Abstract
Haemophilus parasuis is the cause of Glässer's disease and other clinical disorders in pigs. It can also be isolated from the upper respiratory tracts of healthy pigs, and isolates can have significant differences in virulence. In this work, a partial sequence from the 60-kDa heat shock protein (Hsp60) gene was assessed as an epidemiological marker. We analyzed partial sequences of hsp60 and 16S rRNA genes from 103 strains of H. parasuis and other related species to obtain a better classification of the strains and examine the correlation with virulence. The results were compared with those obtained by enterobacterial repetitive intergenic consensus PCR. Our results showed that hsp60 is a reliable marker for epidemiological studies of H. parasuis and that the analysis of its sequence is a better approach than fingerprinting methods. Furthermore, the analysis of the hsp60 and 16S rRNA gene sequences revealed the presence of a separate lineage of virulent strains and indicated the occurrence of lateral gene transfer among H. parasuis and Actinobacillus strains.
Haemophilus parasuis is a gram-negative bacterium of the family Pasteurellaceae and is the etiologic agent of Glässer's disease in pigs, which is characterized by serofibrinous to fibrinopurulent polyserositis, arthritis, and meningitis (34). H. parasuis is also involved in other clinical outcomes, such as pneumonia and sudden death, and causes high morbidity and mortality in naive swine populations (39). Modern production systems based on the early segregation of piglets from the sow seem to have increased the prevalence of Glässer's disease. H. parasuis is frequently isolated from lung tissue, but since the bacterium can also be isolated from the upper respiratory tracts of healthy pigs (21, 31), the meninges, pericardium, pleura, peritoneum, and joints are better samples for clinical diagnosis.
In 1992, Kielstein and Rapp-Gabrielson defined 15 serovars of H. parasuis and demonstrated differences in their virulence, with strains ranging from highly virulent to nonvirulent (24). Strain variability has also been revealed for other phenotypic and genotypic features (2, 3, 7, 8, 29, 32, 33, 35, 38). Since the pig is the only known natural environment for H. parasuis, this high degree of variation in virulence could be an interesting characteristic and might represent different adaptations to colonize and invade different organs of the animal. In agreement with these hypotheses, Oliveira et al. reported the association of serotypes 1, 2, 4, 5, 12, 13, and 14 (and nontypeable isolates) with isolation from systemic sites and of serotype 3 (and nontypeable isolates) with isolation from the upper respiratory tract (35). Unfortunately, there is no clear correlation between serotype and virulence, and even strains belonging to the same serotype exhibit different degrees of virulence. Nevertheless, serotyping has commonly been used to classify H. parasuis strains, although for epidemiological studies it does not provide enough discrimination of isolates, and more importantly, a significant percentage of isolates are nontypeable with this technique. Although information on the genomic sequence of H. parasuis is limited, several groups have attempted to improve the differentiation of field strains by using different genotyping techniques. One of the few known sequences of H. parasuis is the 16S rRNA gene. 16S rRNA gene sequencing is appropriate for species identification and definition (17, 23, 40, 42). This sequence has been used successfully for the classification of the Pasteurellaceae at the species level (14, 30), allowing the differentiation of H. parasuis from other NAD-dependent Pasteurellaceae organisms isolated from swine, mainly Actinobacillus minor, Actinobacillus porcinus, and Actinobacillus indolicus. However, 16S rRNA gene sequences are usually not suitable for strain differentiation due to a lack of variability below the species level. Recently, PCR-restriction fragment length polymorphism (PCR-RFLP) analyses using the sequences of tbpA (12) and aroA (13) have been proposed, but the application of these techniques does not provide sufficient information about the phylogeny between strains. Another approach to differentiating field strains is the use of enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) (41). For strains of H. parasuis, ERIC-PCR fingerprints are highly heterogeneous, and although this method is useful for local epidemiology studies, in particular for assessing different strains circulating in a farm (35, 38), it has no practical application for global studies. In addition, results obtained using ERIC-PCR as well as those obtained by PCR-RFLP from different laboratories are difficult to compare. Thus, an improved method for global studies is needed.
In an attempt to find a more appropriate and reliable epidemiological marker for the classification of H. parasuis, we decided to use partial sequencing of the hsp60 gene (gene encoding the heat shock protein of 60 kDa, or groEL gene). We chose this method for several reasons. First, the results (i.e., the sequences) are easy to compare and reproduce among laboratories. Second, hsp60 is a ubiquitous gene (18), so it must be present in all strains. Additionally, Hsp60 has been demonstrated to play a role in crucial functions of bacteria, such as the pathogenesis of Legionella pneumophila (11, 22), the immune response to Helicobacter pylori (22), and the maintenance of the proteome of symbiotic bacteria such as Buchnera spp. (15, 16). Thus, it is possible that the natural selection on this gene could be different in strains with diverse virulence, providing additional information on the virulence of the strains. Finally, hsp60 of H. parasuis will probably have enough variability below the species level, as demonstrated with other human and pig pathogens (9, 18, 19).
Here, we evaluate the use of the hsp60 sequence as a molecular epidemiological marker for H. parasuis and complete the study of the variation in field strains by using previously described methods.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 103 strains, including 13 H. parasuis reference strains, were used in this study (Table 1). Field strains included clinical isolates, both systemic and respiratory, and nasal isolates from healthy piglets from farms without Glässer's disease. To obtain the nasal isolates, four farms in two separate regions of Spain were selected based on their health status. Eight to 10 nasal swabs were taken from each farm and transported in Amies medium to the laboratory, where they were plated on chocolate agar to isolate colonies. After 2 to 3 days at 37°C with 5% CO2, suspected colonies were selected and subcultured for further analysis. In addition to classical biochemical tests, final identification was performed by 16S rRNA gene sequencing (see below). Clinical isolates were kindly provided by the Department of Infectious Diseases of the Veterinary School of the Universitat Autònoma de Barcelona (Spain), by E. Rodríguez Ferri (Universidad de León, Spain), by Gustavo C. Zielinski (Instituto Nacional de Tecnología Agropecuaria-INTA, Argentina), and by T. Blaha (Federal Institute for Health Protection of Consumers and Veterinary Medicine, Germany). Strains of the closely related species A. minor, A. indolicus, A. porcinus, Actinobacillus pleuropneumoniae, and Pasteurella multocida were also included in the study. All of the strains were maintained in 20% glycerol-brain heart infusion broth at −80°C and routinely cultured in chocolate agar plates at 37°C with 5% CO2.
TABLE 1.
Strains used in this study, sites and countries of isolation, and sequence types for 16S rRNA gene and hsp60 partial sequences
| Strain (virulence) | Isolation site | Country of isolation | 16S rRNA gene ST | hsp60 ST |
|---|---|---|---|---|
| H. parasuis reference strainsa | ||||
| SW140 (virulent) | Unknown (healthy animal) | Japan | B | 15 |
| C5 (moderately virulent) | Unknown | Sweden | C | 5 |
| H465 (nonvirulent) | Trachea | Germany | C | 14 |
| D74 (nonvirulent) | Unknown | Sweden | G | 27 |
| 174 (nonvirulent) | Nasal | Switzerland | G | 26 |
| 84-15995 (virulent) | Lung | United States | H | 15 |
| Nagasaki (highly virulent) | Systemic | Japan | H | 15 |
| 84-22113 (highly virulent) | Systemic | United States | H | 28 |
| SW124 (virulent) | Unknown (healthy animal) | Japan | I | 1 |
| ME4 | Unknown | Unknown | Z | 11 |
| SW114 (nonvirulent) | Unknown (healthy animal) | Japan | AD | 4 |
| 4 (highly virulent) | Unknown (healthy animal) | Japan | AD | 4 |
| H367 (highly virulent) | Unknown | Germany | AF | 34 |
| H. parasuis field strainsb | ||||
| SC14-2 | Nasal | Spain | A | 20 |
| SC14-7 | Nasal | Spain | A | 20 |
| SC18-3 | Nasal | Spain | A | 20 |
| CA36-1 | Nasal | Spain | A | 18 |
| CA37-1 | Nasal | Spain | A | 18 |
| SC18-6 | Nasal | Spain | A | 21 |
| SC14-1 | Nasal | Spain | A | 16 |
| SC12-1 | Nasal | Spain | A | 10 |
| MU26-2 | Nasal | Spain | A | 19 |
| 03/05 | Lung | Portugal | A | 4 |
| 279/03 | Lung | Spain | A | 5 |
| SC18-4 | Nasal | Spain | B | 20 |
| FL8-3 | Nasal | Spain | B | 22 |
| N67-1 | Nasal | Spain | B | 16 |
| N139/05-4 | Nasal | Spain | B | 1 |
| 37 | Unknown (sick animal) | Spain | B | 10 |
| 4959 | Unknown (sick animal) | Germany | B | 14 |
| P555/04 | Systemic | Argentina | B | 9 |
| 2757 | Lung | Germany | C | 43 |
| 7710 | Lung | Germany | C | 14 |
| LH9N-4 | Nasal | Spain | C | 5 |
| 34 | Unknown (sick animal) | Spain | C | 7 |
| 3023 | Lung | Germany | C | 23 |
| CD8-1 | Nasal | Spain | D | 4 |
| CD8-2 | Nasal | Spain | D | 4 |
| CD9-1 | Nasal | Spain | D | 4 |
| CD10-4 | Nasal | Spain | D | 4 |
| CD11-4 | Nasal | Spain | D | 4 |
| 112/02 | Systemic | Spain | D | 16 |
| VB4-1 | Nasal | Spain | E | 6 |
| 32-4 | Nasal | Spain | E | 4 |
| CA32-1 | Nasal | Spain | E | 24 |
| CA36-2 | Nasal | Spain | E | 16 |
| 58g | Unknown (sick animal) | Spain | E | 16 |
| 256/04 | Lung | Portugal | E | 7 |
| 167/03 | Lung | Spain | E | 1 |
| VB5-5 | Nasal | Spain | F | 2 |
| VS6-2 | Nasal | Spain | F | 2 |
| VS6-10 | Nasal | Spain | F | 2 |
| VS7-1 | Nasal | Spain | F | 2 |
| VS7-6 | Nasal | Spain | F | 2 |
| 416-1 | Nasal | Spain | F | 2 |
| IQ8N-6 | Nasal | Spain | G | 25 |
| 4590 | Lung | Germany | G | 27 |
| CA38-4 | Nasal | Spain | H | 15 |
| 23/04 | Systemic | Spain | I | 1 |
| 61/03 | Lung | Spain | I | 29 |
| 66/04-7 | Unknown | United Kingdom | J | 5 |
| 2620 | Systemic | Germany | J | 13 |
| 4857 | Systemic | Germany | J | 30 |
| SC19-1 | Nasal | Spain | K | 17 |
| SC19-2 | Nasal | Spain | K | 17 |
| SC19-4 | Nasal | Spain | K | 17 |
| F9 | Nasal | Spain | L | 2 |
| 393/03-5 | Unknown | Germany | L | 4 |
| CD7-3 | Nasal | Spain | M | 4 |
| IQ9N-3 | Nasal | Spain | M | 7 |
| FL3-1 | Nasal | Spain | N | 31 |
| 233/03 | Lung | Spain | N | 9 |
| N139/05-2 | Nasal | Spain | O | 1 |
| 34/03 | Systemic | Argentina | O | 32 |
| 66/04-1 | Unknown | United Kingdom | P | 12 |
| 66/04-4 | Unknown | United Kingdom | P | 12 |
| JA | Unknown (sick animal) | United Kingdom | Q | 8 |
| 373/03A | Systemic | Spain | Q | 15 |
| MU21-2 | Nasal | Spain | R | 19 |
| MU25-5 | Nasal | Spain | R | 19 |
| FL1-3 | Nasal | Spain | S | 1 |
| 230/03 | Lung | Spain | T | 3 |
| 264/99 | Systemic | Spain | U | 3 |
| 228/04 | Lung | Spain | V | 33 |
| P015/96 | Lung | Argentina | X | 8 |
| 66/04-3 | Unknown | United Kingdom | Y | 9 |
| 66/04-8 | Unknown | United Kingdom | Z | 35 |
| RW | Unknown | United Kingdom | AA | 6 |
| 4503 | Lung | Germany | AB | 13 |
| 393/03-4 | Unknown (sick animal) | Germany | AC | 36 |
| SC11-4 | Nasal | Spain | AE | 16 |
| Other species | ||||
| A. indolicus 37E3 | Unknown | Unknown | ||
| A. porcinus 245/04 | Systemic | Spain | ||
| A. porcinus 4598 | Systemic | Germany | ||
| A. porcinus Sp62 | Unknown | Unknown | ||
| A. porcinus B-20 | Unknown | Unknown | ||
| A. porcinus 27KC10 | Unknown | Unknown | ||
| A. minor 49 | Unknown (sick animal) | Spain | ||
| A. minor 2134 | Unknown (sick animal) | Spain | ||
| A. pleuropneumoniae 262/04 | Lung | Spain | ||
| A. pleuropneumoniae 38 | Unknown (sick animal) | Spain | ||
| Taxon C CAPM5113 | Unknown | Unknown | ||
| P. multocida 251/04 | Lung | Spain |
Virulence was defined as described by Kielstein and Rapp-Gabrielson (24).
Isolates from the same farm can be identified by their having the first two letters of their strain names in common.
DNA extraction, PCRs, and sequencing.
For each strain, a bacterial suspension was made in sterile phosphate-buffered saline and used to extract genomic DNA with a Nucleospin blood kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) following the manufacturer's instructions.
For identification purposes, the 16S rRNA gene was amplified and sequenced. 16S rRNA gene amplification was carried out using 3 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, 5 μl of extracted DNA, 0.5 μM forward primer (16S-up [5′ AGAGTTTGATCATGGCTCAGA 3′]), 0.5 μM reverse primer (16S-dn [5′ AGTCATGAATCATACCGTGGTA 3′]), and 1.5 U EcoTaq polymerase (Ecogen, Madrid, Spain) in a 50-μl reaction mix.
The hsp60 amplicon was obtained with universal degenerate primers for hsp60 by following a previously published protocol (18), with some modifications. The standard PCR mixture for hsp60 contained 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each universal primer, 1.5 U EcoTaq polymerase (Ecogen, Madrid, Spain), and 5 μl of extracted DNA in a 50-μl reaction volume. Amplification was performed for 35 cycles with an annealing temperature of 50°C.
The hsp60 and 16S rRNA gene amplicons were sequenced using a BigDye Terminator v.3.1 kit and an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, Calif.) with the same PCR primers and additional internal primers for the 16S rRNA gene (16SI1 [5′ TTGACGTTAGTCACAGAAG 3′], 16SI2 [5′ TTCGGTATTCCTCCACATC 3′], 16SI3 [5′ TAACGTGATAAATCGACCG 3′], and 16SI4 [5′ TTCACAACACGAGCTGAC 3′]). For identification purposes, sequence database searches were performed using programs based on the BLAST algorithm (1). Both the NCBI (http://www.ncbi.nlm.nih.gov/BLAST) and Ribosomal Database Project (http://rdp.cme.msu.edu) databases were searched.
For ERIC-PCR, purified DNA was quantified by spectrometry, and 100 ng was used as a template. The technique was performed by following a previously published protocol (35), including an extra final extension step of 20 min. Aliquots of 5 μl of PCR product were analyzed by electrophoresis (70 V, 3 h) in a 2% agarose gel. Band patterns were visualized by staining with a 1:10,000 dilution of SYBR gold (Invitrogen S.A., Barcelona, Spain) in 50 mM Tris and 5 mM EDTA buffer (pH 7.4) for 30 min. For normalization purposes, outer lanes contained a Superladder-Mid1 dsDNA marker kit (Eurogentec, Liege, Belgium). Images of the gel were captured with a Bio-Rad (Barcelona, Spain) transilluminator and stored as TIFF files for further analysis. Bands of 100 to 4,000 bp were used in the analysis.
Data analysis.
ERIC-PCR fingerprint analysis, sequence editing and analysis, and similarity matrix calculations were carried out using Fingerprinting II v3.0 software (Bio-Rad). Phylogenetic studies were carried out using the MEGA2 program (27).
ERIC-PCR band patterns were normalized, and Pearson correlation similarity matrixes were calculated. Cluster analysis of ERIC-PCR fingerprints was performed by the unweighted-pair group method using average linkages (UPGMA) as previously recommended (37). Maximum parsimony and neighbor-joining (using the Kimura two-parameter model) consensus trees for hsp60 and 16S rRNA gene partial sequences were constructed with 1,000 bootstrap values, and branches supported by bootstrap values of <50% were collapsed (5, 20).
RESULTS
16S rRNA gene sequencing.
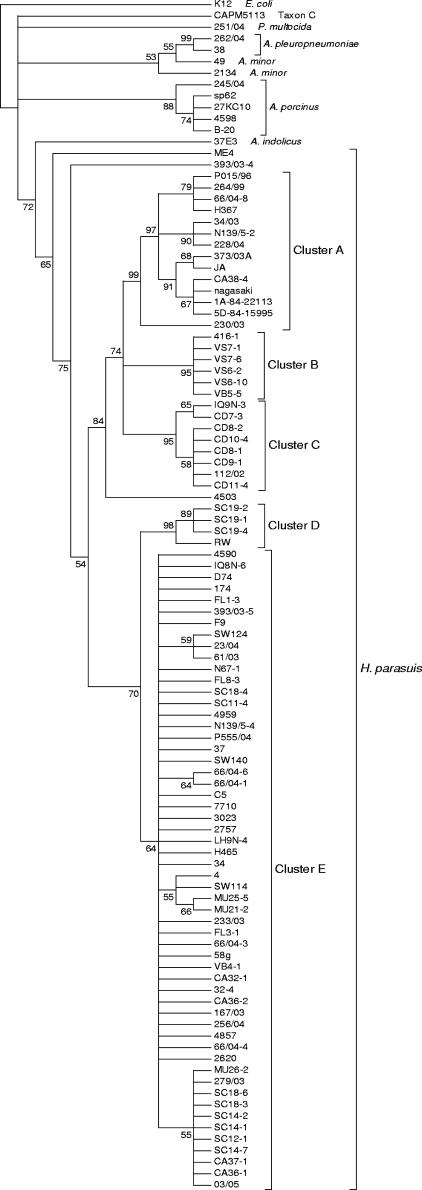
Partial 16S rRNA gene sequences of 1,391 to 1,394 nucleotides in length were obtained for each of the H. parasuis and Actinobacillus strains (GenBank accession numbers DQ228974 to DQ229076). The sequences were aligned with nucleotides 50 to 1448 of the Escherichia coli K-12 16S rRNA gene sequence (rrsH; GenBank accession number NC000913). Six insertion-deletion differences were identified. The aligned sequences showed 251 variable positions out of 1,397 total positions (18%). A pairwise alignment similarity matrix was constructed. The pairwise similarities among the H. parasuis strains ranged from 95.04 to 100%. By taking every different sequence, even if just one nucleotide was different, as a sequence type (ST), 30 different STs were defined for H. parasuis (indicated by consecutive letters A to Z and AA to AF) (Table 1). Interestingly, STs I, J, and Q were associated with clinical isolates, while STs F, K, and M were only found in nasal isolates. Notably, ST H was represented in three virulent reference strains. Maximum parsimony and neighbor-joining analyses were congruent, and the neighbor-joining tree is shown in Fig. 1. This analysis showed a monophyletic cluster containing all of the H. parasuis strains supported by a bootstrap value of 65%. Within the H. parasuis cluster, several subclusters were detected. Cluster A (Fig. 1) was supported by a high bootstrap value (99%) and contained virulent reference strains H367, Nagasaki, 84-22113, and 84-15995, together with clinical isolates (mainly systemic) and just one strain isolated from the nose (CA38-4). It is noteworthy that strain CA38-4 was isolated from a farm with an outbreak of Glässer's disease. Three subclusters showed bootstrap values of 95% or higher, but they were composed of very closely related isolates which were mainly collected from the same farm (clusters B, C, and D) (Fig. 1). Clusters C and D included strains isolated from diseased animals (112/02 and RW), while cluster B was composed of nasal isolates. Finally, a main cluster (cluster E) (Fig. 1) contained the rest of the clinical and nasal isolates and reference strains 4, D74, 174, C5, H465, and SW114.
FIG.1.
Neighbor-joining consensus tree for H. parasuis 16S rRNA gene partial sequences (1,000 bootstraps). The numbers in the nodes indicate the percentages of branching occurrences in 1,000 runs.
hsp60 sequencing.
Once the strains were classified to the species level, we next tested the value of the hsp60 sequence in genotyping H. parasuis isolates. Thus, partial sequences of 596 nucleotides were obtained from the 103 strains tested (GenBank accession numbers DQ198861 to DQ198950 and DQ228961 to DQ228973). The sequences were aligned with nucleotides 254 to 849 of the groEL gene of E. coli K-12 (GenBank accession number NC000913). All the sequences were aligned without gaps, and 228 of 596 (38%) positions were variable, with pairwise similarities ranging from 93.63 to 100%. For the H. parasuis isolates, 36 different STs were identified (indicated by consecutive numbers 1 to 36) (Table 1). Importantly, STs 3, 8, 9, 12, and 13 were associated with clinical isolates, while STs 2, 17, and 19 were only found in nasal isolates. Further examination of the sequences showed that variation was primarily limited to the third codon position (only 24% of amino acid positions were variable), and the average ratio of nonsynonymous to synonymous substitutions (ω) was 0.05. Figure 2 shows the neighbor-joining consensus tree for the sequences. Congruence, calculated as the Pearson product-moment correlation coefficient, between the 16S rRNA gene and hsp60 neighbor-joining trees was 75%. hsp60 sequences grouped all H. parasuis strains in one monophyletic cluster supported by a 99% bootstrap value. Unexpectedly, the following three strains previously classified as Actinobacillus by 16S rRNA gene sequencing were also included in the H. parasuis cluster: A. indolicus reference strain 37E3 and A. minor isolates 49 and 2134 (Fig. 1 and 2). Cluster 1 (Fig. 2) included field isolates, mainly clinical isolates, and virulent reference strains SW140, Nagasaki, 84-15995, and H367. Cluster 2 (Fig. 2) was structured in seven internal branches and included the majority of field isolates and reference strains 84-22311, SW124, C5, H465, D74, 174, 4, and SW114. The second cluster also contained isolate A. minor 49.
FIG. 2.
Neighbor-joining consensus tree for H. parasuis hsp60 partial sequences (1,000 bootstraps). The numbers in the nodes indicate the percentages of branching occurrences in 1,000 runs.
An examination of the hsp60 sequences from Actinobacillus strains available at the NCBI database showed the presence of putative DNA-uptake signal sequences (USS). In the hsp60 gene from A. pleuropneumoniae (accession number U55016), two sequences (AAGTGGCGT at position 226 and AAGTGGCGA at position 1146) very similar to the USS of Haemophilus influenzae (AAGTGCGGT) (4) could be detected. Also, in the Actinobacillus ureae hsp60 partial sequence (accession number AY123720), the sequence AAGTGGCTG was detected. For the H. parasuis and Actinobacillus sequences obtained in this study, the sequence AAGTGGCT/AG was present at position 562 of the amplicons. The presence of these putative USS, together with the different topologies of the 16S rRNA gene and hsp60 trees, supports the occurrence of lateral transfer of the hsp60 gene among the Actinobacillus and Haemophilus strains.
ERIC-PCR fingerprints.
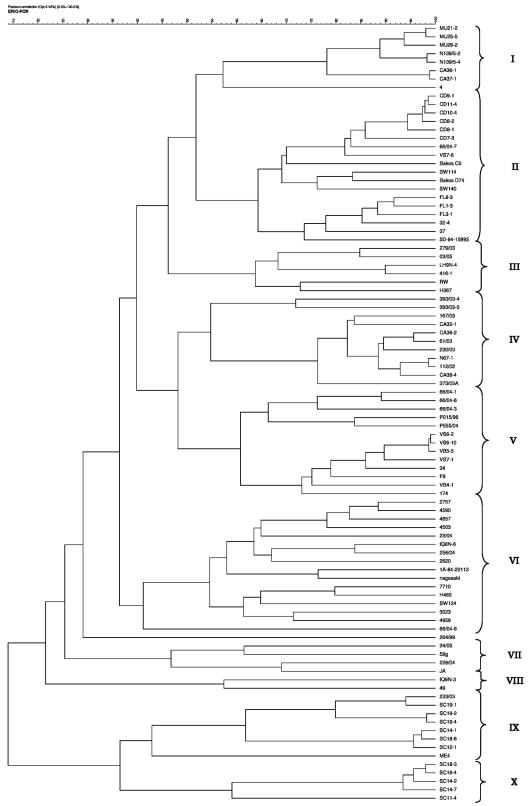
We further compared our data with the previously described ERIC-PCR method for H. parasuis. ERIC-PCR patterns for H. parasuis isolates were highly heterogeneous, and sometimes no common band between different fingerprints could be found. After curve-based Pearson correlation similarity matrix calculation, ERIC-PCR fingerprints led to similarities ranging from 0 to 99.07%. ERIC-PCR fingerprints were more variable and led to less similarity than both hsp60 and 16S rRNA gene sequences. After the UPGMA tree was built, 10 different clusters were defined (I to X) (Fig. 3). Cluster I contained nasal isolates from three different farms in Spain and reference strain 4. Cluster II contained nasal and lung isolates and five reference strains (C5, D74, SW114, SW140, and 84-15995). Clusters III, IV, and V contained isolates from different origins (Spain, Germany, United Kingdom, and Argentina) and several isolates from diseased animals. Reference strain H367 was included in cluster III, and strain 174 was included in cluster V. Notably, cluster VI was formed mainly by virulent reference strains Nagasaki, 84-22113, and SW124 and by isolates from diseased animals. Only the nonvirulent reference strain H465 and nasal isolate IQ8N-6 were also included in cluster VI. Cluster VII was formed by four clinical isolates from Spain, the United Kingdom, and Argentina. Clusters IX and X were mainly nasal isolates from the same farm.
FIG. 3.
UPGMA tree of ERIC-PCR fingerprints for H. parasuis strains.
DISCUSSION
In order to improve the epidemiological study of H. parasuis strains, we employed the hsp60 gene as a marker. This study represents extensive sequencing work on H. parasuis hsp60 and 16S rRNA genes. Also, hsp60 sequences of H. parasuis, A. indolicus, A. porcinus, and A. minor are reported here for the first time. All of the strains tested were sequenced (i.e., typed), including the Actinobacillus strains.
As we expected, sequencing of the hsp60 fragment gave a high level of variation among the strains examined in the study, providing more resolution below the species level than the 16S rRNA gene. The hsp60 sequences were more variable and had fewer pairwise similarities than the 16S rRNA gene sequences, i.e., even though the 16S rRNA gene sequences were longer, they provided a smaller number of alleles than the partial hsp60 sequences. In addition, partial sequencing of hsp60 is less labor-intensive, and in contrast to the case for serotyping, all strains could be typed. Additionally, sequences are easy to compare among different laboratories. All of these features make this method suitable for the unequivocal characterization of H. parasuis strains for global epidemiology.
As mentioned before, ERIC-PCR patterns were highly heterogeneous. ERIC-PCR fingerprints were useful for the discrimination of closely related isolates (i.e., to determine if isolates from the same farm or animal were in fact the same or different strains), but they were too diverse to find relationships between more distant isolates. On the other hand, some clusters of ERIC-PCR fingerprints grouped strains from different countries. This may indicate either that some strains have a very ubiquitous distribution or that the genomic rearrangements producing the fingerprints are entirely random. Since the latter explanation seems improbable, we favor the first one, and it may be explained, at least partially, by globalized pig trading.
The study of strains by sequencing the Hsp60 and 16S rRNA genes yielded a distribution of the strains in several groups. Phylogenetic analysis of hsp60 and 16S rRNA genes led to monophyletic clusters for H. parasuis. Although there was not complete agreement between the gene trees, a clear subcluster of virulent reference strains and systemic isolates was defined in both analyses (cluster A in Fig. 1 and cluster 1 in Fig. 2). This cluster is of particular interest since it could be the first indication of the presence of a highly pathogenic lineage for H. parasuis strains. However, there were also some clinical isolates distributed in other clusters, pointing out the difficulties in reaching a clear conclusion using a monogenic approach. The study of the H. parasuis strains with hsp60 sequences showed two separate clusters (clusters 1 and 2 in Fig. 2). Cluster 1 included several virulent reference strains, and cluster 2 included the majority of H. parasuis strains, showing a clear structure in seven branches. Some disagreements in the topologies of the two trees (16S rRNA gene and hsp60 trees) were detected, involving H. parasuis, A. indolicus, and A. minor strains. This could be due to recent divergence between H. parasuis, A. indolicus, and A. minor (14, 25, 30) or could constitute an indication of horizontal transfer of genes between H. parasuis and Actinobacillus strains. In agreement with the latter explanation, the sequence of the hsp60 gene from A. minor 49 showed a high level of identity (98.15%) with the corresponding gene from H. parasuis ME4. In addition, there were other strains that changed positions between the two trees. This was the case for strains 230/03, 264/99, and 66/04-8, among others. In fact, one of the reasons for phylogenetic tree topology disagreements, unexpected similarities, and unusual phyletic patterns is lateral gene transfer between strains (26). Additional pieces of information that support the idea of lateral gene transfer between these strains are that natural transformation was recently described for H. parasuis (6) and that putative USS could be detected in Actinobacillus and Haemophilus species. Also, a native plasmid has been isolated from H. parasuis (28) which is related to a plasmid found in A. pleuropneumoniae. Thus, it can be hypothesized that these plasmids were also transferred laterally between these species.
Taking into account the large number of different ERIC fingerprints found, the different topologies of the trees, the presence of possible DNA uptake sequences, and the evidence of transformation in H. parasuis, genome rearrangements and lateral gene transfer could be ongoing phenomena in these strains. The presence of lateral gene transfer is noteworthy since it could explain why strains belonging to Actinobacillus species and classified as nonpathogenic commensal biota (10) are isolated from systemic sites in diseased animals. It is possible that those species, which are in contact in the respiratory tract of the pig, share virulence genes.
The large number of strains included in the study and the use of three different markers provided insight into the diversity of H. parasuis. The large numbers of 16S rRNA gene and hsp60 STs found for H. parasuis (30 and 36 STs, respectively) and the ERIC-PCR patterns indicate that H. parasuis is a very heterogeneous species, with a high level of diversity and no clear predominance of a specific ST. The presence of a high level of heterogeneity within this species was already suspected since there are many serologically nontypeable strains and because of the lack of cross-immunization between strains (36).
Although some STs were only found among clinical isolates, no clear relationship between 16S rRNA gene or hsp60 partial sequences or ERIC-PCR fingerprints and the site of isolation (organ or tissue), virulence, or geographical origin was found.
In conclusion, hsp60 sequences can be used as an epidemiological marker for H. parasuis and represent a good alternative to fingerprinting approaches. The possibility of developing molecular diagnostic tools with this sequence, as proposed for other species (18, 19, 43), seems not to be feasible due to the possibility of lateral gene transfer between H. parasuis and related species. In addition, although H. parasuis isolates were clearly monophyletic by their 16S rRNA gene sequences, the bootstrap values were generally low. Thus, other multigenic approaches would be needed in order to clarify the taxonomy of this group of species and to determine the incidence of lateral gene transfer, if any, between isolates.
. . .
Acknowledgments
We acknowledge Nuria Galofré for technical support. Marta Cerdá, Joaquim Segalés, and Lorenzo Fraile are also acknowledged for their helpful discussions.
This work was supported by grant AGL2004-07349 from the Ministerio de Ciencia y Tecnología of Spain. Fellowship support for A.O. from CReSA is also acknowledged.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, H., M. Shibata, N. Kajio, and T. Morozumi. 1996. Pathogenicity of Haemophilus parasuis serovars 4 and 5 in contact-exposed pigs. J. Vet. Med. Sci. 58:559-561. [DOI] [PubMed] [Google Scholar]
- 3.Amano, H., M. Shibata, N. Kajio, and T. Morozumi. 1994. Pathologic observations of pigs intranasally inoculated with serovar 1, 4 and 5 of Haemophilus parasuis using immunoperoxidase method. J. Vet. Med. Sci. 56:639-644. [DOI] [PubMed] [Google Scholar]
- 4.Bakkali, M., T. Y. Chen, H. C. Lee, and R. J. Redfield. 2004. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc. Natl. Acad. Sci. USA 101:4513-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldauf, S. L. 2003. Phylogeny for the faint of heart: a tutorial. Trends Genet. 19:345-351. [DOI] [PubMed] [Google Scholar]
- 6.Bigas, A., M. E. Garrido, A. M. de Rozas, I. Badiola, J. Barbe, and M. Llagostera. 2005. Development of a genetic manipulation system for Haemophilus parasuis. Vet. Microbiol. 105:223-228. [DOI] [PubMed] [Google Scholar]
- 7.Blackall, P. J., D. J. Trott, V. Rapp-Gabrielson, and D. J. Hampson. 1997. Analysis of Haemophilus parasuis by multilocus enzyme electrophoresis. Vet. Microbiol. 56:125-134. [DOI] [PubMed] [Google Scholar]
- 8.Blanco, I., L. Galina-Pantoja, S. Oliveira, C. Pijoan, C. Sanchez, and A. Canals. 2004. Comparison between Haemophilus parasuis infection in colostrums-deprived and sow-reared piglets. Vet. Microbiol. 103:21-27. [DOI] [PubMed] [Google Scholar]
- 9.Brousseau, R., J. E. Hill, G. Prefontaine, S. H. Goh, J. Harel, and S. M. Hemmingsen. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiers, K., F. Haesebrouck, B. Mateusen, I. Van Overbeke, and R. Ducatelle. 2001. Pathogenicity of Actinobacillus minor, Actinobacillus indolicus and Actinobacillus porcinus strains for gnotobiotic piglets. J. Vet. Med. B 48:127-131. [DOI] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331-343. [DOI] [PubMed] [Google Scholar]
- 12.de la Puente Redondo, V. A., J. Navas Mendez, N. Garcia del Blanco, N. Ladron Boronat, C. B. Gutierrez Martin, and E. F. Rodriguez Ferri. 2003. Typing of Haemophilus parasuis strains by PCR-RFLP analysis of the tbpA gene. Vet. Microbiol. 92:253-262. [DOI] [PubMed] [Google Scholar]
- 13.Del Rio, M. L., C. B. Martin, J. Navas, B. Gutierrez-Muniz, J. I. Rodriguez-Barbosa, and E. F. Rodriguez Ferri. 2005. aroA gene PCR-RFLP diversity patterns in Haemophilus parasuis and Actinobacillus species. Res. Vet. Sci. 80:55-61. [DOI] [PubMed] [Google Scholar]
- 14.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 174:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fares, M. A., E. Barrio, B. Sabater-Munoz, and A. Moya. 2002. The evolution of the heat-shock protein GroEL from Buchnera, the primary endosymbiont of aphids, is governed by positive selection. Mol. Biol. Evol. 19:1162-1170. [DOI] [PubMed] [Google Scholar]
- 16.Fares, M. A., A. Moya, and E. Barrio. 2005. Adaptive evolution in GroEL from distantly related endosymbiotic bacteria of insects. J. Evol. Biol. 18:651-660. [DOI] [PubMed] [Google Scholar]
- 17.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 18.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graur, D., and W. H. Li. 1999. Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 21.Harris, D. L., R. F. Ross, and W. P. Switzer. 1969. Incidence of certain microorganisms in nasal cavities of swine in Iowa. Am. J. Vet. Res. 30:1621-1624. [PubMed] [Google Scholar]
- 22.Hoffman, P. S., and R. A. Garduno. 1999. Surface-associated heat shock proteins of Legionella pneumophila and Helicobacter pylori: roles in pathogenesis and immunity. Infect. Dis. Obstet. Gynecol. 7:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda, J. M., and S. L. Abbott. 2002. Bacterial identification for publication: when is enough enough? J. Clin. Microbiol. 40:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielstein, P., and V. J. Rapp-Gabrielson. 1992. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J. Clin. Microbiol. 30:862-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielstein, P., H. Wuthe, O. Angen, R. Mutters, and P. Ahrens. 2001. Phenotypic and genetic characterization of NAD-dependent Pasteurellaceae from the respiratory tract of pigs and their possible pathogenetic importance. Vet. Microbiol. 81:243-255. [DOI] [PubMed] [Google Scholar]
- 26.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Lancashire, J. F., T. D. Terry, P. J. Blackall, and M. P. Jennings. 2005. Plasmid-encoded TetB tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 49:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miniats, O. P., N. L. Smart, and S. Rosendal. 1991. Cross protection among Haemophilus parasuis strains in immunized gnotobiotic pigs. Can. J. Vet. Res. 55:37-41. [PMC free article] [PubMed] [Google Scholar]
- 30.Moller, K., V. Fussing, P. A. Grimont, B. J. Paster, F. E. Dewhirst, and M. Kilian. 1996. Actinobacillus minor sp. nov., Actinobacillus porcinus sp. nov., and Actinobacillus indolicus sp. nov., three new V factor-dependent species from the respiratory tract of pigs. Int. J. Syst. Bacteriol. 46:951-956. [DOI] [PubMed] [Google Scholar]
- 31.Moller, K., and M. Kilian. 1990. V factor-dependent members of the family Pasteurellaceae in the porcine upper respiratory tract. J. Clin. Microbiol. 28:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozumi, T., and J. Nicolet. 1986. Morphological variations of Haemophilus parasuis strains. J. Clin. Microbiol. 23:138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morozumi, T. P., U. Braun, and J. Nicolet. 1986. Deoxyribonucleic acid relatedness among strains of Haemophilus parasuis and other Haemophilus spp. of swine origin. Int. J. Syst. Bacteriol. 36:17-19. [Google Scholar]
- 34.Nicolet, J. 1986. Haemophilus infection, p. 426-436. In A. D. Leman et al. (ed.), Diseases of swine, 6th ed. Iowa State University Press, Ames, Iowa.
- 35.Oliveira, S., P. J. Blackall, and C. Pijoan. 2003. Characterization of the diversity of Haemophilus parasuis field isolates by use of serotyping and genotyping. Am. J. Vet. Res. 64:435-442. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira, S., and C. Pijoan. 2004. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet. Microbiol. 99:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Ooyen, A. V. 2001. New approaches for the generation and analysis of microbial fingerprints. Elsevier, Amsterdam, The Netherlands.
- 38.Rafiee, M., M. Bara, C. P. Stephens, and P. J. Blackall. 2000. Application of ERIC-PCR for the comparison of isolates of Haemophilus parasuis. Aust. Vet. J. 78:846-849. [DOI] [PubMed] [Google Scholar]
- 39.Rapp-Gabrielson, V. J. 1999. Haemophilus parasuis, p. 475-481. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames, Iowa.
- 40.Stackebrandt, E. G., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 41.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevshy, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 43.Wong, R. S., and A. W. Chow. 2002. Identification of enteric pathogens by heat shock protein 60 kDa (HSP60) gene sequences. FEMS Microbiol. Lett. 206:107-113. [DOI] [PubMed] [Google Scholar]