Abstract
Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs, known to play an important role in ecological processes. Conventional light microscopy is the most common method used to detect their presence in planta, but this method fails to discern the presence of multiple AMF species and is not quantitative. These two factors are critically important in ecological studies, where the symbiotic contribution of each isolate needs to be defined. This paper describes the use of quantitative real-time PCR (qRT-PCR) as a detection system to address this issue. We used two Glomus spp., namely, G. intraradices and G. mosseae, to show that it is possible to study the interactions between these two isolates during the cocolonization of a single root system. Three different physiological studies were set up to assess how the interactions affected the occupancy of these fungi intraradically on a temporal basis. These treatments included saline and phosphorus stress, spatial distribution in the root zone, and preference for a particular host. qRT-PCR could prove a valuable tool in the area of AMF field ecology, where such data are critically important for defining the role of each species in the community structure.
Arbuscular mycorrhizal fungi (AMF) colonize roots of a wide range of host plants, whose access to soil resources is thereby facilitated. Apart from the widely documented role of AMF in providing nutritional benefits, many other aspects of this symbiosis have been observed. They include interspecific competition (19, 22), the trajectory of succession (4, 32), stabilization of soil aggregates (33, 39), and determination and promotion of plant diversity (7, 24, 42, 49, 52, 53).
In an ecological context, multiple AMF are routinely observed to colonize a single host plant, which has led to a widely held assumption that a single root system is capable of accommodating more than one AMF isolate (50). This cooccupation phenomenon is poorly understood, and at present we do not know whether such colonization is a result of competitive, synergistic, or antagonistic interaction. From an evolutionary perspective, synergism per se would be the most probable and logical scenario, as such an interaction would allow the host plant to harness multiple benefits, as opposed to the limited benefits a single AMF species can offer.
Our ability to consistently and reliably detect the presence of multiple AMF isolates within a single root fragment by means of light microscopy is severely limited, especially when dealing with field-collected samples. This is primarily because of the extremely limited variety of discernible structures that AMF forms in planta during its ontogenic development, and this problem is more acute when multiple species belonging to a single genus are present. To address this limitation, a wide variety of tools have been examined as means for the reliable and accurate detection of the presence of multiple AMF isolates. Some of these include antibodies (21), isozyme patterns (26, 46), lipid profiles (30), and PCR (13, 34, 38). Of these, conventional PCR-based methods have been acknowledged to be the best because they are sensitive, relatively easy to apply, invariant, and highly robust.
Conventional PCR has been shown to convincingly detect the presence of multiple AMF isolates within a single root system (38). The major drawback of this technique lies in the difficulty in generating quantitative data in relation to the input template because of (i) the exponential kinetics of PCR amplification and (ii) the end-point detection of the amplified product. However, the recent introduction of quantitative real-time PCR (qRT-PCR) technology, a variant of conventional PCR, has largely alleviated this problem. This variant of PCR measures the product formation in real time by means of fluorogenic probes and is acknowledged to be extremely sensitive (23), making it an invaluable tool in all areas of biology where sensitivity and rapidity of detection are critical to the end user (35).
A quantitative measure of the interactions among different AMF isolates as they interact with the host is of immense interest in ecology studies (41), since it enables the functional contribution of each AMF species to be defined. However, to the best of our knowledge, no study has yet quantified the absolute amounts of each AMF isolate within a single root system. In the present study we have sought to examine the infection pattern of two Glomus species with a qRT-PCR protocol that was successfully developed earlier for absolute quantification of G. intraradices DAOM 181602 (2). Our absolute quantification procedure was used to measure the infection of two species of AMF, namely, G. mosseae and G. intraradices, during their colonization of a single root system. The effects of external factors on the colonizing ability of these two Glomus species and on their symbiosis were studied by varying the external phosphorus and salinity levels and by using several different hosts.
MATERIALS AND METHODS
Plant and fungal material.
Plant materials used in the study were the following: Medicago truncatula cv. R108, Lycopersicon esculentum L. cv. Micro-Tom, and Daucus carota L. cv. Red Dragon. The AMF isolates used in the interaction experiments were G. intraradices Schenck & Smith (BEG141) and G. mosseae (Nicolson & Gerdemann) Gerdemann & Trappe (BEG12). Both these isolates were maintained in association with leek plants under standard pot conditions (10). To equalize the concentration of AMF propagules present in each inoculum, the most probable number of propagules (MPN) was determined using the earlier-described method (20). The MPN values for G. intraradices and G. mosseae were 1.0 × 104 and 2.5 × 104.
To verify the working of the G. intraradices- and G. mosseae-specific qRT-PCR primers developed during the present study, other AMF isolates were used for testing. These were G. intraradices (DAOM 181602), G. proliferum Dalpé & Declerck (MUCL 41827), G. lamellosum Dalpé, Koske & Tews (MUCL 43195), and G. cerebriforme McGee (MUCL 43208). All these isolates were obtained by means of in vitro root organ culture methodology with root-inducing transferred DNA-transformed D. carota roots as described previously (5).
Design of PCR primers for qRT-PCR.
All qRT-PCR primers were designed with the Primer Express software (version 2.0; Applied Biosystems, Foster City, CA) using design parameters specified for real-time PCR. The primers were custom synthesized by Sigma-Genosys (Rehovot, Israel). The designs of most of the primers used in the present study were derived from DNA sequences already present in the database (NCBI), except for the G. intraradices-specific primer, which was based on the internal transcribed spacer 1 (ITS1) plus 18S rRNA gene sequence. This region was first amplified by using the NS5/ITS2 primers (51) with genomic DNA extracted from surface-sterilized spores of G. intraradices BEG141. The amplified product was cloned in the pGEM-T vector (Promega, Madison, WI), and the insert was sequenced using standard big dye chemistry (ABI, Foster City, CA). Specificity of all the primers was tested by two methods: first by finding the homologous DNA sequences by means of the GenBank BLAST tool, and second by amplifying the respective target DNA by conventional PCR and visualizing the amplicon on 2.5% agarose gels with ethidium bromide.
Preparation of DNA templates.
Pure genomic DNA from both plant and fungal material was isolated as described previously (2) for constructing the standard calibration curves. Individual spores of the AMF G. intraradices BEG141 and G. mosseae were isolated from pot cultures of a leek host plant, by the wet sieving and a decanting method (17). These spores were surface sterilized with 2% chloramine-T before genomic DNA extraction. The genomic DNA of each preparation was quantified, and its purity was verified spectrophotometrically at 260 nm/280 nm; it was then stored in aliquots at −20°C pending use.
Quantitative real-time-PCR.
The qRT-PCR mixtures were set up with the components supplied in the SYBR green I kit (Eurogentec SA, Seraing, Belgium). A 25-μl aliquot of reaction mixture contained the following components (final concentrations): reaction buffer (1×), MgCl2 (2.5 mM), deoxynucleoside triphosphate (200 μM), forward/reverse primers (0.4 μM each), Hot GoldStar enzyme (0.2 U), 0.75 μl SYBR green (1:2,000 dilution in dimethyl sulfoxide), 5 μl DNA template, and PCR-grade water to make up the final volume. Duplicate reactions were routinely used for each sample, and each set included template controls containing water to check for contamination in the reaction components. The qRT-PCR was carried out in an ABI Prism 7000 instrument (Perkin-Elmer Biosystems, Norwalk, CT), with the following cycling program: 10 min at 95°C, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. This program was common to all the primers except when G. mosseae-specific primers were used, for which the annealing temperature was kept at 62°C. The threshold cycle (Ct) was calculated by the ABI Prism 7000 sequence detection software to indicate significant fluorescence signals rising above background during the early cycles of the exponential amplification phase of the PCR amplification process. The dissociation curve of the amplified product was examined for each reaction to rule out the possibility of primer-dimers contributing to the amplification signal.
The qRT-PCR signal was calibrated on genomic DNA of L. esculentum, M. truncatula, and D. carota roots colonized by isolates of both Glomus spp. used in the present study. From the standard calibration curves, the amounts of plant and fungal DNA were calculated, and the values were expressed in absolute terms as described by the following equation: (amount of fungal DNA, in ng)/(amount of plant DNA, in ng).
Interaction of G. mosseae and G. intraradices during symbiotic colonization.
Three different physiological studies were set up to study the interactions of G. mosseae and G. intraradices BEG141 during their colonization of a host plant. Working inocula for the physiological experiments were prepared by mixing a defined amount of the same with sterilized sand on a weight basis. For example, a 1% G. intraradices inoculum was prepared by mixing 1 g of G. intraradices BEG141 inoculum in 99 g of sterilized sand. A pregerminated seedling of the host plant was planted in a 60-ml pot containing sterile sand (amended with 15 ppm of superphosphate and K2SO4) and inoculum, watered once every 2 days, and fertilized twice a week with half-strength Johnson's solution minus phosphorus (29). The host plants were harvested after fixed time intervals, according to the treatment, and the roots were cut into small pieces (1 to 2 cm) and thoroughly homogenized. One aliquot of this sample was immersed in a storage solution (a 3:1 [vol/vol] mixture of 99% ethanol and 60% acetic acid) and stored at 4°C pending subsequent microscopic analysis, and the remainder was flash-frozen in liquid nitrogen and then stored at −80°C for qRT-PCR analysis. Trypan blue dye was used to visualize the intraradical fungal structures as described earlier (18). A subsample of the stained roots was examined for the amount of fungal presence using the magnified grid line intersect method (31). The colonization was expressed as total colonization (as a percentage), which was a sum of the values obtained from the vesicular, arbuscular, and hyphal parameters. The three physiological studies were as follows.
(i) Direction of spread of G. mosseae BEG12 and G. intraradices BEG141 in the root system as a function of inoculation.
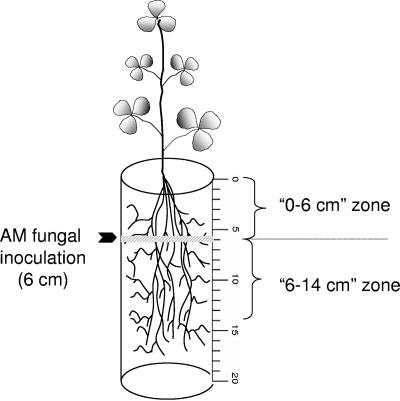
To determine the direction of spread of AMF within the root zone, the inocula were placed at a fixed point in the root zone and the spread in the roots was detected by using qRT-PCR. The AMF inocula were placed at a fixed depth of 6 cm (1% [wt/wt]) as a single band in the conical containers (dimensions, 20 cm by 4.75 cm) (Fig. 1). To facilitate data interpretation, the region above the inoculation zone was designated the 0- to 6-cm zone and that below it as the 6- to 14-cm zone. Three different experimental treatments were set up: (i) 1% G. intraradices BEG141; (ii) 1% G. mosseae; and finally (iii) 1% G. intraradices BEG141 plus 1% G. mosseae. Four weeks postinoculation, the roots were cut every 2 cm (0 to 2, 2 to 4, 4 to 6, 6 to 8, 8 to 10, 10 to 12, 12 to 14, and 14 to 16 cm) from the start of the shoot portion (Fig. 1). Segments from each 2-cm zone were washed separately and processed for microscopic and qRT-PCR analyses as described above.
FIG. 1.
Schematic depiction of the spatial distribution experiment of AMF in the root zone. The fungal inoculum was placed as a narrow band at a depth of 6 cm from the top of the cone. The region above the inoculum was referred to as the 0- to 6-cm zone and that below it was referred to as the 6- to 14-cm zone.
(ii) Preference for a particular host plant by G. mosseae and G. intraradices BEG141.
The preferences of G. mosseae and G. intraradices BEG141 for a particular host plant were assessed by inoculating them into three different host plants, namely, L. esculentum, M. truncatula, and D. carota. Each host plant was coinoculated with these two AMF species as well as inoculated singly at various inoculation ratios. These ratios were on a weight basis (wt/wt) and were as follows: 1% G. intraradices BEG141, 1% G. mosseae, 1% G. intraradices BEG141 plus 1% G. mosseae, 1% G. intraradices BEG141 plus 5% G. mosseae, and finally, 5% G. intraradices BEG141 plus 1% G. mosseae. Plants were harvested at the fourth and sixth week postinoculation and were prepared for microscopic and qRT-PCR analyses as described above.
(iii) Effects of increasing levels of salinity and phosphorus on fungal spread.
The effects of high levels of salinity and phosphorus on the interaction between G. intraradices BEG141 and G. mosseae during their intraradical spread in M. truncatula were studied. This host was chosen on the basis of the results obtained in the host specificity experiments (described below), in which M. truncatula was found to harbor both AM fungal species equally. Coinoculation of this host was done with 1% G. intraradices BEG141 plus 1% G. mosseae essentially as described in the previous section. Both saline and phosphorus stresses were imposed by fertilizing the plants with water spiked (described below) with various concentrations of NaCl and K2HPO4, respectively. The volume of the externally added solutions was approximately equal to the volume of the pots (60 ml), which were allowed to drain freely, similar to routine pot culture maintenance.
Two weeks postinoculation, saline stress was imposed on the mycorrhizal and control plants by watering them every 2 days with saline (NaCl) solutions of four different concentrations: 0, 25, 50, and 75 mM. Prior to the termination of the experiment, electrical conductance of the soil for each saline application was measured. The values for the 0, 25, 50, and 75 mM saline treatments were found to be 0, 3.5, 5.0, and 8.5 dS/m, respectively. Similarly, in a separate set of inoculated and control plants, phosphorus stress was imposed by amending the nutrient solution with four different levels of phosphorus (as K2HPO4): 0, 0.1, 0.5, and 1.0 mM. In the fifth, sixth, and seventh weeks following initiation of the stress regimen, the host plants were harvested and prepared for microscopic and qRT-PCR analysis as described above.
Nucleotide sequence accession number.
The sequence for ITS1 plus the 18S rRNA gene was deposited in GenBank under the accession number DQ028775.
RESULTS
Primer design and standard curve preparation.
The qRT-PCR primer for G. mosseae BEG12 was designed on the basis of the 28S rRNA gene sequence previously deposited in GenBank (AY541918). To design a specific primer for the G. intraradices BEG141 isolate, first the ITS1 plus 18S rRNA region was amplified by PCR and the resulting fragment of about 777 bp was sequenced. BLAST (NCBI) analysis of this sequence showed that it was of G. intraradices origin and therefore it was used for designing the qRT-PCR primers.
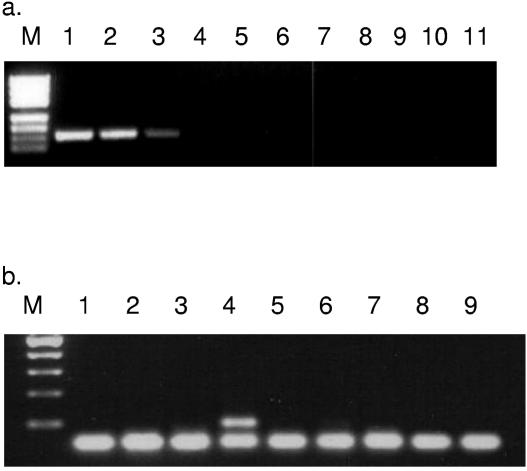
Both the plant and fungal primers developed in the present study had annealing temperatures (Tm) in the range of 60 to 62°C, in keeping with the design requirements of qRT-PCR (Table 1). The G. intraradices BEG141-specific primer pair, VC-F/R, specifically amplified a 101-bp fragment from the target isolate that included two different strains of G. intraradices (Fig. 2a). However, no product was amplified under similar reaction conditions when other nonspecific templates were tested. Similarly, the G. mosseae-specific primer pair Mos-F/R amplified an expected 101-bp fragment from spores of G. mosseae but failed to amplify fragments from other nontarget templates, including plant DNA (Fig. 2b). The primers specific for M. truncatula and L. esculentum that were used in the present study were the same as those described by us previously (2). The D. carota-specific primer was designed on the basis of the chalcone synthase 9b gene (GenBank accession number DCU52848) which, empirically, amplified only from its target (data not shown).
TABLE 1.
List of PCR primers designed in qRT-PCR analysis for the present study
| Target organism | Primer designation | Primer sequence | Annealing temp (°C) | Amplicon size (bp) | Target sequence (GenBank accession no.) | Reference |
|---|---|---|---|---|---|---|
| G. mosseae BEG12 | Mos-F | GAAGTCAGTCATACCAACGGGAA | 62 | 101 | 28S rRNA gene (AY541918) | Present work |
| Mos-R | CTCGCGAATCCGAAGGC | |||||
| G. intraradices BEG141 | VC-F | GAGACCATGATCAGAGGTCAGGT | 60 | 101 | ITS1 + 18S rRNA gene (DQ028775) | Present work |
| VC-R | GGTCATTTAGAGGAAGTAAAAGTCGTAAC | |||||
| M. truncatula L. cv. R108 | Mt-F | GGTGATGGTTGTCAGAGTCAATG | 60 | 101 | Chitinase 1 (Y10373) | Alkan et al. (2) |
| Mt-R | CTGTCGTCACGGTGCTTGAG | |||||
| L. esculentum L. cv. Micro-Tom | Lechs2-F | TTCGGTTAAGCGGCTCATGA | 60 | 101 | Chalcone synthase 2 (X55195) | Alkan et al. (2) |
| Lechs2-R | CTCGAGCACCCTTGTTGTTCTC | |||||
| D. carota L. cv. Red Dragon | Dcchs9b-F | GACTTTCGCCTCACCAAGCTC | 60 | 101 | Chalcone synthase 9b (DCU52848) | Present work |
| Dcchs9b-R | GCCAGACGAAGAACCGTGC |
FIG. 2.
Verification of AMF-specific qRT-PCR primers designed in the present study. (a) VC-F/R primer pair specific for G. intraradices. Lanes: M, pUC 19 marker; 1, G. intraradices BEG141; 2, G. intraradices DAOM 181602; 3, G. intraradices MUCL 43194; 4, G. mosseae; 5, G. cerebriforme MUCL 43208; 6, G. lamellosum MUCL 43195; 7, G. proliferum MUCL 41827; 8, M. truncatula; 9, L. esculentum; 10, D. carota; 11, negative control. (b) Mos-F/R primer pair specific for G. mosseae. Lanes: M, 100-bp ladder; 1, G. intraradices BEG141; 2, G. intraradices DAOM 181602; 3, G. intraradices MUCL 43194; 4, G. mosseae BEG141; 5, G. cerebriforme MUCL 43208; 6, M. truncatula; 7, L. esculentum; 8, D. carota; 9, negative control.
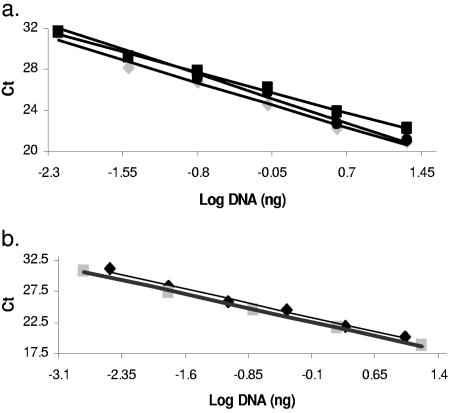
The aforementioned specific primer pairs were used to generate a standard curve to test the linearity of the signal. A linear relationship was obtained between the Ct of the qRT-PCR amplification reaction and DNA concentration for both the plant-specific (Fig. 3a) and the AMF-specific (Fig. 3b) primers. The primer pair specific for G. intraradices BEG141 and G. mosseae was linear between the 3.2 × 10−3 and 10 ng and between 1.6 × 10−3 and 16 ng ranges, respectively. In the case of host-specific primers, the linear range was found empirically and found to lie within 6.4 × 10−3 to 20 ng. The regression equations of this relationship were as follows: G. intraradices (y = −3.0x + 22.9; R2 = 1); G. mosseae (y = −3.0x + 22.2; R2 = 0.9); M. truncatula (y = −2.9x + 24.3; R2 = 0.9); L. esculentum DNA (y = −2.6x + 25.6; R2 = 0.9); D. carota (y = −3.2x + 24.9; R2 = 0.9).
FIG. 3.
Standard curves obtained by plotting Ct against genomic DNA concentration. (a) Host-specific primers: M. truncatula DNA (diamond; y = −2.9x + 24.3; R2 = 0.9); L. esculentum DNA (square; y = −2.6x + 25.6; R2 = 0.9); D. carota (circle; y = −3.2x + 24.9; R2 = 0.9). (b) AMF-specific primers: G. intraradices BEG141 (diamond; y = −3.06x + 22.9; R2 = 1.0); G. mosseae BEG12 (square; y = −3.45x + 22.2; R2 = 0.9).
Interaction between G. mosseae and G. intraradices. (i) Spatial distribution of AM fungi in the root zone.
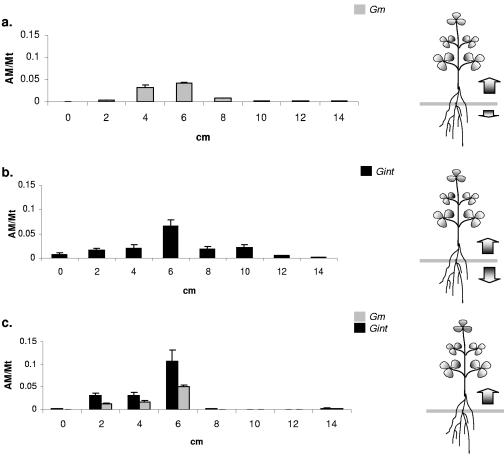
The distribution of the spread of both of the isolates in the host root was assessed 4 weeks postinoculation. When either AM fungal isolate was used alone, the maximal fungal density was detected in the vicinity of the inoculation zone (6 cm) (Fig. 4a and b), and the fungal occupancy (as indicated by the qRT-PCR signal) in the roots diminished with increasing distance of the analyzed root segment from the zone of inoculation. Although both these isolates spread in each direction when they were inoculated singly, they differed drastically in the intensity of their spread: G. intraradices BEG141 spread very uniformly in each direction (Fig. 4b), whereas G. mosseae proliferated predominantly in the root zones that were close to the shoots (Fig. 4a). Under coinoculation conditions, the direction of fungal spread from the inoculation zone was found to be completely unidirectional: the qRT-PCR signal for each of the isolates was detected exclusively in the 0- to 6-cm zone, i.e., in the region near the shoot, and no signal was detected in the 6- to 14-cm zone (Fig. 4c). The highest colonization level of each of the isolates was about 10%.
FIG. 4.
Colonization of AM fungi along the root zone when the inocula were placed at a fixed depth (6 cm; gray bar) in the potting system. Distributions shown are for G. mosseae (a), G. intraradices BEG141 (b), and both isolates inoculated together (c). The AM/Mt on the y axis refers to the ratio between the amount of fungal DNA and the amount of M. truncatula DNA, and the x axis refers to the distance (in cm) from the shoot end. The error bars represent standard deviations of means between four replicate qRT-PCR signal values.
(ii) Host preference.
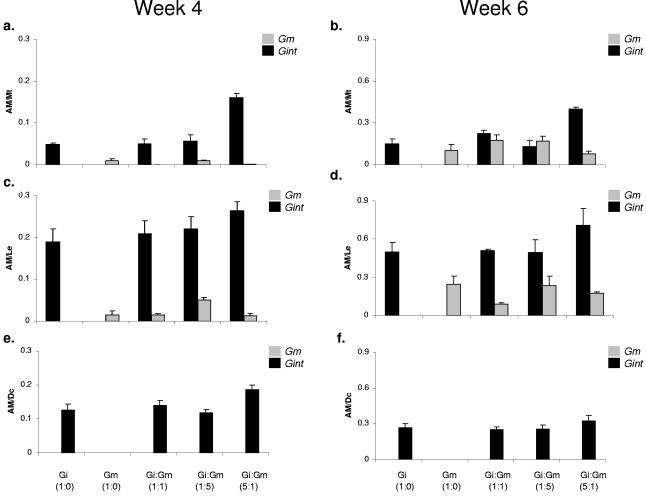
Both G. intraradices BEG141 and G. mosseae showed a preference for one of the three hosts tested: L. esculentum, M. truncatula, and D. carota. At the first sampling date (fourth week postinoculation), G. intraradices BEG141 was found to be the predominant symbiont in each of the three hosts (Fig. 5a, c, and e), whereas the G. mosseae signal was detected in L. esculentum (Fig. 5c) and M. truncatula (Fig. 5a) roots but no measurable signal of this isolate could be seen in D. carota roots (Fig. 5e). During the sixth week, there was a significant increase in the colonization intensity of both G. mosseae and G. intraradices BEG141 in L. esculentum (Fig. 5d) and M. truncatula (Fig. 5b), but no detectable signal for G. mosseae was observed in D. carota roots (Fig. 5f). Among the three hosts plants used in this study, L. esculentum showed the highest colonization rates: 25% at week 4 and 30 to 65% at week 6 for both the isolates. M. truncatula was more uniformly colonized by both AM fungal isolates. The colonization of M. truncatula and D. carota ranged from 10 to 20% during week 4 and from 25 to 45% during week 6.
FIG. 5.
Host specificity of G. mosseae (Gm) and G. intraradices BEG141 (Gi) and mixed inocula (Gi:Gm) for three different hosts, M. truncatula (a and b), L. esculentum (c and d), and D. carota (e and f), during the fourth and sixth weeks postinoculation. AM/Mt, AM/Le, and AM/Dc on the y axis refer to the ratios between the amounts of fungal DNA (in ng) and those of M. truncatula, L. esculentum, and D. carota DNA (in ng) in the respective samples. The error bars represent standard deviations of means from four replicate qRT-PCR signal values.
(iii) Effect of salinity and phosphorus stress.
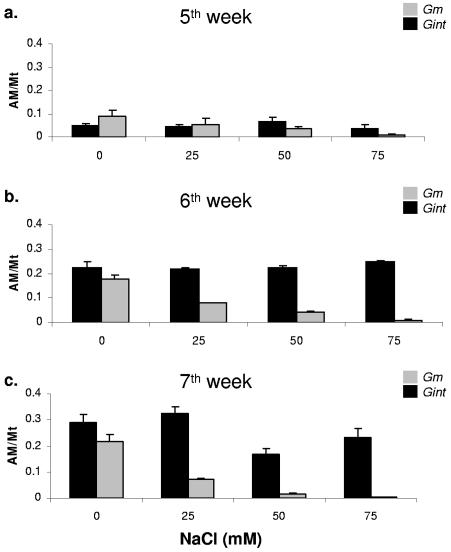
Altering the salinity levels via the fertilization solution also altered the relative spread of both the isolates in the M. truncatula roots. During the initial phase of the symbiotic establishment (week 5), both G. mosseae and G. intraradices BEG141 achieved approximately equal occupancy of the roots under all the salinity regimes (Fig. 6a). However, as the symbiosis progressed, there was a gradual shift in the fungal occupancy ratios, and the difference increased with increasing salinity. The levels of G. mosseae were found to decrease dramatically with increasing concentration of NaCl, specifically at the 50 and 75 mM levels, during the later stages of symbiotic interactions, specifically in the sixth (Fig. 6b) and seventh weeks. The occupancy of G. intraradices was consistently higher in all the saline fertilization treatments, indicating insensitivity to external saline stress. Nevertheless, a moderate decrease in G. intraradices BEG141 occupancy at higher NaCl levels was observed during the later stages of symbiosis (Fig. 6b and c). In the control treatment (zero NaCl), the occupancy by both AMF, G. mosseae and G. intraradices BEG141, was found to be approximately equal at all three sampling points, which suggests that salinity was a major factor influencing the intraradical spread of these two symbionts.
FIG. 6.
Effect of salinity level on fungal competition in M. truncatula roots. Four levels of salinity were used, and the colonization by each fungus was quantified at the fifth (a), sixth (b), and seventh (c) weeks following initiation of the stress treatment. AM/Mt on the y axis refers to the ratio between amount of fungal DNA (in ng) and that of M. truncatula DNA (in ng) in the sample. The error bars represent standard deviations of means from four replicate qRT-PCR signal values.
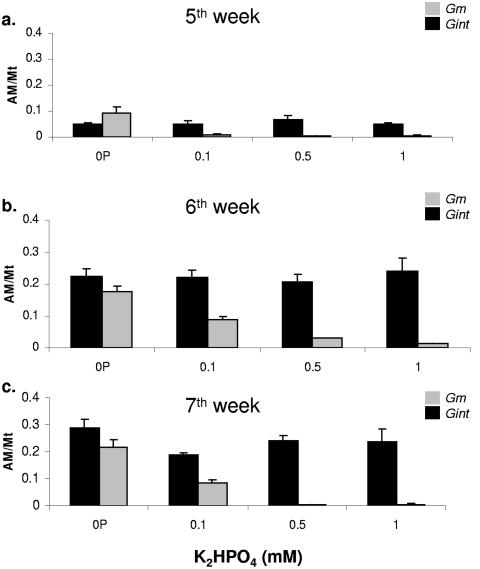
Increasing the levels of external phosphorus impaired the ability of G. mosseae to colonize M. truncatula (Fig. 7). Under the no-phosphorus fertilization regimen, the growth of both isolates was similar at all three sampling points (Fig. 7a, b, and c). As the phosphorus levels were increased, there was a drastic reduction in the intraradical spread of G. mosseae, and no signal was detected when the P level was >0.5 mM (Fig. 7a, b, and c), especially during the later phase of symbiosis. In contrast, high levels of phosphorus did not have any effect on the intraradical spread of G. intraradices BEG141 at any of the three sampling points: its colonization rate under both NaCl and P stresses ranged from 10 to 20% during week 5, from 25 to 40% in week 6, and from 25 to 55% in week 7.
FIG. 7.
Effect of phosphorus stress on the colonization ability of G. intraradices BEG141 and G. mosseae in M. truncatula roots. Four different phosphorus levels were used: 0, 0.1, 0.5, and 1.0 mM. Plants were harvested at the fifth (a), sixth (b), and seventh (c) weeks following initiation of the stress treatment. AM/Mt on the y axis refers to the ratio between the amount of fungal DNA (in ng) and that of M. truncatula DNA (in ng) in the sample. The error bars represent standard deviations of means from four replicate qRT-PCR signal values.
DISCUSSION
Previously, it was not generally considered that a single root system could be colonized by multiple AMF species, since it was believed that AMF “competed” for a root zone occupancy (1, 25, 36, 52). This view was later shown found to be incorrect, and the occurrence of multiple occupancy of a single root segment by AMF of diverse genera and/or species is now commonly accepted. This has necessitated the adoption of new techniques in AMF biology, the qRT-PCR technology being one of them. The initial application of this technique has been very successful and has enabled some fundamental questions to be answered; they concern, for example, enumeration of AMF propagules in the soil (15, 16) and quantification of AMF in planta (2, 13, 27). Under natural conditions, multiple occupancy appears to be more beneficial to the host plant, since it enables it to harness a wider spectrum of benefits than colonization by a single AMF isolate. However, the techniques formerly available to detect the presence of more than one species of AMF within a single root system made it difficult to develop a quantitative understanding of AMF-host symbiosis.
Following a single-isolate inoculation, the fungal spread was largely uniform in both directions when G. intraradices BEG141 was used. However, G. mosseae showed a higher tendency to spread towards the shoot regions (Fig. 4a and b). Interestingly, when these isolates were coinoculated, the fungal growth was exclusively upwards (into the 0- to 6-cm zone), with no qRT-PCR signal observed in the root segments below the inoculation zone (in the 6- to 14-cm zone) (Fig. 4c). We hypothesize that the directional movement of the fungi is governed by the dynamics of carbon synthesis and translocation in the host plant, which is a key feature of this symbiotic association. It has been firmly established that AMF enhance the “sink” demand for carbon in the host plant roots (11, 44, 53, 54), of which as much as 10 to 23% may be transferred from the host to the fungus (28). Radiotracer studies have shown that the leaves are the site of enhanced carbon fixation activity, the products of which are translocated to the roots, thus creating a “concentration gradient” which the fungi would experience intraradically while accessing these photosynthates (8). In our present study, the coinoculation conditions would have dramatically increased the fungal carbon demand, resulting in the preferential spread of the AMF into the 0- to 6-cm zone, where the carbon levels would be higher than in the 6- to 14-cm zone. Interestingly it was observed earlier (6) that active AMF propagules under field conditions are more active in the top soil, corroborating our experimental observations.
The observation that about 160 species of AM fungi colonize an estimated 225,000 terrestrial plant species (43) has led to a conclusion that some AM fungi possess a broad host range. However, recent empirical evidence has strongly discounted this generalized perception, and now a new concept of “host preference” is increasingly being accepted. This characteristic has been found to be an important factor in determining the plant community structure and ecosystem functioning (42, 47-49). From the present study it's very clear that both of the isolates demonstrate some sort of host preference in their colonization process (Fig. 5). Important clues as to the possible “stimulative” influence of one isolate of AM fungi over another as observed here in the case of G. mosseae cannot be ruled out (37).
Soil salinity is an increasing problem worldwide, particularly in arid areas, but research findings have emerged that show that AMF have the potential to ameliorate the effect of salinity on plant growth (3, 9, 40). However, there is still a lack of information as to whether salinity directly affects the growth of the AMF, of the plant, or the interaction between them. The presence of salts in the growth substrate has been shown to delay germination of spores and to limit the growth of AMF hyphae (14), thus causing a loss in symbiotic efficiency, but these data were obtained in closed, plant-free systems in which the salt concentrations were assumed to remain static over the course of the experiments. However, in the field and in pots containing plants, the concentrations of salts in the soil solution may change markedly across very small distances and will inevitably change with time as the water content of the soil changes. Our present studies showed that we could track the progress of a single isolate under a saline condition over a temporal course, in order to assess how each isolate would perform under a practical field condition. In our study G. intraradices was found to be highly superior to G. mosseae in its ability to tolerate salinity, which showed that this species could be used under practical field conditions.
Sylvia and Schenck (45) first observed that AM fungal species differed in their ability to tolerate phosphorus. In our present study, we consistently observed the high sensitivity of G. mosseae to increasing levels of P (Fig. 7), which was consistent with one previous finding (12) where it was shown that to achieve a successful G. mosseae symbiosis, it is imperative that the external P be kept at very low levels (absolute zero P levels).
Summarily, the application of external saline and phosphorus shows that both of the AMF isolates are sensitive towards them. Whether NaCl or KH2PO4 directly or indirectly (e.g., pH change) affect the growth and intraradical spread of both of the AMF isolates used in the present study remains to be conclusively demonstrated. Notwithstanding the high costs of the qRT-PCR technology, our work demonstrates that it is possible to gain new insights in the area of AMF-host interactions. In the future, when this technology becomes affordable, it could become an ideal tool and provide a faster source of data for detecting the early stages of AMF-host interactions without recourse to performing laborious microscope-based colonization assays.
Acknowledgments
The present work was supported by financial grants from the Israeli Ministry of Agriculture and Rural Development to Y.K.
The technical assistance of Bruria Ben-Dor is gratefully acknowledged.
REFERENCES
- 1.Abbott, L. K., and A. D. Robson. 1984. Colonization of the root system of subterranean clover by three species of vesicular arbuscular mycorrhizal fungi. N. Phytol. 96:275-281. [Google Scholar]
- 2.Alkan, N., V. Gadkar, J. Coburn, O. Yarden, and Y. Kapulnik. 2004. Quantification of the arbuscular mycorrhizal fungus Glomus intraradices in host tissue using real-time polymerase chain reaction. N. Phytol. 161:877-885. [DOI] [PubMed] [Google Scholar]
- 3.Al-Karaki, G. N., R. Hammad, and M. Rusan. 2001. Response of two L. esculentum cultivars differing in salt tolerance to inoculation with mycorrhizal fungi under salt stress. Mycorrhiza 11:43-47. [Google Scholar]
- 4.Allen, M. F., and E. B. Allen. 1990. The mediation of competition by mycorrhizae in successional and patchy environments, p. 367-389. In J. B. Grace and D. Tilman (ed.), Perspectives on plant competition. Academic Press, New York, N.Y.
- 5.Bècard, G., and Y. Piché. 1992. Establishment of vesicular-arbuscular mycorrhiza in root organ culture: review and proposed methodology. Methods Microbiol. 24:89-108. [Google Scholar]
- 6.Bellgard, S. E. 1993. The topsoil as the major store of propagules of vesicular-arbuscular mycorrhizal fungi in southeast Australian sandstone soils. Mycorrhiza 3:19-24. [Google Scholar]
- 7.Bidartondo, M. I., D. Redecker, I. Hijri, A. Wiemken, T. D. Bruns, L. Domínguez, A. Sérsic, J. R. Leake, and D. J. Read. 2002. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature 419:389-392. [DOI] [PubMed] [Google Scholar]
- 8.Black, K. G., D. T. Mitchell, and B. A. Osborne. 2000. Effect of mycorrhizal-enhanced leaf photosynthates on carbon partitioning, translocation and photosynthesis in cucumber. Plant Cell Environ. 23:797-809. [Google Scholar]
- 9.Copeman, R. H., C. A. Martin, and J. C. Stutz. 1996. L. esculentum growth in response to salinity and mycorrhizal fungi from saline or nonsaline soils. Hort. Sci. 31:341-344. [Google Scholar]
- 10.David-Schwartz, R., V. Gadkar, S. Wininger, A. A. Levy, G. Galili, and Y. Kapulnik. 2003. Isolation of a pre-mycorrhizal infection (pmi2) mutant of L. esculentum, resistant to arbuscular mycorrhizal fungal colonization. Mol. Plant-Microbe Interact. 16:382-388. [DOI] [PubMed] [Google Scholar]
- 11.Douds, D. D., Jr., C. R. Johnson, and K. E. Koch. 1988. Carbon cost of the fungal symbiont relative to net leaf P accumulation in a split-root VA mycorrhizal symbiosis. Plant Physiol. 86:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douds, D. D., Jr. 1997. A procedure for the establishment of Glomus mosseae in dual culture with Ri T-DNA-transformed D. carota roots. Mycorrhiza 7:57-61. [Google Scholar]
- 13.Edwards, S. G., A. H. Fitter, and J. P. W. Young. 1997. Quantification of an arbuscular mycorrhizal fungus, Glomus mosseae, within plant roots by competitive polymerase chain reaction. Mycol. Res. 101:1440-1444. [Google Scholar]
- 14.Estaun, M. V. 1989. Effect of sodium chloride and mannitol on germination and hyphal growth of the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. Agric. Ecosyst. Environ. 29:123-129. [Google Scholar]
- 15.Filion, M., M. St. Arnaud, and S. H. Jabaji-Hare. 2003. Direct amplification of fungal DNA from soil substrate using real-time PCR. J. Microbiol. Methods 53:67-76. [DOI] [PubMed] [Google Scholar]
- 16.Filion, M., M. St. Arnaud, and S. H. Jabaji-Hare. 2003. Quantification of Fusarium solani f. sp. phaseoli in mycorrhizal bean plants and surrounding mycorrhizosphere soil using real-time polymerase chain reaction and direct isolations on selective media. Phytopathology 93:229-235. [DOI] [PubMed] [Google Scholar]
- 17.Gerdemann, J. W., and T. H. Nicholson. 1963. Spores of mycorrhizal endogone species extracted from soil by wet sieving and decanting. Trans. Br. Mycol. Soc. 46:235-244. [Google Scholar]
- 18.Giovannetti, M., and B. Mosse. 1980. An evaluation of technique for measuring vesicular mycorrhizal infection in roots. N. Phytol. 84:489-500. [Google Scholar]
- 19.Grime, J. P., J. M. Macky, S. H. Hillier, and D. J. Read. 1987. Mechanisms of floristic diversity: evidence from microcosms. Nature 328:420-422. [Google Scholar]
- 20.Haas, J. H., and J. Krikun. 1985. Efficacy of endomycorrhizal-fungus isolates and inoculum quantities required for growth response. N. Phytol. 100:613-621. [Google Scholar]
- 21.Hahn, A., P. Bonfante, K. Horn, F. Pausch, and B. Hock. 1993. Production of monoclonal antibodies against surface antigens of spores from arbuscular mycorrhizal fungi by an improved immunization and screening procedure. Mycorrhiza 4:69-78. [Google Scholar]
- 22.Hartnett, D. C., B. A. D. Hetrick, G. W. T. Wilson, and D. J. Gibson. 1993. Mycorrhizal influence on intra- and inter-specific neighbor interactions among co-occurring prairie grasses. J. Ecol. 81:787-795. [Google Scholar]
- 23.Heid, C. A., J. Stevens, K. J. Livak, and M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 24.Helgason, T., J. W. Merryweather, J. Denison, P. Wilson, J. P. W. Young, and A. H. Fitter. 2002. Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J. Ecol. 90:371-384. [Google Scholar]
- 25.Hepper, C. M., C. Azcón-Aguilar, S. Rosendhal, and R. Sen. 1988. Competition between three species of Glomus used as spatially separated introduced and indigenous mycorrhizal inocula for leek (Allium porrum L.). N. Phytol. 110:207-215. [Google Scholar]
- 26.Hepper, C. M., R. Sen, C. Azcón-Aguilar, and C. Grace. 1988. Variation in certain isozymes among different geographical isolates of the vesicular-arbuscular mycorrhizal fungi Glomus clarum, Glomus monosporum, and Glomus mosseae. Soil Biol. Biochem. 20:51-59. [Google Scholar]
- 27.Isayenkov, S., T. Fester, and B. Hause. 2004. Rapid determination of fungal colonization and arbuscule formation in roots of Medicago truncatula using real-time (RT) PCR. J. Plant Physiol. 61:1379-1383. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsen, I., and L. Rosendahl. 1990. Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. N. Phytol. 115:77-83. [Google Scholar]
- 29.Johnson, C. M., P. R. Stout, J. C. Beyer, and A. B. Carlson. 1957. Comparative chlorine requirements of different species. Plant Soil 8:337. [Google Scholar]
- 30.Madan, R., C. Pankhurst, B. Hawke, and S. Smith. 2002. Use of fatty acids for identification of AM fungi and estimation of the biomass of AM spores in soil. Soil Biol. Biochem. 34:125-128. [Google Scholar]
- 31.McGonigle, T. P., M. H. Miller, D. G. Evans, G. L. Fairchild, and J. A. Swan. 1990. A new method, which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. N. Phytol. 115:495-501. [DOI] [PubMed] [Google Scholar]
- 32.Medve, R. J. 1984. The mycorrhizae of pioneer species in disturbed ecosystems in western Pennsylvania. Am. J. Bot. 71:787-794. [Google Scholar]
- 33.Miller, M. A., and J. D. Jastrow. 2000. Mycorrhizal fungi influence soil structure, p. 3-18. In D. D. Douds, Jr., and Y. Kapulnik (ed.), Arbuscular mycorrhizas: molecular biology and physiology. Kluwer Academic Press, Dordrecht, The Netherlands.
- 34.Millner, P. D., W. W. Mulbry, and S. L. Reynolds. 2001. Taxon-specific oligonucleotide primers for detection of Glomus etunicatum. Mycorrhiza 10:259-265. [DOI] [PubMed] [Google Scholar]
- 35.Orlando, C., P. Pinzani, and M. Pazzagli. 1998. Developments in quantitative PCR. Clin. Chem. Lab. Med. 36:255-269. [DOI] [PubMed] [Google Scholar]
- 36.Pearson, J. N., L. K. Abbott, and D. A. Kasper. 1993. Mediation of competition between two colonizing VA mycorrhizal fungi by host plants. N. Phytol. 123:93-98. [Google Scholar]
- 37.Pinior, A., U. Wyss, Y. Piché, and H. Vierheilig. 1999. Plants colonized by AM fungi regulate further root colonization by AM fungi through altered root exudation. Can. J. Bot. 77:891-897. [Google Scholar]
- 38.Redecker, D. 2000. Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10:73-80. [Google Scholar]
- 39.Rillig, M. C. 2004. Arbuscular mycorrhizae and terrestrial ecosystem processes. Ecol. Lett. 7:740-754. [Google Scholar]
- 40.Ruiz-Lozano, J. M., R. Azcón, and M. Gomez. 1996. Alleviation of salt stress by arbuscular mycorrhizal Glomus species in Lactuca sativa plants. J. Physiol. Plant 98:767-772. [Google Scholar]
- 41.Sanders, I. R., and A. H. Fitter. 1992. Evidence for differential responses between host-fungus combinations of vesicular-arbuscular mycorrihizas from a grassland. Mycol. Res. 96:415-419. [Google Scholar]
- 42.Sanders, I. R. 2003. Preference, specificity and cheating in the arbuscular mycorrhizal symbiosis. Trends Plant Sci. 8:143-144. [DOI] [PubMed] [Google Scholar]
- 43.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis, 2nd ed. Academic Press, London, United Kingdom.
- 44.Snellgrove, R. C., W. E. Splittstoesser, D. P. Stibley, and P. B. Tinker. 1982. The distribution of carbon and the demand of the fungal symbiont in leek plants with vesicular-arbuscular mycorrhizas. N. Phytol. 92:75-87. [Google Scholar]
- 45.Sylvia, D. M., and N. C. Schenck. 1983. Application of superphosphate to mycorrhizal plants stimulates sporulation of phosphorus-tolerate vesicular-arbuscular mycorrhizal fungi. N. Phytol. 95:655-661. [Google Scholar]
- 46.Thingstrup, I., and S. Rosendahl. 1994. Quantification of fungal activity in arbuscular mycorrhizal symbiosis by polyacrylamide gel electrophoresis and densitometry of malate dehydrogenate. Soil Biol. Biochem. 26:1483-1489. [Google Scholar]
- 47.Vandenkoornhuyse, P., R. Husband, T. J. Daniell, I. J. Watson, J. M. Duck, A. H. Fitter, and J. P. W. Young. 2002. Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol. Ecol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 48.Vandenkoornhuyse, P., K. P. Ridgway, I. J. Watson, A. H. Fitter, and J. P. W. Young. 2003. Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol. Ecol. 12:3085-3095. [DOI] [PubMed] [Google Scholar]
- 49.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 50.van Tunien, D., E. Jacquot, B. Zhao, and V. Pearson. 1998. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA targeted nested PCR. Mol. Ecol. 7:879-887. [DOI] [PubMed] [Google Scholar]
- 51.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 52.Wilson, J. M., and M. J. Trinick. 1983. Infection development and interaction between vesicular arbuscular mycorrhizal fungi. N. Phytol. 93:543-553. [Google Scholar]
- 53.Wright, D. P., J. D. Scholes, and D. J. Read. 1998. Effects of VA mycorrhizal colonization on photosynthesis and biomass production of Trifolium repens L. Plant Cell. Environ. 21:209-216. [Google Scholar]
- 54.Wright, D. P., J. D. Scholes, and D. J. Read. 1998. Mycorrhizal sink strength influences whole carbon balance of Trifolium repens L. Plant Cell Environ. 21:881-891. [Google Scholar]