Abstract
The most important antigen component of a promising multicomponent group B meningococcal recombinant protein vaccine is based on genome-derived neisserial antigen 1870, which recently was renamed factor H-binding protein (FHBP) to reflect one of its critical functions as a complement regulatory protein. Neisseria meningitidis strains can be subdivided into three FHBP variant groups based on divergence of FHBP amino acid sequences. Within each variant group, amino acid sequences are >90% conserved. To develop an FHBP-based group B vaccine, it is important to know the distribution of FHBP variant 1, 2, and 3 strains in different geographic regions, since antibodies against FHBP are bactericidal against strains within the homologous group but show minimal activity against strains from other groups. We have devised a high-throughput, quantitative PCR-based method that allows rapid and precise assignment of FHBP genes into each of the three major variant lineages. Among 48 group B isolates from patients hospitalized in California in 2003 to 2004, 83%, 13%, and 4%, respectively, had variant 1, 2, and 3 genes. Thus, a vaccine based on the variant 1 protein has the potential to prevent the majority of cases of group B disease. The quantitative PCR-based method will be useful for determining and monitoring the prevalence of meningococcal isolates with genes encoding different FHBP variant proteins. The technique also is suitable for monitoring variation of genes encoding other protein antigens targeted for vaccination.
Neisseria meningitidis is an important cause of bacterial sepsis and meningitis worldwide. Nearly all pathogenic meningococci comprise one of five capsular groups. Safe and effective vaccines are available that target the capsular polysaccharides of groups A, C, W-135, and Y. However, there is no broadly effective vaccine against group B, which is responsible for approximately 30% of meningococcal disease in the United States (17) and 50% to 90% of disease in Europe (2).
The group B capsular polysaccharide is an autoantigen and is therefore poorly immunogenic. Also, there are potential safety issues that are difficult to resolve for a vaccine that elicits autoantibodies. An alternative approach that avoids these concerns targets noncapsular antigens. A number of genes that encode new meningococcal vaccine candidates was identified from the genome sequences of several N. meningitidis strains (13, 20). Bioinformatic analysis was used to predict proteins that were conserved and potentially exposed on the surface of the bacteria (15). One of the proteins identified, designated genome-derived neisserial antigen 1870 (GNA1870) (also described by other authors as lipoprotein 2086 [15]), is a 29-kDa lipoprotein that recently was renamed factor H-binding protein (FHBP) to reflect a critical function of this molecule in down-regulation of the alternative complement pathway and enhancement of serum resistance (9). By flow cytometry, FHBP is surface exposed on live encapsulated N. meningitidis cells, and recombinant FHBP induces serum bactericidal antibodies in mice (11, 23). The mouse sera also passively conferred protection against meningococcal bacteremia in an infant rat model (11, 23).
Three FHBP variant groups (1, 2, and 3) have been identified based on divergence of their deduced amino acid sequences. FHBP from strains within these variant groups exhibits >90% amino acid identity, and there is approximately 63% to 85% identity in FHBP among variant groups (11). Vaccination of mice with each of the three FHBP variant recombinant proteins induced antibodies that were bactericidal against strains of the same FHBP variant group but much less so against strains in the other groups (11). Since the original publication by Masignani et al. (11), two additional studies have provided experimental support for cross-reactive protective activity of antibodies among strains expressing subvariants within a variant group (6, 23).
A promising multicomponent group B recombinant protein vaccine that is under development uses a variant 1 FHBP protein as one of its principal antigens (11, 23). In previous studies, approximately half to two-thirds of disease-producing group B isolates expressed variant 1 FHBP proteins (11, 23). Data on the prevalence and diversity of FHBP variant groups among strains from different geographic regions have important implications for monitoring the potential effectiveness of an FHBP-based vaccine. Herein we describe a high-throughput quantitative PCR (QPCR)-based method that allows rapid and precise assignment of FHBP genes into each of the three major variant lineages.
MATERIALS AND METHODS
Cloning of FHBP and construction of knockout strains.
The FHBP variant genes from three prototype strains, MC58, 961-5945, and M1239 (variants 1, 2, and 3, respectively), were cloned into an Escherichia coli expression plasmid, pET21b (Novagen, Madison, WI), as previously described (11). FHBP knockout strains were constructed as previously described (11).
Growth of meningococci.
N. meningitidis strains were stored in sterile, 10% nonfat milk at −80°C prior to use. The strains were subcultured on chocolate agar plates (Remel, Lenexa, KS) and incubated for ∼16 h at 37°C in 5% CO2. Liquid cultures were grown in Mueller-Hinton broth (BD Biosciences, Franklin Lakes, NJ) supplemented with 0.25% d-glucose (Sigma-Aldrich, St. Louis, MO) to an optical density at 620 nm of 0.6. Cultures were either heat killed at 56°C for 1 h or used directly for genomic DNA preparation.
DNA purification.
One milliliter of N. meningitidis culture (∼109 CFU/ml), grown as described above, was used for isolation of genomic DNA. The bacteria were processed using the DNeasy tissue kit (QIAGEN, Chatsworth, CA). The purified DNA was eluted in a 200-μl volume of elution buffer AE. Five microliters of the eluate was tested for sterility by being plated on chocolate agar and incubated as described above. DNA concentrations were determined by measuring the absorbance at 260 nm using an extinction coefficient of 20 g−1 · cm−1 · liter.
Plasmid DNA was isolated from 100-ml cultures of E. coli grown to stationary phase. Plasmid DNA was purified using the Hi-Speed Midi kit (QIAGEN). DNA was eluted in a 500-μl volume of elution buffer (10 mM Tris · Cl, pH 8.0), and the concentration was determined as described above.
DNA sequencing.
FHBP DNA sequences were determined commercially, using primer sequences that had been reported previously (11). Sequencing templates were PCR products amplified from genomic DNA or heat-killed liquid cultures.
Conventional and quantitative PCR. (i) Primer design.
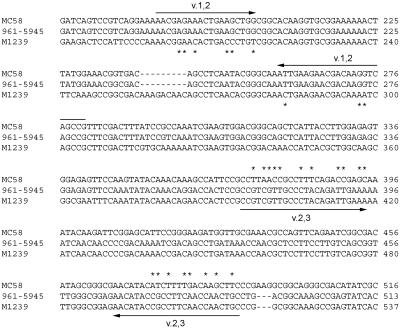
Oligonucleotide primers were designed using Primer3 software (Whitehead Institute, Cambridge, MA) to yield thermal transition midpoint values of 60°C and amplicon sizes of 100 to 200 bp. Through alignment of all N. meningitidis FHBP sequences in GenBank (n = 71), we performed sequence alignments using ClustalW (3) to identify regions that were absolutely conserved within each variant group. Based on these alignments, oligonucleotide primers were designed to recognize either variants 1 and 2 (v.1 and v.2, also called primer pair v.1,2) or variants 2 and 3 (v.2 and v.3, also called primer pair v.2,3) (Fig. 1). Two primer pairs is the minimum needed to discriminate among three variant groups, thus maximizing the number of strains that can be examined in one grouping experiment.
FIG. 1.
DNA sequence alignment of FHBP from prototype strains. FHBP from strains MC58 (variant 1; GenBank accession no. AE002098), 961-5945 (variant 2; DQ523568), and M1239 (DQ523569) were aligned using ClustalW (3). The variant 1- and 2-specific (v.1,2) primers are shown above the alignment, and the variant 2- and 3-specific (v.2,3) primers are shown below the alignment. Where the primer sequences differ from one of the FHBP sequences, an asterisk is shown adjacent to the divergent sequence. Only the regions used for primer design are shown; the numbering with respect to the full-length open reading frames is shown at the right.
The oligonucleotides used were v.1,2_f (5′-AAACGAGAAACTGAAGCTGGCGG), v.1,2_r (5′ CGGCTGACCTTGTCGTTCTTCAAT), v.2,3_f (5′-CCGTCGTTGCCCTACAGATTGAAA), and v.2,3_r (5′-GCAGTTGGTTGAAGGCGGTATGTT). The primers used for the 16S rRNA control reactions were 16S_f (5′-TGCTTGGTAGCGTAGCTAACGC) and 16S_r (5′-TTAATCCACATCATCCACCGC).
(ii) Conventional PCR.
PCR was performed using plasmid (1 ng) or genomic DNA (10 ng) templates. PCRs consisted of 1× ThermoPol buffer (New England Biolabs, Beverly, MA), 200 μM deoxynucleoside triphosphates, 200 nM each forward and reverse primers, and Taq DNA polymerase (New England Biolabs). Cycling conditions were 95°C for 10 min and 30 cycles of 94°C for 15 s, followed by 60°C for 30 s and then 72°C for 5 min. Products were separated on a 2% agarose gel in sodium borate buffer (1) and visualized by staining with ethidium bromide.
(iii) Quantitative PCR.
QPCR was performed using an ABI 7900HT instrument (Applied Biosystems, Foster City, CA) and SYBR green detection. Reactions consisted of 1× SYBR green master mix (Applied Biosystems) and 400 nM each forward and reverse primers. QPCR templates were either plasmid (1 ng) or genomic DNA (10 ng) or ∼106 CFU equivalents of a liquid culture (optical density, 0.6) washed in phosphate-buffered saline. For colony QPCR, an ∼1-mm-diameter colony was picked into 100 μl of 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and the cells were killed by incubating them for 1 h at 56°C. Reactions were assembled in a 22-μl volume and then distributed into quadruplicate 4.5-μl reaction mixtures in a 384-well optical plate (Applied Biosystems) and sealed with optical tape (Applied Biosystems). Cycling parameters were as described above for conventional PCR, followed by a dissociation step for measurement of thermal melting transition temperatures. Amplification data were exported and reduced using Microsoft Excel. All data shown are the averages of two to five replicates.
RESULTS
FHBP molecular grouping of prototype strains.
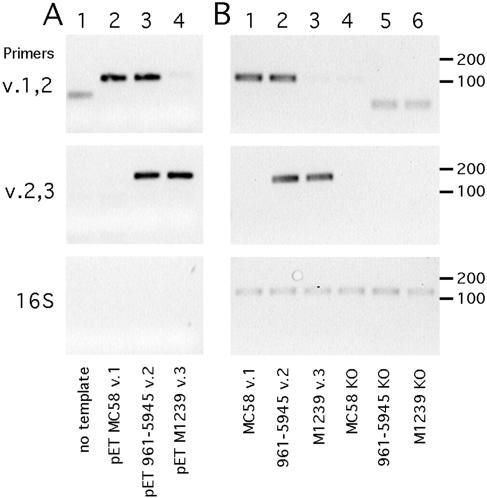
To demonstrate the ability of the molecular grouping method to discriminate among variants, we first established a conventional PCR approach. Primer pairs specific for variants 1 and 2 (v.1,2) or variants 2 and 3 (v.2,3) were used to amplify plasmid clones of FHBP (Fig. 2A). The v.1,2 primer pair efficiently amplified FHBP from strains MC58 (variant 1; upper panel, lane 2) and 961-5945 (variant 2; lane 3), but not M1239 (variant 3; lane 4) or a no-template control reaction (lane 1). Similarly, the v.2,3 primer pair efficiently amplified FHBP from strains 961-5945 and M1239 but not from strain MC58 or a no-template control reaction (Fig. 2A, middle panel). Primers specific for 16S rRNA failed to amplify any of the plasmid templates (Fig. 2A, lower panel).
FIG. 2.
Results of a conventional PCR-based assay for FHBP grouping. Three primer pairs, specific for FHBP variants 1 and 2 (v.1,2), variants 2 and 3 (v.2,3), and 16S rRNA genes, were used to amplify cognate and noncognate templates. The positions of DNA size standards in base pairs are shown at the right. (A) Variant-specific detection of plasmid templates. Lane 1, no template control; 2, pET21b-FHBP (strain MC58, variant 1); 3, pET21b-FHBP (961-5945, variant 2); 4, pET21b-FHBP (M1239, variant 3). (B) Detection of genomic DNA templates. Lane 1, MC58 wild type (WT); 2, 961-5945 WT; 3, M1239 WT; 4, MC58 FHBP knockout (KO); 5, 961-5945 FHBP KO; 6, M1239 FHBP KO.
Using genomic DNA templates, similar results were obtained. The v.1,2 primer pair specifically amplified its cognate targets (Fig. 2B, upper panel, lanes 1 to 2) but not a noncognate target (lane 3) or isogenic FHBP knockout strains (lanes 4 to 6). Similar results were obtained for the v.2,3 primer pair (Fig. 2B, middle panel). Primers specific for 16S rRNA amplified all genomic DNA templates with similar efficiencies (Fig. 2B, lower panel). Some products were observed for FHBP variant-specific primers with no template or noncognate templates; however, these products were faint and smaller in size than specific products, and they were easily distinguished from specific products.
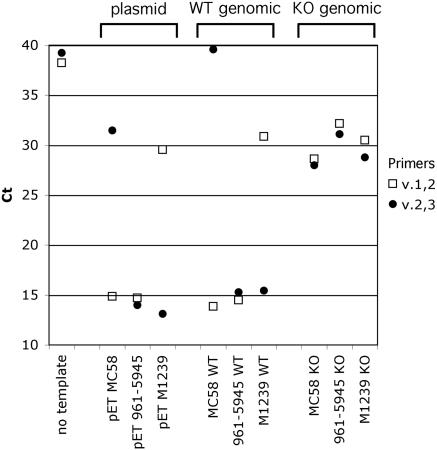
To enhance the speed, throughput, and quantification of the FHBP grouping method, we implemented a QPCR approach. The same templates used above were amplified in QPCR reactions using SYBR green fluorescence to detect double-stranded DNA products (Fig. 3). The MC58 FHBP (variant 1) plasmid clone gave a low cycle threshold (CT) value (indicative of a high signal) for the v.1,2 primer pair but a high CT (low signal) for the v.2,3 primer pair. The 961-5945 FHBP (variant 2) plasmid gave low CT values (high signals) for both v.1,2 and v.2,3 primer pairs. The M1239 FHBP (variant 3) plasmid gave a high CT for the v.1,2 primer pair and a low CT for the v.2,3 primer pair. A no-template control reaction gave high CT values for both primer pairs (Fig. 3). Overall, the QPCR approach gave similar results compared with the conventional PCR approach.
FIG. 3.
Results of a quantitative PCR-based assay for FHBP grouping of prototype variant 1, 2, and 3 strains. The CT is plotted versus template source. A low CTcorresponds with a high PCR amplification signal. Plasmid samples are pET plasmid FHBP clones from strains MC58 (variant 1), 961-5945 (variant 2), and M1239 (variant 3); WT genomic samples are wild-type (WT) genomic DNA from the same strains as the respective plasmid samples; KO genomic samples are FHBP knockout (KO) genomic DNA from the same respective strains as plasmid and WT genomic samples.
The same respective patterns of CT values were seen for the three FHBP variants when genomic DNA was used as the template for QPCR (Fig. 3, middle). For the isogenic strains bearing genetic knockouts of FHBP, the 16S rRNA primers yielded an amplicon at a low CT value (data not shown), but neither of the variant-specific primer pairs did so (Fig. 3, right). As with the conventional PCR approach, small amounts of product were detected for noncognate primer-template combinations, which is likely a result of weak cross-reactivity between the noncognate primers and the genomic DNA template from the strain. However, the values obtained with the noncognate primers were typically >12 CT cycles higher than for the respective cognate combinations. The smallest difference observed was 5 CT cycles, which still reflects a 32-fold difference in QPCR product. Thus, there was no difficulty in distinguishing positive from negative QPCR results.
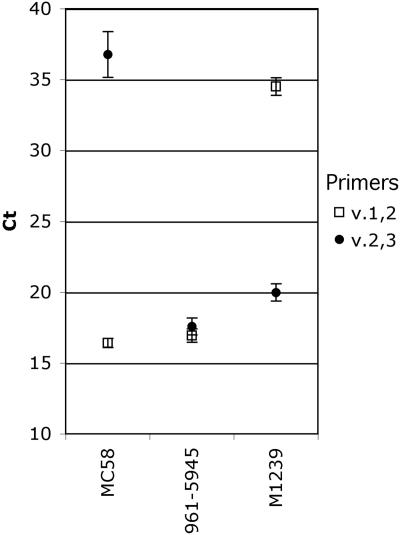
Next we tested the feasibility of performing quantitative PCR directly from bacterial colonies, which enhances potential throughput of the method. From an overnight culture on chocolate agar, we suspended five colonies of each of the three prototype variant 1, 2, and 3 control strains into a small volume of buffer solution. The respective results obtained from colony QPCR were similar to those of the other template sources tested (plasmid or genomic DNA or liquid bacterial culture). The mean values for positive signals from the bacterial colony suspensions were between 16.1 and 20.0, and the values for negative signals were 34.5 to 36.8 (Fig. 4). The standard deviation (1σ) was between 0.3 and 1.6.
FIG. 4.
Results of a QPCR using bacterial colony suspensions as templates. A single colony from each prototype strain was suspended in buffer in five replicates, which were used as templates in QPCR with v.1,2 and v.2,3 primer pairs. Data points represent the mean CTvalues, and error bars correspond to the standard deviations (1σ).
FHBP grouping of diverse strains.
To examine the performance of FHBP variant-specific primers on a larger scale, we performed QPCR-based FHBP grouping of 14 strains of a known FHBP variant group that represented the genetic diversity of each of the variant groups based on the phylogeny of the 71 FHBP DNA sequences represented in GenBank. The sources and characteristics of the 14 strains are summarized in Table 1. We performed the QPCR reactions using heat-killed N. meningitidis cells as PCR templates.
TABLE 1.
Meningococcal strains used in this study
| Strain (reference[s]) | Country of origin | Yr isolated | FHBP variant group | Serologic classification | Electrophoretic type (sequence type)a |
|---|---|---|---|---|---|
| MC58 (12, 20) | United Kingdom | 1985 | 1 | B:15:P1.7,16 | ET5 complex (74) |
| MC58ΔFHBP (11) | United Kingdom | 1985 | Not applicable | B:15:P1.7,16 | ET5 complex (74) |
| 4243 (14) | United States | 1994 | 1b | C:2a:P1.5,2 | ET37 complex (11) |
| M6190 | United States | 1999 | 1b | B:2a:P1.5,2 | ET37 complex (1988) |
| NZ98/254 (10) | New Zealand | 1998 | 1b | B:4:P1.4 | Lineage 3 (42) |
| M1390 | United States | 1995 | 1b | B:15:P1.7,4 | Lineage 3 (41) |
| M4105 | United States | 1996 | 1b | B:4,7:P1.7,4 | Lineage 3 (154) |
| Z2491 (5) | The Gambia | 1983 | 1b | A:4:P1.7 | Subgroup IV-1 (4) |
| RM1090 | United States | Pre-1995 | 2b | C:2a:P1.5-1,2-2 | Unknown |
| 961-5945 | Austria | 1996 | 2 | B:2b:P1.21,16 | A4 (153) |
| 2996 (22) | United Kingdom | 1975 | 2 | B:2b:P1.19,15 | Unknown (540) |
| 8047 | United States | 1978 | 2 | B:2b:P1.5-1,2-2 | Unknown |
| M1239 | United States | 1994 | 3 | B:14:P1.23,14 | Lineage 3 (437) |
| M01-0240364 | United Kingdom | 2001 | 3 | B:2a:P1.5,2 | ET37 complex (11) |
Sequence type was determined previously by multilocus sequence typing (http://www.mlst.net).
Subvariants of the primary group as defined by the FHBP gene sequence from MC58 (variant 1) or 2996 (variant 2).
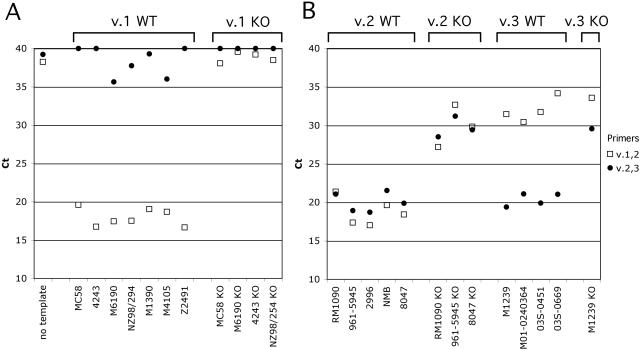
The FHBP variant grouping of these 13 strains and the respective isogenic FHBP knockout mutants of 8 of these strains is shown in Fig. 5. All of the strains harboring an FHBP variant 1 gene gave the characteristic pattern of a low CTvalue for v.1 and v.2 primers and a high CTfor v.2 and v.3 primers (Fig. 5A, left). In contrast, the isogenic FHBP knockout mutants gave similar respective CTvalues for 16S rRNA (data not shown) but high CTvalues with both of the v.1,2 and v.2,3 FHBP primer pairs (Fig. 5A, right).
FIG. 5.
QPCR grouping using as PCR template heat-killed N. meningitidis cells from strains with FHBP variant group defined by DNA sequence (Table 1). (A) QPCR data for FHBP variant 1 strains. Left, variant 1 wild-type (WT) strains; right, variant 1 knockout (KO) strains. (B) QPCR data for FHBP variant 2 and 3 strains. Descriptions correspond to those for panel A. Two disease isolates from California, 03S-0451 and 03S-0669, are shown as additional examples of variant 3 strains (confirmed by DNA sequence determination; Table 2).
The five strains bearing an FHBP variant 2 gene gave the characteristic pattern of low CTvalues for v.1,2 and v.2,3 primer pairs (Fig. 5B, first set). The isogenic knockout mutants gave the expected pattern of high CTvalues for both primer pairs (second set). The four FHBP variant 3 strains gave a low CTfor v.2 and v.3 primers but a high CTfor v.1 and v.2 primers (third set). The M1239 FHBP variant 3 knockout control gave high CTvalues for both of the variant-specific primer pairs (fourth set).
FHBP grouping of disease isolates from California.
We next applied the FHBP molecular grouping method to a collection of capsular group B isolates submitted to the California State Health Department between 2003 and 2004. The isolates were from 48 patients with meningococcal disease who were hospitalized in 22 counties. By QPCR-based grouping, the majority of the isolates had FHBP variant group 1 genes (83%), with a smaller representation of variant 2 (13%) and variant 3 genes (4%).
To validate the QPCR approach on these unknown strains, we determined the sequences of the FHBP genes of 15 of the California isolates. These isolates included the eight that had been identified by QPCR as having FHBP variant 2 or 3 genes and a subset of 7 of the 40 isolates identified by QPCR as having variant 1 genes. The deduced FHBP amino acid sequences of the 15 isolates were compared to the variant 1, 2, and 3 prototype strains to determine the sequence identity (Table 2). This analysis showed that the QPCR grouping method accurately identified the variant group of each strain as judged from DNA sequencing.
TABLE 2.
FHBP-deduced amino acid sequence identity among selected isolates from patients in Californiaa
| Strain | QPCR variant | FHBP amino acid sequence identity (%)
|
||
|---|---|---|---|---|
| MC58 (variant 1) | 961-5945 (variant 2) | M1239 (variant 3) | ||
| 03S-0355b | 1 | 100.0 | 73.1 | 55.1 |
| 03S-0408 | 1 | 96.4 | 73.5 | 55.1 |
| 03S-0444 | 1 | 93.1 | 74.2 | 54.4 |
| 03S-0501 | 1 | 93.5 | 74.2 | 54.8 |
| 04S-0013 | 1 | 97.8 | 73.8 | 55.8 |
| 04S-0017 | 1 | 93.1 | 73.5 | 54.4 |
| 04S-0058b | 1 | 100.0 | 73.1 | 55.1 |
| 03S-0150 | 2 | 72.4 | 98.5 | 77.0 |
| 03S-0321 | 2 | 72.7 | 100.0 | 78.4 |
| 03S-0658 | 2 | 70.5 | 94.5 | 80.5 |
| 03S-0673 | 2 | 70.5 | 94.5 | 80.5 |
| 03S-0691 | 2 | 72.7 | 100.0 | 78.4 |
| 04S-0062 | 2 | 69.8 | 95.6 | 80.5 |
| 03S-0451 | 3 | 54.8 | 78.9 | 91.5 |
| 03S-0669 | 3 | 55.5 | 79.4 | 95.7 |
FHBP DNA sequencing was performed on all of the California isolates grouped as variant 2 or 3 by QPCR and on 7 of the 40 isolates identified as variant 1. The closest match to one of the three FHBP prototype sequences is shown in boldface.
Positive for reactivity with the MAb JAR-1 (23), which was raised against the FHBP from strain MC58.
DISCUSSION
Previously, DNA sequence typing has been used for well-characterized meningococcal antigens, such as the major porin PorA (19) as well as FetA (21), and more recently for FHBP (6, 11). A conventional PCR-based approach also has been used to assign capsular groups (16) and PorB variable region types (18).
DNA sequencing of the genes encoding the new meningococcal vaccine antigen, designated GNA1870 or FHBP, is useful to identify common or representative versions of the three variant groups of this promising vaccine candidate. Since DNA sequence determination of one or more genes from many strains is resource intensive, we developed a rapid, high-throughput assay to distinguish strains from different FHBP variant groups. We considered using the same respective primers for performing conventional PCR and identifying specific products by electrophoretic mobility in agarose gels. However, compared with the typing methods for PorB and capsular group mentioned above or conventional PCR, the QPCR method has advantages of being more rapid, relatively inexpensive, and scalable to grouping a large number of isolates in a single day.
A number of monoclonal antibodies (MAbs) have been raised against FHBP variant 1 (7, 23). However, to date, there are no MAbs that recognize all strains within a variant group. One of the MAbs, JAR-1, binds to all strains expressing the canonical variant 1 protein (examples are strains MC58, H44/76, CU385, and others from the hypervirulent lineage associated with electrophoretic type 5) (23). In the present study, all of the California meningococcal isolates recognized by MAb JAR-1 in a whole-cell dot blot assay (n = 35) had FHBP variant 1 genes by QPCR. Five additional California isolates identified as having FHBP variant 1 genes by QPCR were negative for binding to MAb JAR-1. By DNA sequencing, these five isolates were confirmed as having genes encoding FHBP variant 1 (Table 2). FHBP group assignments of all of the isolates that were identified as variant 2 or 3 by QPCR also were confirmed by DNA sequencing of the FHBP gene. Thus, the QPCR method provided a precise grouping designated for FHBP variant 1, 2, and 3 strains.
To develop an FHBP-based group B vaccine, it is important to know the distribution of variant 1, 2, and 3 FHBP strains in different geographic regions, since antibodies raised to the variant 1 protein are bactericidal against most FHBP variant 1 strains but show little functional activity against variant 2 or 3 strains (6, 23). In a previous study by Masignani et al., FHBP variant 1 genes constituted 54% of strains examined (11). These data suggested that at least two FHBP proteins (variants 1 and 2) might be required for an FHBP-based vaccine to provide broad coverage against strains causing most group B meningococcal disease. However, the strains in the study by Masignani et al. were selected to be representative of the known genetic diversity among N. meningitidis strains and, therefore, most likely included a disproportionate number of rare isolates chosen on the basis of genetic diversity rather than disease prevalence.
In the present study, FHBP variant 1 was present in 83% of group B isolates obtained during 2003 to 2004 from patients residing in 22 counties in California. By DNA sequencing, variant 1 also was present in 69% of recent isolates from Swedish patients (8). Thus, the variant 1 protein is the most prevalent of the three variants, and a single FHBP protein has the potential to prevent the majority of cases of disease. In devising a multicomponent group B recombinant protein vaccine, it therefore may be preferable to include FHBP variant 1 and other unrelated, promising vaccine antigens, such as GNA2132 (15) and/or NadA (4). Adding FHBP variant 2 and/or variant 3 proteins may provide less of a benefit, since the immune response to several antigenically unrelated proteins may help avoid selection of escape mutants. Such a strategy, however, underscores the need to determine the prevalence of different variants of FHBP among strains from additional geographic areas and to monitor carefully for emergence of FHBP variant 2 or 3 N. meningitidis strains after a vaccine is introduced. The QPCR method described herein is rapid and precise and can be applied to large numbers of isolates in a single day. The method also is potentially suitable for monitoring genetic variation of the other proteins targeted for vaccination.
Acknowledgments
This work was supported by Public Health Service grants R01 AI46464 and R21 AI061533 from the National Institute of Allergy and Infectious Diseases, NIH. The investigation was conducted in a facility constructed with support from Research Facilities Improvement Program grant number C06 RR16226 from the National Center for Research Resources, NIH.
We thank Will Probert, Chief of Enterics and Special Pathogens Section, California Department of Health Services, Richmond, CA, for providing the group B isolates from patients hospitalized in California; Oliver Koeberling for the pET21b-FHBP (strain 961-5945) plasmid; Jo Anne Welsch and Maggie Ching for cell suspensions of the group B disease isolates; and Carlos Grenier for preparation of DNA sequencing samples.
REFERENCES
- 1.Brody, J. R., and S. E. Kern. 2004. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques 36:214-216. [DOI] [PubMed] [Google Scholar]
- 2.Cartwright, K., N. Noah, and H. Peltola. 2001. Meningococcal disease in Europe: epidemiology, mortality, and prevention with conjugate vaccines. Report of a European advisory board meeting Vienna, Austria, 6-8 October, 2000. Vaccine 19:4347-4356. [DOI] [PubMed] [Google Scholar]
- 3.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowe, B. A., R. A. Wall, B. Kusecek, B. Neumann, T. Olyhoek, H. Abdillahi, M. Hassan-King, B. M. Greenwood, J. T. Poolman, and M. Achtman. 1989. Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in The Gambia, West Africa. J. Infect. Dis. 159:686-700. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher, L. D., L. Bernfield, V. Barniak, J. E. Farley, A. Howell, M. Knauf, P. Ooi, R. P. Smith, P. Weise, M. Wetherell, X. Xie, R. Zagursky, Y. Zhang, and G. W. Zlotnick. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 72:2088-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giuliani, M. M., L. Santini, B. Brunelli, A. Biolchi, B. Arico, F. Di Marcello, E. Cartocci, M. Comanducci, V. Masignani, L. Lozzi, S. Savino, M. Scarselli, R. Rappuoli, and M. Pizza. 2005. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect. Immun. 73:1151-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobsson, S., S. Thulin, P. Molling, M. Unemo, M. Comanducci, R. Rappuoli, and P. Olcen. 2006. Sequence constancies and variations in genes encoding three new meningococcal vaccine candidate antigens. Vaccine 24:2161-2168. [DOI] [PubMed] [Google Scholar]
- 9.Madico, G., J. A. Welsch, L. A. Lewis, A. McNaughton, D. H. Perlman, C. E. Costello, J. Ngampasutadol, U. Vogel, D. M. Granoff, and S. Ram. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 10.Martin, D. R., S. J. Walker, M. G. Baker, and D. R. Lennon. 1998. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J. Infect. Dis. 177:497-500. [DOI] [PubMed] [Google Scholar]
- 11.Masignani, V., M. Comanducci, M. M. Giuliani, S. Bambini, J. Adu-Bobie, B. Arico, B. Brunelli, A. Pieri, L. Santini, S. Savino, D. Serruto, D. Litt, S. Kroll, J. A. Welsch, D. M. Granoff, R. Rappuoli, and M. Pizza. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuinness, B. T., I. N. Clarke, P. R. Lambden, A. K. Barlow, J. T. Poolman, D. M. Jones, and J. E. Heckels. 1991. Point mutation in meningococcal porA gene associated with increased endemic disease. Lancet 337:514-517. [DOI] [PubMed] [Google Scholar]
- 13.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 14.Pastor, P., F. B. Medley, and T. V. Murphy. 2000. Meningococcal disease in Dallas County, Texas: results of a six-year population-based study. Pediatr. Infect. Dis. J. 19:324-328. [DOI] [PubMed] [Google Scholar]
- 15.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 16.Pollard, A. J., G. Probe, C. Trombley, A. Castell, S. Whitehead, J. M. Bigham, S. Champagne, J. Isaac-Renton, R. Tan, M. Guiver, R. Borrow, D. P. Speert, and E. Thomas. 2002. Evaluation of a diagnostic polymerase chain reaction assay for Neisseria meningitidis in North America and field experience during an outbreak. Arch. Pathol. Lab. Med. 126:1209-1215. [DOI] [PubMed] [Google Scholar]
- 17.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 18.Sacchi, C. T., A. P. de Lemos, A. M. Whitney, C. E. Melles, C. A. Solari, C. E. Frasch, and L. W. Mayer. 1998. The use of oligonucleotide probes for meningococcal serotype characterization. Rev. Inst. Med. Trop. Sao Paulo 40:113-117. [DOI] [PubMed] [Google Scholar]
- 19.Sacchi, C. T., A. P. Lemos, M. E. Brandt, A. M. Whitney, C. E. Melles, C. A. Solari, C. E. Frasch, and L. W. Mayer. 1998. Proposed standardization of Neisseria meningitidis PorA variable-region typing nomenclature. Clin. Diagn. Lab. Immunol. 5:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149:1849-1858. [DOI] [PubMed] [Google Scholar]
- 22.Van der Ley, P., and J. T. Poolman. 1992. Construction of a multivalent meningococcal vaccine strain based on the class 1 outer membrane protein. Infect. Immun. 60:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsch, J. A., R. Rossi, M. Comanducci, and D. M. Granoff. 2004. Protective activity of monoclonal antibodies to genome-derived neisserial antigen 1870, a Neisseria meningitidis candidate vaccine. J. Immunol. 172:5606-5615. [DOI] [PubMed] [Google Scholar]