio, and J. C. Reyes. 2002. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43:247-256. [DOI] [PubMed] [Google Scholar]
- 77.Luque, I., J. A. Ochoa de Alda, C. Richaud, G. Zabulon, J. C. Thomas, and J. Houmard. 2003. The NblAI protein from the filamentous cyanobacterium Tolypothrix PCC 7601: regulation of its expression and interactions with phycobilisome components. Mol. Microbiol. 50:1043-1054. [DOI] [PubMed] [Google Scholar]
- 78.Luque, I., G. Zabulon, A. Contreras, and J. Houmard. 2001. Convergence of two global transcriptional regulators on nitrogen induction of the stress-acclimation gene nblA in the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol. 41:937-947. [DOI] [PubMed] [Google Scholar]
- 79.Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie.2003. . Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741. [DOI] [PubMed] [Google Scholar]
- 80.Marin, K., I. Suzuki, K. Yamaguchi, K. Ribbeck, H. Yamamoto, Y. Kanesaki, M. Hagemann, and N. Murata. 2003. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803.Proc. Natl. Acad. Sci. USA 100:9061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 82.Mary, I., and D. Vaulot. 2003. Two-component systems in Prochlorococcus MED4: genomic analysis and differential expression under stress. FEMS Microbiol. Lett. 226:135-144. [DOI] [PubMed] [Google Scholar]
- 83.Masepohl, B., K. Gorlitz, and H. Bohme. 1996. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta 1307:26-30. [DOI] [PubMed] [Google Scholar]
- 84.Mazel, D., J. Houmard, A. M. Castets, and N. Tandeau de Marsac.1990. . Highly repetitive DNA sequences in cyanobacterial genomes. J. Bacteriol. 172:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McAllister-Lucas, L. M., T. L. Haik, J. L. Colbran, W. K., Sonnenburg, D. Seger, I. V. Turko, J. A. Beavo, S. H. Francis, and J. D. Corbin. 1995. An essential aspartic acid at each of two allosteric cGMP-binding sites of a cGMP-specific phosphodiesterase.J. Biol. Chem. 270:30671-30679. [DOI] [PubMed] [Google Scholar]
- 86.Mika, F., and R. Hengge. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev. 19:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mikami, K., Y. Kanesaki, I. Suzuki, and N. Murata. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC 6803. Mol. Microbiol. 46:905-915. [DOI] [PubMed] [Google Scholar]
- 88.Millard, A., M. R. Clokie, D. A. Shub, and N. H. Mann. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains.Proc. Natl. Acad. Sci. USA 101:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizuno, T., T. Kaneko, and S. Tabata. 1996. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803.DNA Res. 3:407-414. [DOI] [PubMed] [Google Scholar]
- 90.Morrison, S. S., C. W. Mullineaux, and M. K. Ashby. 2005. The influence of acetyl phosphate on DspA signalling in the cyanobacterium Synechocystis sp. PCC6803.BMC Microbiol. 5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muro-Pastor, A. M., E. Olmedo-Verd, and E. Flores. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC7120. FEMS Microbiol. Lett. 256:171-177. [DOI] [PubMed] [Google Scholar]
- 92.Mutsuda, M., K. P. Michel, X. Zhang, B. L. Montgomery, and S. S. Golden. 2003. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 278:19102-19110. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura, N., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9:123-130. [DOI] [PubMed] [Google Scholar]
- 94.Nakamura, Y., T. Kaneko, S. Sato, M. Mimoro, H. Miyashita, T. Tsuchiya, S. Sasamoto, A. Watanabe, K. Kawashima, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, C. Takeuchi, M. Yamada, and S. Tabata. 2003. Complete genome structure of Gloeobacter violaceus PCC7421, a cyanobacterium that lacks thylakoids. DNA Res. 10:137-145. [DOI] [PubMed] [Google Scholar]
- 95.Narikawa, R., S. Okamoto, M. Ikeuchi, and M. Ohmori. 2004. Molecular evolution of PAS domain-containing proteins of filamentous cyanobacteria through domain shuffling and domain duplication.DNA Res. 11:69-81. [DOI] [PubMed] [Google Scholar]
- 96.Nikolskaya, A. N., A. Y. Mulkidjanian, I. B. Beech, and M. Y. Galperin. 2003. MASE1 and MASE2: two novel integral membrane sensory domains. J. Mol. Microbiol. Biotechnol. 5:11-16. [DOI] [PubMed] [Google Scholar]
- 97.Ning, D., and X. Xu. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447-453. [DOI] [PubMed] [Google Scholar]
- 98.Ochoa de Alda, J. A. G., and J. Houmard.2000. . Genomic survey of cAMP and cGMP signalling components in the cyanobacterium Synechocystis PCC 6803.Microbiology 146:3183-3194. [DOI] [PubMed] [Google Scholar]
- 99.Ochoa de Alda, J. A. G., A. del Pico, A. Pedraza, and J. Houmard. 2005. Caracterización, clasificación y filogenia de adenilil y guanilil ciclasas de cianobacterias. Oppidum 1:311-356. [Google Scholar]
- 100.Ogawa, T., D. H. Bao, H. Katoh, M. Shibata, H. B. Pakrasi, and M. Bhattacharyya-Pakrasi. 2002. A two-component signal transduction pathway regulates manganese homeostasis in Synechocystis 6803, a photosynthetic organism.J. Biol. Chem. 277:28981-28986. [DOI] [PubMed] [Google Scholar]
- 101.Ohmori, M., and S. Okamoto. 2004. Photoresponsive cAMP signal transduction in cyanobacteria. Photochem. Photobiol. Sci. 3:503-511. [DOI] [PubMed] [Google Scholar]
- 102.Ohmori, M., M. Ikeuchi, S. Sato, C. P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabata. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 103.Okamoto, S., M. Ikeuchi, and M. Ohmori. 1999. Experimental analysis of recently transposed insertion sequences in the cyanobacterium Synechocystis sp. PCC 6803. DNA Res. 6:265-273. [DOI] [PubMed] [Google Scholar]
- 104.Okamoto, S., M. Kasahara, A. Kamiya, Y. Nakahira, and M. Ohmori.2004. . A phytochrome-like protein AphC triggers the cAMP signaling induced by Far-red light in the cyanobacterium Anabaena sp. strain PCC7120. Photochem. Photobiol. 80:429-433. [DOI] [PubMed] [Google Scholar]
- 105.Paithoonrangsarid, K., M. A. Shoumskaya, Y. Kanesaki, S. Satoh, S. Tabata, D. A. Los, V. V. Zinchenko, H. Hayashi, M. Tanticharoen, I. Suzuki, and N. Murata.2004. . Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis.J. Biol. Chem. 279:53078-53086. [DOI] [PubMed] [Google Scholar]
- 106.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 107.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 109.Phalip, V., J. H. Li, and C. C. Zhang.2001. . HstK, a cyanobacterial protein with both a serine/threonine kinase domain and a histidine kinase domain: implication for the mechanism of signal transduction. Biochem. J. 360:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 111.Porter, S. L., and J. P. Armitage. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J. Mol. Biol. 324:35-45. [DOI] [PubMed] [Google Scholar]
- 112.Porter, S. L., and J. P. Armitage. 2004. Chemotaxis in Rhodobacter sphaeroides requires an atypical histidine protein kinase. J. Biol. Chem. 279:54573-54580. [DOI] [PubMed] [Google Scholar]
- 113.Quail, P. H. 1991. Phytochrome: a light-activated molecular switch that regulates plant gene expression. Annu. Rev. Genet. 25:389-409. [DOI] [PubMed] [Google Scholar]
- 114.Rantala, A., D. P. Fewer, M. Hisbergues, I. Rouhiainen, J. Vaitomaa, T. Borner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Richaud, C., G. Zabulon, A. Joder, and J. C. Thomas.2001. . Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J. Bacteriol. 183:2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rippka, R., R. W. Castenholz, and M. Herdman. 2001. The archaea and the deeply branching and phototropic bacteria, p.562 -566. In G. Garrity, D. R. Boone, and R. W. Castenholz (ed.),Bergey's manual of systematic bacteriology , 2nd ed.,vol. 1 . Springer-Verlag, New York, N.Y. [Google Scholar]
- 117.Robinson, N. J., P. J. Robinson, A. Gupta, A. J. Bleasby, B. A. Whitton, and A. P. Morby.1995. . Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 23:729-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 119.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 120.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scanlan, D. J. 2003. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microbiol. Physiol. 47:1-64. [DOI] [PubMed] [Google Scholar]
- 122.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky.2005. . The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schmitz, O., M. Katayama, S. B. Williams, T. Kondo, and S. S. Golden. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289:765-768. [DOI] [PubMed] [Google Scholar]
- 124.Schwartz, S. H., T. A. Black, K. Jager, J. M. Panoff, and C. P. Wolk. 1998. Regulation of an osmoticum-responsive gene in Anabaena sp. strain PCC 7120.J. Bacteriol. 180:6332-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schwarz, R., and A. R. Grossman. 1998. A response regulator of cyanobacteria integrates diverse signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shoumskaya, M. A., K. Paithoonrangsarid, Y. Kanesaki, D. A. Los, V. V. Zinchenko, M. Tanticharoen, I. Suzuki, and N. Murata. 2005. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 280:21531-21538. [DOI] [PubMed] [Google Scholar]
- 127.Singh, A. K., and L. A. Sherman. 2005. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp. strain PCC 6803 and its requirement for heterotrophic growth. J. Bacteriol. 187:2368-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sonnhammer, E. L., and E. V. Koonin. 2002. Orthology, paralogy and proposed classification for paralog subtypes.Trends Genet. 18:619-620. [DOI] [PubMed] [Google Scholar]
- 129.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327-2335. [DOI] [PubMed] [Google Scholar]
- 130.Steegborn, C., T. N. Litvin, L. R. Levin, J. Buck, and H. Wu. 2004. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment.Nat. Struct. Mol. Biol. 12:32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stewart, V. 2003. Nitrate- and nitrite responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 132.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction.Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 133.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 134.Suzuki, I., Y. Kanesaki, H. Hayashi, J. J. Hall, W. J. Simon, A. R. Slabas, and N. Murata. 2005. The histidine kinase hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis.Plant Physiol. 138:1409-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Suzuki, I., D. A. Los, Y. Kanesaki, K. Mikami, and N. Murata.2000. . The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Suzuki, S., A. Ferjani, I. Suzuki, and N. Murata. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279:13234-13240. [DOI] [PubMed] [Google Scholar]
- 137.Terauchi, K., B. L. Montgomery, A. R. Grossman, J. C. Lagarias, and D. M. Kehoe. 2004. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol. Microbiol. 51:567-577. [DOI] [PubMed] [Google Scholar]
- 138.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria.Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tu, C.-J., J. Shrager, R. L. Burnap, B. L. Postier, and A. R. Grossman. 2004. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803. J. Bacteriol. 186:3889-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.van Waasbergen, L. G., N. Dolganov, and A. R. Grossman. 2002. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942.J. Bacteriol. 184:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vioque, A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 25:3471-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein.1992. . KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Webre, D. J., P. M. Wolanin, and J. B. Stock. 2003. Bacterial chemotaxis. Curr. Biol. 13:R47-R49. [DOI] [PubMed] [Google Scholar]
- 146.Wuichet, K., and I. B. Zhulin. 2003. Molecular evolution of sensory domains in cyanobacterial chemoreceptors.Trends Microbiol. 11:200-210. [DOI] [PubMed] [Google Scholar]
- 147.Yakunin, A. F., M. Proudfoot, E. Kuznetsova, A. Savchenko, G. Brown, C. H. Arrowsmith, and A. M. Edwards.2004. . The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279:36819-36827. [DOI] [PubMed] [Google Scholar]
- 148.Yamaguchi, K., I. Suzuki, H. Yamamoto, A. Lyukevich, I. Bodrova, D. A. Los, I. Piven, V. Zinchenko, M. Kanehisa, and N. Murata.2002. . A two-component Mn2+-sensing system negatively regulates expression of the mntCAB operon in Synechocystis. Plant Cell 14:2901-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yeh, K.-C., and J. C. Lagarias. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95:13976-13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yeh, K.-C., S.-H. Wu, J. T. Murphy, and J. C. Lagarias. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277:1505-1508. [DOI] [PubMed] [Google Scholar]
- 151.Yoshihara, S., X. Geng, and M. Ikeuchi. 2002. PilG gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 43:513-521. [DOI] [PubMed] [Google Scholar]
- 152.Yoshihara, S., F. Suzuki, H. Fujita, X. X. Geng, and M. Ikeuchi.2000. . Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 41:1299-1304. [DOI] [PubMed] [Google Scholar]
- 153.Zarembinski, T. I., L. W. Hung, H. J. Mueller-Dieckmann, K. K. Kim., H. Yokata, R. Kim, and S. H. Kim. 1998. Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc. Natl. Acad. Sci. USA 95:15189-15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou, R., and C. P. Wolk. 2003. A two-component system mediates developmental regulation of biosynthesis of a heterocyst polysaccharide. J. Biol. Chem. 278:19939-19946. [DOI] [PubMed] [Google Scholar]
- 155.Zhu, Y., and M. Inouye. 2004. The HAMP linker in histidine kinase dimeric receptors is critical for symmetric transmembrane signal transduction. J. Biol. Chem. 279:48152-48158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.