Abstract
The roles of slow antibiotic penetration, oxygen limitation, and low metabolic activity in the tolerance of Pseudomonas aeruginosa in biofilms to killing by antibiotics were investigated in vitro. Tobramycin and ciprofloxacin penetrated biofilms but failed to effectively kill the bacteria. Bacteria in colony biofilms survived prolonged exposure to either 10 μg of tobramycin ml−1or 1.0 μg of ciprofloxacin ml−1. After 100 h of antibiotic treatment, during which the colony biofilms were transferred to fresh antibiotic-containing plates every 24 h, the log reduction in viable cell numbers was only 0.49 ± 0.18 for tobramycin and 1.42 ± 0.03 for ciprofloxacin. Antibiotic permeation through colony biofilms, indicated by a diffusion cell bioassay, demonstrated that there was no acceleration in bacterial killing once the antibiotics penetrated the biofilms. These results suggested that limited antibiotic diffusion is not the primary protective mechanism for these biofilms. Transmission electron microscopic observations of antibiotic-affected cells showed lysed, vacuolated, and elongated cells exclusively near the air interface in antibiotic-treated biofilms, suggesting a role for oxygen limitation in protecting biofilm bacteria from antibiotics. To test this hypothesis, a microelectrode analysis was performed. The results demonstrated that oxygen penetrated 50 to 90 μm into the biofilm from the air interface. This oxic zone correlated to the region of the biofilm where an inducible green fluorescent protein was expressed, indicating that this was the active zone of bacterial metabolic activity. These results show that oxygen limitation and low metabolic activity in the interior of the biofilm, not poor antibiotic penetration, are correlated with antibiotic tolerance of this P. aeruginosa biofilm system.
One explanation for the chronic nature of some infections involving the opportunistic pathogen Pseudomonas aeruginosa is that this organism is adept at forming biofilms in which the bacteria are protected from host defenses and from killing by antibiotics (6). The protection of bacteria growing in biofilms from antibiotics probably depends on multiple factors (3, 19, 27, 28). Two explanations have long dominated the debate for this reduced antibiotic susceptibility. The first and most intuitive is that the antibiotic fails to physically penetrate the biofilm. The second explanation is that nutrient limitation leads to slow growth or stationary phase existence for many of the cells in a biofilm, reducing their antimicrobial susceptibility (3, 25). This article addresses the relative contributions of poor antibiotic penetration versus oxygen limitation and reduced metabolic activity in the protection of P. aeruginosa biofilms from killing by tobramycin and ciprofloxacin.
Several articles report measurements of antibiotic penetration in P. aeruginosa biofilms (15, 18, 24, 30, 31, 35). The general consensus of these studies is that fluoroquinolones, such as ofloxacin and ciprofloxacin, penetrate P. aeruginosa biofilms readily while aminoglycosides, such as tobramycin and gentamicin, are retarded in their delivery. This conclusion is supported by a number of other investigations suggesting that aminoglycosides diffuse more slowly because they bind to extracellular polymers such as alginate (13, 14, 20). There are conflicting results regarding the penetration of a β-lactam antibiotic, piperacillin (15, 24).
A difficulty in the studies of antibiotic penetration into P. aeruginosa biofilms is in determining whether slow antibiotic penetration contributes to reduced antibiotic susceptibility of bacteria in the biofilm mode of growth. While it is interesting that fluoroquinolones permeate biofilms more rapidly than aminoglycosides, this observation by itself does not tell us whether the fluoroquinolones penetrate rapidly enough so that transport limitation can be ruled out as a protective mechanism. Likewise, the relatively slow penetration of an aminoglycoside does not necessarily indicate that delivery was slow enough to contribute to the reduced susceptibility of the biofilm. In the present study, to test the relationship between antibiotic penetration and biofilm susceptibility, we compared the time course of antibiotic penetration with the time course of bacterial killing by a fluoroquinolone and by an aminoglycoside.
A second difficulty of antibiotic penetration studies is the possibility that the antibiotics move through gaps or channels in the biofilm without reaching into the interiors of cell clusters. Many biofilms are now recognized to be heterogeneous structures through which fluid-filled channels run, so this is a reasonable concern (5, 29). We chose a biofilm model that does not have water channels to help eliminate this possibility. We also addressed this issue by visualizing the action of the antibiotic locally within the biofilm by microscopic techniques (37, 38).
The purpose of the work reported in this article was to pair complementary measurements relevant to bacterial susceptibility in a biofilm. In this study, the same experimental system was used to measure the time course of antibiotic penetration, the time course of bacterial killing, the spatial pattern of antibiotic action within the biofilm, the local availability of oxygen, and the spatial pattern of protein synthesis within the biofilm. Comparing these data allowed us to infer the relative importance of slow antibiotic penetration and low metabolic activity in protecting bacteria in the biofilm.
MATERIALS AND METHODS
Strains, media, and antibiotics.
Pure cultures of the mucoid P. aeruginosa strain FRD1 (22) were used throughout these studies. This bacterium was originally isolated from a pulmonary infection of a cystic fibrosis patient, and it produces large amounts of the extracellular polysaccharide alginate. P. aeruginosa FRD1 was grown in tryptic soy broth (TSB) or tryptic soy agar (TSA) supplied by Difco (Detroit, Mich.). To visualize gene expression with epifluorescence microscopy, we used P. aeruginosa FRD1 containing the plasmid pAB1. Plasmid pAB1 contains the gene for the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible green fluorescent protein (GFP) with the mut2 mutation (4). The gfp-mut2 allele produces a stable GFP. Plasmid pAB1 was constructed by digesting plasmid pMF230 (21) with XbaI and HindIII, releasing the 0.8-kb gfp-mut2 gene fragment. The fragment was ligated into the XbaI-HindIII sites of plasmid pMF54 (12), with gfp being placed behind the trc promoter. The trc promoter is a chimera of the trp and lac promoters. pAB1 contained the lacI repressor, and therefore induction of gfp could be controlled through the addition of IPTG to the medium. pAB1 also contained the oriT site, allowing introduction of the plasmid into P. aeruginosa by triparental mating with the conjugation helper plasmid pRK2013 (11). pAB1 was stably maintained in P. aeruginosa, since it contained the stable replication DNA fragment (12). Escherichia coli ATCC 25922 was used as an antibiotic-sensitive indicator organism for zone-of-inhibition bioassays. E. coli was grown in Luria-Bertani (LB) broth (Difco), and the zone-of-inhibition bioassay was performed with Mueller-Hinton agar plates (Difco). All bacteria were grown and experiments were conducted at 37°C. Tobramycin sulfate was obtained from Sigma (St. Louis, Mo.), and ciprofloxacin hydrochloride was a gift of the Bayer Corporation (Leverkusen, Germany). Viable cell numbers were determined by colony formation on R2A agar (Difco). MICs were determined by using Etest strips (AB Biodisk, Solna, Sweden).
Biofilm preparation.
Colony biofilms (1) were prepared by growing bacteria on top of polymer membranes resting on TSA plates. Polycarbonate membrane filters (diameter, 25 mm; Poretics, Livermore, Calif.) were sterilized by exposure to UV light, placed on a TSA plate, and seeded with a 5-μl drop of an overnight culture of P. aeruginosa FRD1. The overnight cultures were grown in TSB and diluted in the same medium prior to spotting on the membrane to an optical density at 600 nm of 0.13. The plates were inverted and incubated at 37°C for 48 h. Each colony biofilm was transferred to a fresh TSA plate every 24 h. Further details of the colony biofilm growth procedure are described by Anderl et al. (1). Biofilm thickness was determined by image analysis of frozen sections stained with 4′,6-diamidino-2-phenylindole (Sigma) and examined by fluorescence microscopy (16). The average specific growth rate of bacteria in colony biofilms was calculated by a least-squares linear regression of the natural log of cell numbers against time to compute the slope of the line.
Biofilm susceptibility.
The killing of bacteria in colony biofilms was measured by transferring colony biofilms to agar plates containing antibiotics. After 48 h of growth in the absence of antibiotics, colony biofilms were transferred to TSA plates with either 10 μg of tobramycin ml−1 or 1.0 μg of ciprofloxacin ml−1. Plates were incubated at 37°C. Each colony biofilm was transferred to a fresh antibiotic-containing plate every 24 h. The biofilms were sampled by placing the membrane and associated bacteria into a tube containing 9 ml of phosphate-buffered water (pH 7.2) (9) and vortexing at high speed for 1 min. The resulting cell suspension was serially diluted, and viable bacteria were enumerated by drop-plating (five 10-μl drops) on R2A. Killing was reported as a log reduction or, when growth was observed, as a log increase. The log reduction was calculated relative to the cell count at time zero. Experiments were performed in triplicate.
Planktonic susceptibility.
The susceptibility of bacteria in suspension was measured by using 15 ml of an overnight culture of P. aeruginosa FRD1 in TSB that was mixed with 5 ml of fresh TSB. This culture was divided into two equal volumes, and antibiotic (10 μl) was added to one aliquot and an equal volume of water was added to the second aliquot. Both cultures were placed in an orbital shaker at 37°C and sampled every hour for 4 h. Bacteria were pelleted in a microcentrifuge, rinsed, and then resuspended in fresh phosphate-buffered water. The bacteria were pelleted and washed a second time to remove residual antibiotic. After the final spin, 1.5 ml of phosphate-buffered water was added to the centrifuge tube and the tube was vortexed to resuspend the bacteria. The resulting cell suspension was serially diluted, and viable bacterial numbers were determined by plating on R2A. Experiments were performed in triplicate.
Bacteria resuspended from biofilms were also examined for antibiotic sensitivity. Developed colony biofilms were removed from TSA plates and placed in 9 ml of phosphate-buffered water. The biofilms were dispersed by vortexing the tube for 2 min. This suspension of bacteria was then processed as described above for the planktonic overnight culture to measure antibiotic killing. Experiments were performed in triplicate.
Antibiotic penetration.
Antibiotic penetration through 48-h P. aeruginosa colony biofilms was measured by the technique of Anderl et al. (1). This involved placing a second, smaller (13-mm-diameter) membrane on top of a bacterial colony and then placing a wetted paper disk (6-mm-diameter), of the type used in zone-of-inhibition bioassays, on top of the smaller membrane. This formed a diffusion cell in which the colony biofilm was sandwiched between the two membranes. These assemblies were transferred to separate TSA plates containing either 10 μg of tobramycin ml−1 or 1.0 μg of ciprofloxacin ml−1 and incubated at 37°C. Antibiotic had to diffuse out of the agar, through the lower membrane, through the bacterial colony, and through the upper membrane to reach the moistened disk. Disks were removed at intervals, sealed in paraffin film, and stored at 4°C until all the samples had been collected. Disks were then placed on Mueller-Hinton plates spread with 100 μl of an E. coli suspension diluted to an optical density at 600 nm of 0.05. The diameter of the zone of inhibition around the disk was measured after 8 h of incubation. Parallel experiments were performed with sterile control assemblies. These consisted of the two membranes and the concentration disk without bacteria. Experiments were performed in triplicate.
A standard relationship between antibiotic concentration and zone of inhibition was obtained by loading known concentrations of tobramycin or ciprofloxacin onto fresh disks and performing the bioassay. Zones of inhibition measured in penetration experiments were converted to concentrations of the respective antibiotic by using these standard curves. Antibiotic standards were determined in duplicate.
Transmission electron microscopy.
Evidence of local antibiotic action against bacteria in colony biofilms was obtained by transmission electron microscopy. Colony biofilms were fixed with 5% glutaraldehyde for 12 h, stained with osmium tetroxide (1%), and washed. Specimens were then dehydrated in an ethanol series and stained with 1% uranyl acetate-1% phosphotungstic acid. The dehydrated samples were embedded in Spurr's epoxy resin, which was polymerized for 14 h at 70°C. Thin sections were cut, stained with Reynolds lead acetate, and examined by using a JEOL JEM-100CX electron microscope.
The samples prepared for electron microscopy included a 48-h biofilm not exposed to antibiotic (time zero control), a 48-h biofilm transferred to TSA without antibiotic for an additional 12 h (12-h control), a 48-h biofilm transferred to TSA containing 10 μg of tobramycin ml−1 for 36 h (36-h tobramycin), and a 48-h biofilm transferred to TSA containing 1.0 μg of ciprofloxacin ml−1 for 12 h (12-h ciprofloxacin).
Oxygen penetration.
Oxygen concentration profiles in colony biofilms were measured by using a dissolved oxygen microelectrode. The oxygen microelectrode is based on the principle of the common amperometric Clark oxygen electrode and is described in more detail by Jorgensen and Revsbech (17). It consists of an outer casing sealed at the sensor tip with an oxygen-permeable silicone membrane. The casing is fabricated from a Pasteur pipette that is tapered down to an active sensor tip of 15 μm. A carbonate buffer electrolyte fills the internal cavity. Three electrodes occupy the internal cavity: a gold-tipped, glass-encased platinum cathode where oxygen diffusing through the silicone membrane is reduced, a silver-silver chloride counterelectrode which serves as the current return, and a guard electrode which reduces unwanted oxygen entering from the back of the electrode. A potential of −0.8 VDC is applied between the cathode and the common electrode. The current from the cathode, which is proportional to the concentration of oxygen in the bulk external solution, is measured with a picoammeter in the range of 0 to 3 nA. The same potential is applied between the guard electrode and the common electrode to reduce the background signal while measuring low concentrations of oxygen.
The oxygen microelectrode was lowered into the biofilm by a computer-controlled stepping motor with equipment that has been detailed elsewhere (23). Forty-eight-hour colony biofilms were transferred to fresh TSA and probed 2.5 h later. The entire apparatus was housed in an incubator at 37°C. The electrode was calibrated in air, and a zero level was obtained by placing the electrode tip in 0.5% agar containing a suspension (0.2%) of freshly prepared ferrous sulfide (2).
Spatial pattern of protein synthetic activity.
The distribution of protein synthetic activity in the biofilm was visualized with P. aeruginosa FRD1 (pAB1). When the inducing agent IPTG was added to the medium, bacteria carrying the plasmid expressed gfp-mut2 and exhibited intense green fluorescence within a few hours. Because only cells that are capable of de novo protein synthesis can exhibit this response to the inducer, this construct can be used to map the spatial pattern of synthetic activity within the biofilm. Colony biofilms were grown for 48 h on TSA lacking IPTG and then transferred to TSA supplemented with 1 mM IPTG for 4 h. An uninduced control was transferred to TSA without inducer for 4 h, and a positive control was prepared by growing the colony biofilm for 52 h in the continuous presence of IPTG. Colony biofilms were cryoembedded, and frozen sections were examined with a Nikon E800 microscope by using epifluorescent illumination (36).
RESULTS AND DISCUSSION
After 48 h of growth as a colony biofilm, P. aeruginosa FRD1 organisms accumulated to levels of 10.4 ± 0.2 log10 CFU per membrane (n = 11). Since the area covered by the colony was approximately 1 cm2, the areal cell density in these biofilms was of the order of magnitude of 1010 CFU cm−2. The mean biofilm thickness was 233 ± 26 μm. P. aeruginosa colony biofilms were relatively homogeneous with respect to thickness and cell distribution, with no indication of gaps or water channels.
Because the mucoid phenotype of FRD1 is unstable under some conditions, the frequency of reversion to the nonmucoid form was assayed by dispersing untreated 48-h biofilms, plating them, and scoring the resulting colonies for mucoid character. The frequency of reversion to a nonmucoid phenotype was less than one percent over the time course of these experiments.
Tobramycin at 10 μg ml−1 or ciprofloxacin at 1.0 μg ml−1 rapidly killed growing planktonic bacteria (Fig. 1). For comparison, the MICs on TSA were 0.1 and 0.01 μg ml−1 for tobramycin and ciprofloxacin, respectively. MICs on Mueller-Hinton agar were the same. After 4 h of antibiotic exposure, tobramycin-treated P. aeruginosa showed a 5.2 ± 0.2 log reduction and ciprofloxacin-treated cells experienced a 5.9 ± 0.1 log reduction. In the same 4-h period, an untreated control exhibited a log increase of 0.85 ± 0.01, corresponding to a specific growth rate of 0.49 ± 0.02 h−1. Tobramycin at 5 μg ml−1 also effectively killed planktonic bacteria; the log reduction after 4 h of exposure to this concentration of tobramycin was 5.2. The initial cell density in planktonic experiments was 10.0 ± 0.1 log10 CFU ml−1.
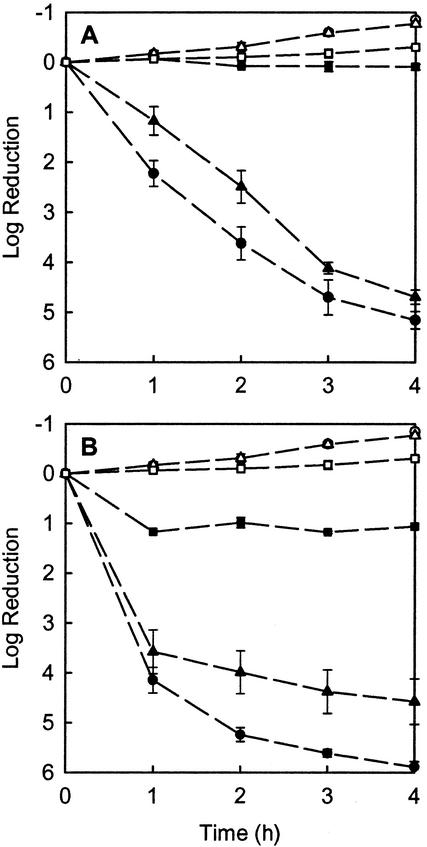
FIG. 1.
Killing of P. aeruginosa by tobramycin (A) or ciprofloxacin (B). Symbols denote planktonic cells (circles), resuspended biofilm cells (triangles), and colony biofilms (squares). Resuspended biofilm cells were dispersed from biofilms immediately prior to antibiotic treatment. Filled symbols indicate antibiotic treatment and open symbols indicate the corresponding untreated controls. Error bars indicate the standard errors of the means. The initial cell densities were 10.4 ± 0.2 log10 CFU per membrane for biofilms, 10.0 ± 0.1 log10 CFU ml−1 for planktonic cells, and 9.9 ± 0.1 log10 CFU ml−1 for resuspended biofilm cells.
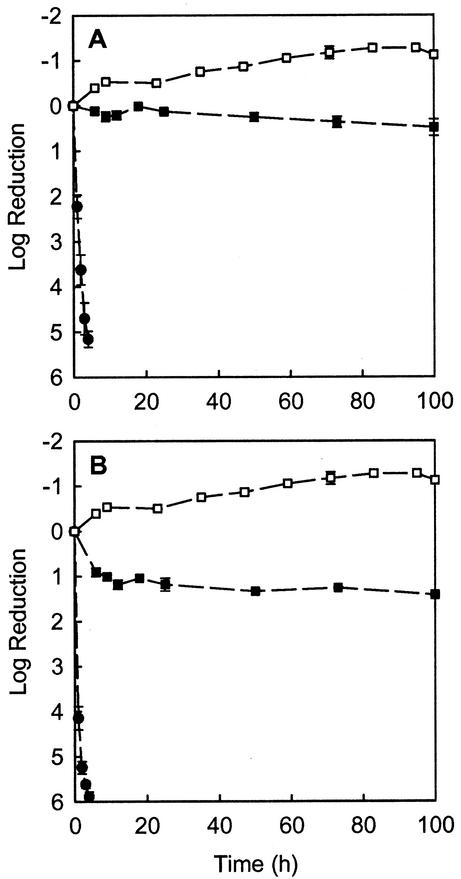
Both antibiotics had little effect on the viability of P. aeruginosa in colony biofilms (Fig. 1). After 4 h of exposure to 10 μg of tobramycin ml−1, the measured change in viable cell numbers was a negligible log increase of 0.09 ± 0.06. Treatment with 1.0 μg of ciprofloxacin ml−1 for 4 h resulted in a log reduction of viable cells of 1.06 ± 0.01. Untreated colony biofilms exhibited a log increase of 0.30 ± 0.03 in 4 h. Even after prolonged exposure to the antibiotics for up to 100 h, there was little additional killing of biofilm bacteria (Fig. 2). After 100 h of treatment in which colony biofilms were moved to fresh antibiotic-containing plates each day, the log reduction in viable cell numbers was 0.49 ± 0.18 for tobramycin at 10 μg ml−1 and 1.42 ± 0.03 for ciprofloxacin at 1.0 μg ml−1.
FIG. 2.
Killing of P. aeruginosa in colony biofilms (squares) upon prolonged exposure to tobramycin (A) or ciprofloxacin (B). Filled symbols indicate antibiotic treatment and open symbols indicate the untreated controls. Killing of planktonic bacteria (filled circles) is shown for reference. Error bars indicate the standard errors of the means. The initial cell densities were 10.4 ± 0.2 log10 CFU per membrane for biofilms and 10.0 ± 0.1 log10 CFU ml−1 for planktonic cells.
When the 48-h colony biofilms, which had not yet been exposed to antibiotics, were resuspended and treated with 10 μg of tobramycin ml−1 or 1.0 μg of ciprofloxacin ml−1, bacterial sensitivity was rapidly restored (Fig. 1). Resuspended biofilm bacteria treated for 4 h with 10 μg of tobramycin ml−1 experienced a 4.7 ± 0.1 log reduction. Resuspended biofilm bacteria treated for 4 h with 1.0 μg of ciprofloxacin ml−1 experienced a 4.6 ± 0.8 log reduction. Resuspended biofilm bacteria that were untreated exhibited a log increase of 0.77 ± 0.01 in the 4-h period.
The preceding results show that the colony biofilm model captures the antibiotic-tolerant phenotype of biofilms and that this tolerance is reversible. When bacteria are dispersed from colony biofilms, their antibiotic sensitivity returns. Two possible protective mechanisms are consistent with these observations. First, the antibiotics may fail to permeate throughout the biofilm. Alternatively, oxygen depletion locally within the biofilm could directly antagonize action of the antibiotic or cause bacteria in the anoxic zone to enter a stationary-phase state in which they are less antibiotic susceptible.
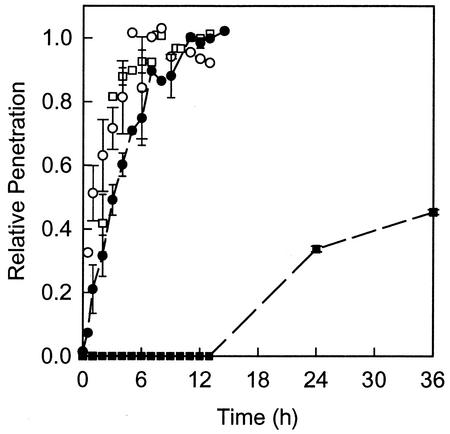
Antibiotic penetration through colony biofilms was measured by using a diffusion cell in which the biofilm was sandwiched between two plastic membranes. A concentration disk of the type used in zone-of-inhibition assays was placed on top of this stack, and the entire assembly was then transferred to an agar plate containing the antibiotic. Penetration data are presented as the ratio of the concentration of antibiotic measured in the disk on the far side of the biofilm to the concentration of antibiotic measured in the agar source (Fig. 3). Control experiments conducted in the presence of biofilm but with no antibiotic yielded no zones of inhibition, showing that P. aeruginosa does not produce inhibitory compounds that interfere with the bioassay. Control experiments in the absence of biofilm and antibiotic also yielded no zones of inhibition, showing that the membranes do not carry residual inhibitory substances. Colony biofilms incubated on antibiotic-containing plates were not able to degrade or otherwise neutralize either of the antibiotics to a significant degree. This was demonstrated by removing colony biofilms from the plate and spreading the plate with a sensitive strain of E. coli. These indicator bacteria were unable to grow on any part of the plate, including the location that had been beneath the colony biofilm.
FIG. 3.
Penetration of tobramycin (squares) and ciprofloxacin (circles) through colony biofilms of P. aeruginosa. Filled symbols indicate an experiment with colony biofilm present and open symbols indicate the sterile controls. Relative penetration is the ratio of the antibiotic concentration measured in the disk on the far side of the membrane assembly to the concentration of antibiotic in equilibrium with the agar. Error bars indicate the standard errors of the means.
Both antibiotics penetrated quickly through the stacked membranes when no biofilm was present (Fig. 3). The concentration of ciprofloxacin in the test disk on the distal side of the colony biofilm reached 50% of the agar concentration within about 1 h in the sterile control, and the concentration of tobramycin reached 50% of the agar concentration within about 2 h in the same type of control experiment. When biofilm was present, ciprofloxacin still penetrated relatively quickly (Fig. 3). Within 3 h, the concentration of ciprofloxacin in the disk on the far side of the colony biofilm attained 50% of the agar concentration. By 12 h, the concentration of ciprofloxacin on the far side of the biofilm was within a few percent of the agar concentration of the antibiotic. Tobramycin, on the other hand, was clearly retarded in its delivery through the biofilm. No tobramycin could be detected in the first 12 h of the experiment. After 36 h, the concentration of tobramycin measured in disks on the far side of the colony biofilm had reached approximately 45% of the agar concentration of the antibiotic.
The slower penetration of tobramycin in comparison to ciprofloxacin is in agreement with published studies of antibiotic permeation of P. aeruginosa biofilms (24, 35). The retarded penetration of tobramycin is likely due to greater binding of this cationic molecule to biofilm components. Adsorption of an antibiotic to the biofilm matrix, even if the antibiotic is not deactivated, will retard its penetration (26). It is possible that the relatively retarded penetration of tobramycin contributes to the protection of the biofilm by allowing more time for bacteria to implement adaptive stress responses (32).
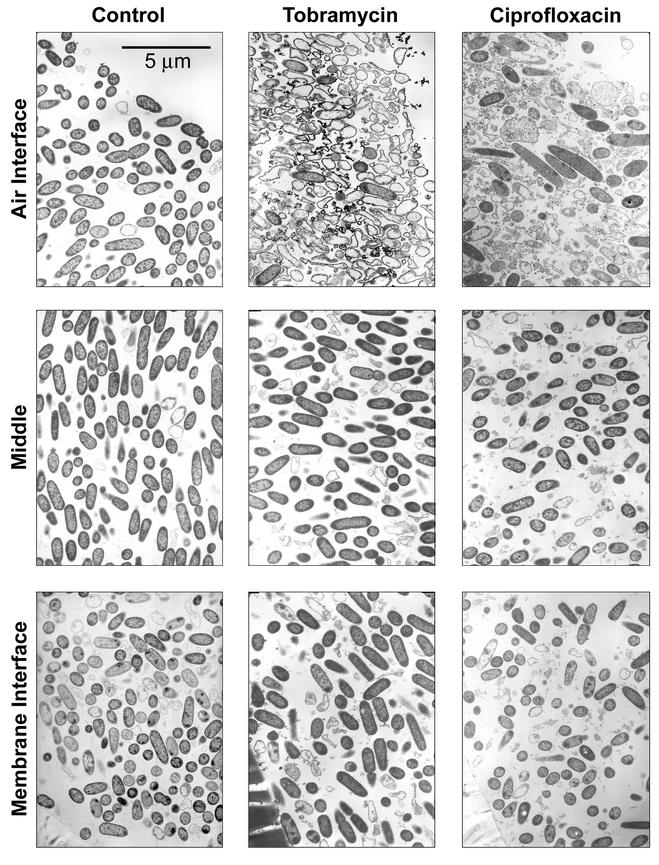
The effect of penetration of tobramycin and ciprofloxacin through P. aeruginosa colony biofilms was confirmed by examining antibiotic-treated and untreated colony biofilms by transmission electron microscopy. In the 12-h control colony biofilm, bacteria were evenly distributed throughout the colony and exhibited the typical rod-shaped morphology that is expected of P. aeruginosa (Fig. 4, left column). The cell shape varied little with location in the biofilm. Bacterial cells in the time zero control were similar in appearance to those in the 12-h control (data not shown). Antibiotic action was evident in both ciprofloxacin- and tobramycin-treated colony biofilms near the air-biofilm interface (Fig. 4). The zone of obvious antibiotic action was approximately 30 to 80 μm thick. This zone of the ciprofloxacin-treated biofilm contained bloated cells, vacuolated cells, and cell debris. Filamentation, which is characteristic of the mode of action of the fluoroquinolones, was also observed near the air-biofilm interface but not in the interior of the biofilms. Near the air interface in the tobramycin-treated biofilm, lysed cells and cell ghosts predominated. As with the ciprofloxacin-treated biofilms, there was little discernible visual evidence of antibiotic action near the polymer membranes or in the middle of tobramycin-treated colony biofilms. These results confirm that the antibiotic was able to penetrate through the entire extent of the colony biofilm. However, the antibiotics acted on the bacteria only at the distal edges of the biofilm while leaving the remainder of the bacteria, including those directly adjacent to the membrane surface, unaffected. No water channels were observed in the colony biofilms, allaying concerns that the antibiotic might have penetrated through such pores without accessing all of the bacteria. This was also confirmed by the fact that antibiotic action was apparent along the full length of the air interface of the colony biofilm.
FIG. 4.
Transmission electron micrographs of P. aeruginosa colony biofilms. The left column is the 12-h untreated control. The middle column shows a biofilm treated with 10 μg of tobramycin ml−1 for 36 h. The right column shows a biofilm treated with 1.0 μg of ciprofloxacin ml−1 for 12 h. As labeled, the rows indicate different locations within the colony.
Though there were differences in the rates of transport, both tobramycin and ciprofloxacin were able to penetrate P. aeruginosa biofilms. Bacterial killing did not accelerate coincidently with the penetration of the antibiotic to the interior of the biofilm. These results demonstrate that poor antibiotic penetration is not the most important protective mechanism in this system.
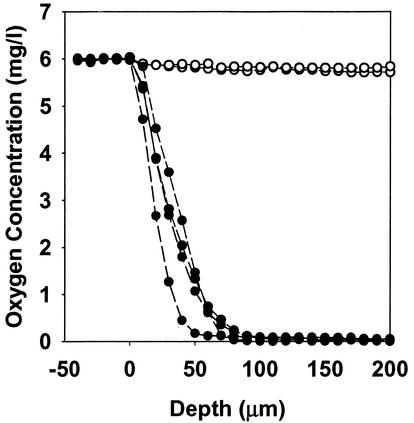
Oxygen limitation and the resulting low metabolic activity may contribute to antibiotic tolerance in these biofilms. The electron microscopic evidence presented above, showing that antibiotic action is focused near the air-biofilm interface, implies a role for oxygen limitation in the observed antibiotic resistance. Therefore, we used oxygen microelectrodes to measure oxygen concentration profiles in these colony biofilms. The results indicated that oxygen penetrated from 50 to 90 μm into colony biofilms formed by P. aeruginosa FRD1 (Fig. 5). The oxygen-replete zone coincides with the region of active metabolism in the biofilm. Spatial patterns of protein synthetic activity in colony biofilms were visualized by using epifluorescence microscopy of frozen biofilm sections following the induction of the GFP. A band of bright fluorescence, 52 ± 5 μm thick, was apparent along the air interface of the biofilm (Fig. 6B). No such band was evident in the uninduced control (Fig. 6A). Since the GFP requires small amounts of oxygen for activity, it is possible that the zone of active fluorescence was limited by oxygen availability. However, a control, in which the biofilm was grown in the continuous presence of the inducer, exhibited bright fluorescence throughout the depth of the biofilm (Fig. 6C).
FIG. 5.
Oxygen concentration profiles in P. aeruginosa colony biofilms. Data for four replicate colonies are shown (filled symbols) along with duplicate profiles in a sterile agar control (open symbols).
FIG. 6.
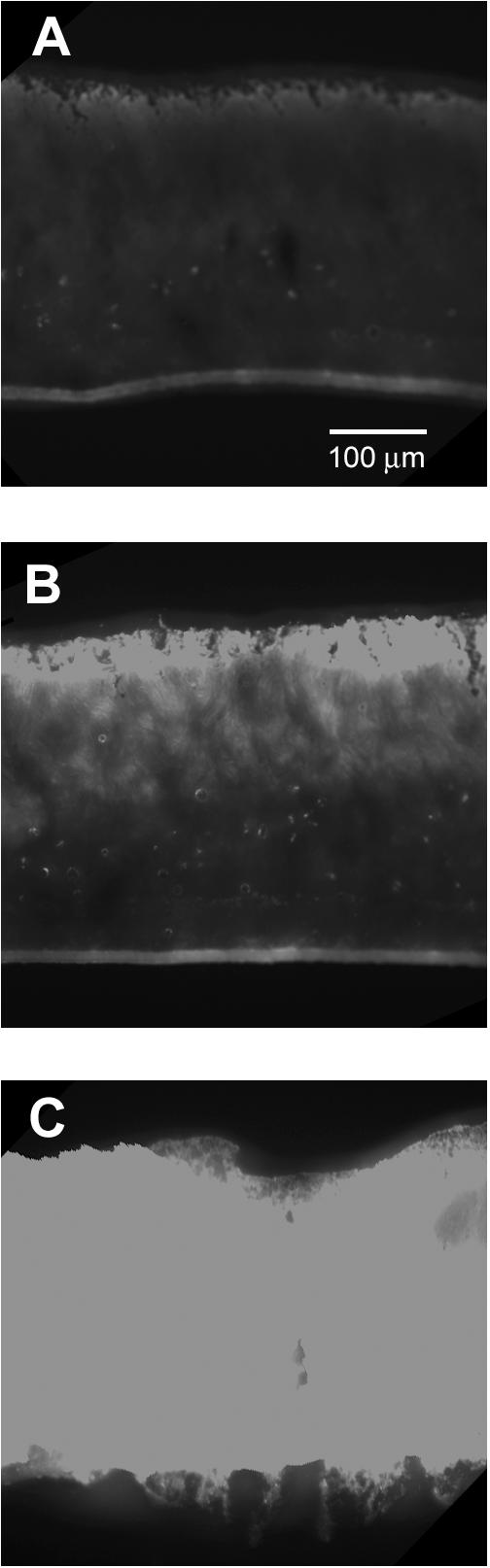
Visualization of the spatial pattern of protein synthetic activity in P. aeruginosa frozen sections of colony biofilms. Strain FRD1(pAB1) was grown for a total of 52 h with no IPTG present (A), for 48 h without IPTG followed by 4 h with IPTG (B), or for 52 h in the continuous presence of IPTG (C). The exposure time was the same for all three panels.
These images of stratified metabolic activity are supported by measurements of the average specific growth rate of bacteria in colony biofilms. In the first 6 h of colony biofilm development, bacteria accumulated rapidly, with a specific growth rate of 0.66 ± 0.04 h−1. For comparison, the maximum specific growth rate during exponential phase in planktonic culture was 0.80 ± 0.04 h−1. In the last 24 h of biofilm development, the average specific growth rate was only 0.013 ± 0.009 h−1. Therefore, the average specific growth rate of biofilm bacteria prior to antibiotic treatment was only about 2% of their growth rate in the initial stages of biofilm formation, suggesting that only those bacteria at the distal edges of the biofilm were replicating.
In summary, tobramycin and ciprofloxacin penetrate biofilms of P. aeruginosa but fail to kill the bacteria. Antibiotic tolerance in this system is likely due to oxygen limitation which restricts bacterial metabolic activity to a narrow zone adjacent to the air interface. Outside this zone, bacteria are not easily killed by the antibiotics. Oxygen gradients are a common feature of life in biofilms (8), and there is some evidence in the literature that anaerobic conditions or slow growth antagonize antibiotic action against P. aeruginosa (7, 10). Other work in our laboratory has shown that P. aeruginosa biofilms exhibit stratified patterns of growth and metabolic activity (33, 34). These results suggest that these stratified patterns may contribute to the antibiotic tolerance of some biofilms.
Acknowledgments
This work was supported through cooperative agreement EEC-8907039 between the National Science Foundation and Montana State University, by the industrial partners of the Center for Biofilm Engineering, by an award from the W. M. Keck Foundation, and in part by Public Health Service grant AI-46588 (M.J.F.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brock, T. D., and K. O'Dea. 1977. Amorphous ferrous sulfide as a reducing agent for culture of anaerobes. Appl. Environ. Microbiol. 33:254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, M. R. W., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-783. [DOI] [PubMed] [Google Scholar]
- 4.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., Z. Lewandowski, D. de Beer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Davey, P., M. Barza, and M. Stuart. 1988. Tolerance of Pseudomonas aeruginosa to killing by ciprofloxacin, gentamicin and imipenem in vitro and in vivo. J. Antimicrob. Chemother. 21:395-404. [DOI] [PubMed] [Google Scholar]
- 8.de Beer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structure on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 9.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 10.Evans, D. J., D. G. Allison, M. R. W. Brown, and P. Gilbert. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27:177-184. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, M. J., and D. E. Ohman. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, C. A., N. A. Hodges, and C. Marriott. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J. Antimicrob. Chemother. 22:667-674. [DOI] [PubMed] [Google Scholar]
- 14.Hatch, R. A., and N. L. Schiller. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:974-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyle, B. D., J. Alcantara, and J. W. Costerton. 1992. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob. Agents Chemother. 36:2054-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, C.-T., P. S. Stewart, and G. A. McFeters. 1998. The study of biofilms by classical fluorescent microscopy, p. 411-429. In M. H. F. Wilkinson and F. Schut (ed.), Digital image analysis of microbes. John Wiley & Sons, Chichester, United Kingdom.
- 17.Jorgensen, B. B., and N. P. Revsbech. 1988. Microsensors. Methods Enzymol. 167:639-659. [Google Scholar]
- 18.Kumon, H., K. Tomochika, T. Matunaga, M. Ogawa, and H. Ohmori. 1994. A sandwich cup method for the penetration assay of antimicrobial agents through Pseudomonas exopolysaccharides. Microbiol. Immunol. 38:615-619. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols, W. W., S. M. Dorrington, M. P. E. Slack, and H. L. Walmsley. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob. Agents Chemother. 32:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen, K., and Z. Lewandowski. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59:302-309. [DOI] [PubMed] [Google Scholar]
- 24.Shigeta, M., G. Tanaka, H. Komatsuzawa, M. Sugai, H. Suginaka, and T. Usui. 1997. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy 43:340-345. [DOI] [PubMed] [Google Scholar]
- 25.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart, P. S. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 29.Stoodley, P., D. de Beer, and Z. Lewandowski. 1994. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suci, P., M. W. Mittelman, F. P. Yu, and G. G. Geesey. 1994. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:2125-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrany, J. D., P. S. Stewart, and P. A. Suci. 1997. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa biofilms displaying rapid-transport characteristics. Antimicrob. Agents Chemother. 41:1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 33.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 34.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasuda, H., Y. Ajiki, T. Koga, H. Kawada, and T. Yokota. 1993. Interaction between biofilms formed by Pseudomonas aeruginosa and clarithromycin. Antimicrob. Agents Chemother. 37:1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, F. P., G. M. Callis, P. S. Stewart, T. Griebe, and G. A. McFeters. 1994. Cryosectioning of biofilms for microscopic examination. Biofouling 8:85-91.
- 37.Zahller, J., and P. S. Stewart. 2002. A transmission electron microscopic study of antibiotic action on Klebsiella pneumoniae biofilm. Antimicrob. Agents Chemother. 46:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng, Z., and P. S. Stewart. 2002. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 46:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]