Abstract
A standardized real-time reverse transcription-PCR (RT-PCR) assay has been developed for an accurate estimation of the number of genome copies of hepatitis A virus (HAV) in clinical and shellfish samples. Real-time procedures were based on the amplification of a fragment of the highly conserved 5′ noncoding region and detection through an internal fluorescent probe, including TaqMan and beacon chemistries, in one- and two-step RT-PCR formats. The best performance in terms of sensitivity and reproducibility was achieved by a one-step TaqMan RT-PCR, with a sensitivity enabling the detection of 0.05 infectious unit and 10 copies of a single-stranded RNA (ssRNA) synthetic transcript. Standard reagents, such as a mengovirus strain and an ssRNA transcript, were employed as controls of nucleic acid extraction and RT-PCR, respectively. The test proved to be highly specific after a broad panel of enteric viruses was tested. Sequence alignment of target regions of the primers and probe proved them to be adequate for the quantification of all HAV genotypes. In addition, a quasispecies analysis of the mutant spectrum indicated that these regions are not prone to variability, thus confirming their robustness.
Hepatitis A virus (HAV) infection is the leading cause of acute viral hepatitis throughout the world (11). The distribution patterns of hepatitis A in different geographical areas of the world are closely related to socioeconomic development (11). The endemicity is low in developed regions and high in underdeveloped countries. HAV infection is propagated mainly via the fecal-oral route, with person-to-person contact being the most common mode of transmission (18). Transmission through the parental route may also occur (19, 30). While in approximately 40% of the reported cases of hepatitis A the source of infection cannot be identified, waterborne (3) and food-borne (8, 25, 29) outbreaks of the disease have been reported. Within these categories, shellfish grown and harvested from waters receiving urban contaminants is a cause of large outbreaks of infectious hepatitis (10, 29). However, due to technical limitations, the roles of lightly contaminated blood products, water, and food in the burden of sporadic cases remain unknown. The development of sensitive, reliable techniques for the accurate quantification of HAV in these types of samples is required to ensure the safety of these products.
HAV is the prototype of the Hepatovirus genus within the Picornaviridae family. Its 7.5-kb single-stranded RNA (ssRNA) genome bears different regions: the 5′ and 3′ noncoding regions (NCRs); the P1 region, which encodes the structural proteins VP1, VP2, VP3, and a putative VP4; and the P2 and P3 regions, encoding nonstructural proteins associated with replication (11). Immunological evidence has determined the existence of a single serotype of HAV (16), although sequencing of the putative VP1/2A junction allows the differentiation of six genotypes (6, 27), based on nucleotide homologies lower than 85%. However, all six genotypes are very closely related in the 5′NCR, which is the most conserved region of the genome due to its functional structure in the processes of translation and replication and a maximum nucleotide divergence of less than 5%. Consequently, the 5′NCR is a very convenient region for the design of an accurate real-time quantification technique.
MATERIALS AND METHODS
Cells and virus.
The cytopathogenic pHM175 43c strain of HAV (a gift of T. Cromeans, Centers for Disease Control and Prevention, Atlanta, Ga.) was used throughout this study. Virus replication in FRhK-4 cells was performed as previously described (2). Infectious mengovirus was obtained after transfection of a cDNA clone, pMC0 (kindly provided by Ann Palmenberg, University of Wisconsin), into HeLa cells as previously described (17), and viral stocks were thereafter produced in the same cells. Additionally a broad collection of enteric viruses, including different members of the Picornaviridae family as well as rotavirus, norovirus, adenovirus, astrovirus, and hepatitis E virus, were tested for their amplification/detection by using the HAV-derived primers and probe.
Selection of primers and probe.
Multiple-sequence alignments with the HAV sequences available at GenBank, including strains belonging to all human genotypes and subgenotypes so far described, were carried out with the ClustalW program (European Bioinformatics Institute). Two software programs, Primer Express from Applied Biosystems and Primer 3 from Stratagene, were used for the search for optimal primer and probe targets.
Quasispecies analysis.
For the analysis of the quasispecies distribution, the pHM175 43c HAV strain was plaque purified three times in FRhK-4 cells as previously described (28). A biological clone (pHM175 43c P0) was serially passaged 50 times in the same cell line as previously described (2), and two populations, pHM175 43C P26 and pHM175 43C P50, were analyzed. RNA extracted from both viral populations was retrotranscribed to a cDNA with the Moloney murine leukemia virus reverse transcriptase (Roche) using the primer HAV295 (5′-TGCTAATCATGGAGTTGACC-3′), and the cDNA was copied and amplified with the thermostable Pwo polymerase (Roche) from Pyrococcus woesei, which has proofreading activity, by adding the primer HAV30BamHI (5′-CGGGATCCCTCTTGGAAGTCCATG-3′) to the reaction mix. The latter primer incorporates a BamHI restriction site to facilitate its cloning into the pGEM-3Zf(+) vector. The synthesis of the cDNA was performed as previously described (28). Since the DNA fragments produced by Pwo polymerase are blunt ended, a single digestion with the BamHI enzyme was necessary for the amplimer, while the plasmid vector was digested with both BamHI and HincII restriction enzymes. Digested DNAs were purified with the High Pure PCR product purification kit (Roche) according to the directions of the manufacturer. DNA ligations were performed overnight at 16°C using T4 DNA ligase (Roche), and transformant clones were screened by the standard white/blue galactosidase colorimetric reaction. Plasmid DNA from each of 100 clones was purified by using the Wizard Plus SV Minipreps kit (Promega). Nucleotide sequencing was carried out in an ABI PRISM 377 automated DNA sequencer, the ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems), and vector-derived primers. All mutations were confirmed by sequencing both strands of DNA.
Real-time reverse transcription-PCR (RT-PCR) TaqMan assays.
The TaqMan probe was labeled with 6-carboxyfluorescein at the 5′ end and was modified with a minor-groove binder at the 3′ end. The real-time instruments used were the ABI PRISM 7700 (Applied Biosystems) and the Mx3000P (Stratagene). Kits from different companies, including Applied Biosystems, QIAGEN, Invitrogen, and Stratagene, were evaluated. However, the one-step Brilliant QRT-PCR Core Reagent kit from Stratagene and a two-step assay with a home-designed RT reaction combined with the FullVelocity QPCR master mix from Stratagene and the Mx3000P apparatus were employed for the final development of the technique. For the one-step reaction, the concentrations of primers, probe, and Mg2+ were optimized at 0.6 μM, 0.25 μM, and 3 mM, respectively, in a final volume of 25 μl containing 5 μl of sample. The temperature-time program was as follows: 1 h at 50°C for the RT reaction, 10 min at 95°C as a hot start, and 45 cycles of 15 s at 95°C for denaturation, 1 min at 60°C for annealing, and 1 min at 70°C for extension. The fluorescence was measured at the end of each cycle. In the two-step assay the RT reaction was performed at 45°C for 1 h, using the Expand reverse transcriptase (Roche) with 0.5 μM of the reverse primer, 0.2 mM of nucleotides, and 5 mM of dithiothreitol in a final volume of 25 μl containing 10 μl of sample. The optimized composition for the PCR mix using the FullVelocity QPCR kit was 0.9 μM of the reverse primer, 0.5 μM of the forward primer, and 0.25 μM of probe in a final volume of 25 μl, in which 5 μl of the RNA template had been added. The amplification program consisted of preheating for 2 min at 95°C and 40 cycles of 10 s at 95°C for denaturation and 1 min at 60°C for annealing-extension. The fluorescence was read at the end of each cycle.
Construction of an RNA standard.
A synthetic RNA molecule to be used as an internal standard was obtained by in vitro transcription of a cloned cDNA corresponding to the amplimer resulting from the RT-PCR with the reverse primer HAV240 (5′-GGAGAGCCCTGGAAGAAAG-3′) and the forward primer HAV68BamHI (5′TCACCGCCGTTTGCCTAGGCTATAGGCTAAATTTTCCCTTTCGGATCCCCC3′) of the pHM175 43c HAV strain. This amplimer corresponds to the one synthesized by the real-time techniques presented here but includes a newly generated BamHI restriction site in order to be distinguished from the viral amplimer in case of cross-contamination. The cloned amplimer was synthesized in an RT-PCR with the Expand reverse transcriptase enzyme and the Pwo DNA polymerase (Roche), which generates blunt-ended termini. The pGEM-3Zf(+) vector was digested with the enzyme HincII. After purification, both amplimer and vector were ligated overnight at 16°C using T4 DNA ligase. Ligation products were transformed in Escherichia coli DH5α, and transformant clones were screened by the standard white/blue galactosidase colorimetric reaction. Several positive clones were analyzed, and one containing the cDNA under the control of the SP6 polymerase was selected. The in vitro transcription was performed on linearized plasmid samples by using the Riboprobe in vitro transcription system SP6 kit (Promega), following the manufacturer's instructions. The concentration of RNA transcripts was measured by optical density at 260 nm (OD260) after their purification with the RNeasy Plant minikit (QIAGEN), following the recommended procedure.
Mengovirus control.
The infectious clone mentioned above (pMC0), which lacks the poly(C) tract (17) in comparison to the wild-type mengovirus, gives rise to a mutant virus strain, vMC0, with growth properties identical to those of the wild-type virus but with an avirulent phenotype (17). vMC0 may be thus considered a good candidate for controlling the process of nucleic acid extraction based on the fact that it belongs to the same Picornaviridae family as HAV. For this purpose a real-time TaqMan technique has been developed based on the amplification of a target sequence structurally related to that of HAV.
Clinical samples, shellfish samples, and viral RNA extraction.
Clinical samples included stool and serum samples from patients affected by hepatitis A. Additionally, stool and serum samples from patients not affected by hepatitis A were experimentally contaminated with the pHM175 43c HAV strain. Stools were suspended (10%, wt/vol) in phosphate-buffered saline containing 2 M NaNO3, 1% bovine serum albumin (BSA) (fraction V), and 0.1% Triton X-100 (pH 7.2) and pelleted at 1,000 × g for 5 min, and nucleic acids were extracted from 150 μl of the resulting supernatant by using the RNeasy Plant minikit (QIAGEN). Nucleic acids were purified from 150 μl of serum by using the NucleoSpin RNA virus (Macherey-Nagel).
Frozen samples of clams directly associated with an outbreak of hepatitis A (29) were analyzed. Processing of shellfish was performed essentially by the method described by Atmar et al. (1). Briefly, the stomachs and digestive diverticula were dissected from the clams and subjected to high-speed homogenization (Sorval OCI Omni mixer; Omni International, Waterbury, Conn.). Viruses were extracted from the homogenates (corresponding to 1.5 g of shellfish tissue) by sequential extractions with chloroform-butanol and Cat-Floc T (Calgon Corp., Elwood, Pa.) and concentrated by polyethylene glycol precipitation. Nucleic acids from these concentrates were extracted with the RNeasy Plant minikit (QIAGEN) according to the manufacturer's instructions.
RNA extraction from viruses harvested from cell cultures was performed with the NucleoSpin RNA virus (Macherey-Nagel).
RESULTS
Primer-probe set selection.
The development of an optimal real-time RT-PCR methodology relies on the proper selection of the target sequences for primers and probe annealing. Despite the existence of different software programs to design these optimized primers and oligoprobes, the level of genetic conservation among different viral strains is not accurately analyzed, although it probably is as important as the nucleotide composition for an efficient quantitation. In this context, the selection of a highly conserved primer-probe set should be the first step in the design of a real-time RT-PCR for viral quantitation.
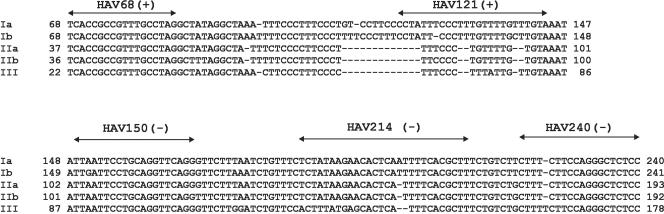
The most conserved region of HAV corresponds to the 5′NCR, being the regions spanning positions 66 to 95 and 227 to 262 (numbering corresponds to strain HM175, accession number M14707) (11). Although some HAV strains include some deletions in the region from position 99 to 207, it is otherwise quite well conserved between nucleotides 150 and 190. Taking this information into account, a standard RT-PCR-Southern blot was previously designed (29) based on the reverse primer HAV240 (5′-GGAGAGCCCTGGAAGAAAG-3′), the forward primer HAV68 (5′-TCACCGCCGTTTGCCTAG-3′), and the probe HAV150 (5′-TTAATTCCTGCAGGTTCAGG-3′) and proved to be successful with both serum and shellfish samples.
The adaptation of this technique to the real-time format could be straightforward or intricate depending on the software used for the design of primers and probe. The sequences for the reverse primer and the probe suggested by the Primer 3 software mostly matched the HAV240 and HAV150 regions. In contrast, the sequence advised for the forward primer was located exactly 20 nucleotides to the 5′ end of the HAV68 primer, probably making the length of the amplicon too long for an efficient assay. The Primer Express software suggested a complete adjustment for the primer sequences with no change of the probe sequence. However, the proposed regions were not highly conserved among the HAV sequences available in GenBank in comparison to the HAV240 and HAV68 primers. In the HAV240 primer region, the single mutation detected was a point insertion in genotype III (Fig. 1), while in the HAV68 primer region, no mutations were observed among the available consensus sequences of the five human genotypes (Fig. 1). In contrast, six mutations were detected in each of the regions recommended for the reverse HAV214 (5′-AAGCGTGAAATGAGTGTTCTTATAGA-3′) and forward HAV121 (5′-TCCTATTCCCTTTGTTTTGCTTGTA-3′) primers. Most important, however, was the occurrence of important deletions in genotypes IIa, IIb, and III in the HAV121 primer region.
FIG. 1.
Consensus sequence alignment of human HAV genotypes in the amplified region, including the primer and probe target sequences. Nucleotide numbering is according to the HM-175 strain. Terminal 5′ end nucleotides in the available sequences from genotypes II and III are missing.
Concerning the probe target, a negative-strand oligoprobe, HAV150(−) (5′CCTGAACCTGCAGGAATTAA3′), was synthesized in order to increase the C/G ratio. Finally, the potential loss of sensitivity due to the extended length of the amplified fragment could be amended by modifying the amplification program, as discussed below.
Quasispecies analysis of the primer-probe annealing regions.
To further analyze the degree of variability of the target sequences, an analysis of the quasispecies distribution of the pHM175 43c strain of HAV was undertaken. One hundred molecular clones were analyzed at each of two passages (passage 26 [P26] and P50) of replication in the FrhK4 cell line. Regarding the target sequences for the HAV240 reverse primer, no mutations were detected at either P26 or P50. In contrast, one point mutation was observed at P50 in the region of the suggested primer HAV214. The numbers of mutations found in the forward primer regions were one at P50 in the HAV68 target sequence and two at P26 in the HAV121 region. Finally, the target region for the probe showed a point mutation at P50.
Specificity of the chosen primer-probe set.
In order to confirm the specific detection of HAV, a wide variety of enteric viruses were tested for their amplification/detection by using the HAV240 and HAV68/HAV150 primer-probe set. The assayed viruses included 10 different picornaviruses: Poliovirus (serotype 1 vaccine strain), Human Enterovirus B (echovirus 1, echovirus 11, echovirus 30, and coxackievirus B5), Human Enterovirus C (coxackievirus A24), Human Enterovirus D (enterovirus 70), Bovine Enterovirus, Porcine teschovirus (porcine enterovirus 1), and Encephalomyocarditis virus. Other enteric viruses, such as Hepatitis E virus, human and porcine Rotavirus (group A), Norovirus (Norwalk-like virus), Mamastrovirus (human astrovirus type 1), and Human adenovirus F (enteric adenovirus type 40), were also employed. As expected, none of the tested virus gave positive results at either high concentrations (106 to 108 50% tissue culture infective doses [TCID50]/ml or undiluted 10% fecal suspensions) or low concentrations (104 50% TCID50/ml or 1/10 dilutions of 10% fecal suspensions). The HAV strain pHM175 43c, belonging to genotype IB, was used as a positive control.
Best standard molecules for generation of standard curves.
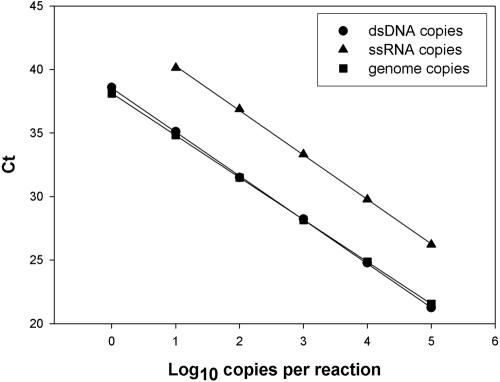
One of the crucial steps in a real-time quantification is the choice of the most proper molecule for the generation of the standard curve. There are three candidates that are able to be amplified and detected by means of the selected primers and probe and whose concentration may be determined: a double-stranded DNA (dsDNA) molecule corresponding to the actual amplimer, an ssRNA molecule obtained after in vitro transcription of a cloned cDNA corresponding to the amplimer, and the actual virus (pHM175 43c strain) genome. The concentration of each of these molecules was estimated by determining the OD260 in the two former cases and by infectivity in the third case. In this last case, however, the final concentration was refined by applying a factor of ×60 to the infectious titer in order to achieve a reliable number of physical genomes (7, 13). As an example, a representative set of standard curves for a one-step TaqMan reaction is depicted in Fig. 2. The estimated regression lines for the dsDNA, ssRNA, and viral genome molecules in distilled water were y = 38.56 − 3.46x (r2 = 0.999), y = 43.75 − 3.50x (r2 = 0.999), and y = 38.13 − 3.32x (r2 = 0.999), respectively (Fig. 2). Although the slopes of the three curves are similar, the intercept value in the case of the ssRNA molecule is significantly higher. From these results it is advisable to recommend the use of either the dsDNA molecule or the actual viral genome for the construction of the standard curve. Using the pHM175 43c strain, a linear range of detection from 1 physical particle/reaction or 200 physical particles/ml to 105 physical particles/reaction or 2 × 107 physical particles/ml was achieved. With the dsDNA molecule, the upper limit was extended to at least 2 × 1010 molecules/ml.
FIG. 2.
Standard curves for the HAV real-time TaqMan RT-PCR assay. Three molecules were employed: a dsDNA, an ssRNA, and the strain pHM175 43c viral genome. The concentrations of the synthetic molecules were estimated by determining the OD260. Virus titer was determined by infectivity, and physical genomes were estimated by applying a factor of ×60 to the infectious titer (7, 13).
Limits of detection for different real-time RT-PCR formats.
Two real-time RT-PCR formats based on the detection of amplicons through two different fluorogenic chemistries were assayed for the development of an HAV quantification technique. A TaqMan probe on the one hand and a beacon probe on the other were assayed. Additionally, in the case of the TaqMan probe, several one-step and two-step formats were also evaluated. In each case standard curves were drawn using the ssRNA molecule, the dsDNA amplicon, and the actual virus. Once optimized (i.e., adjusting primers and probe concentration for each kit), all of them gave very good detection limits in both one- and two-step formats (10 ssRNA molecules, 1 viral RNA molecule, and 0.05 infectious virus per reaction), with the sole exception of the Applied Biosystems one-step kit, which showed a significantly lower sensitivity. However the beacon probe format was neither as sensitive nor as reproducible as the TaqMan probe format (10 ssRNA molecules, 10 viral RNA molecules, and 0.5 infectious virus per reaction). Since the use of a two-step format did not increase the sensitivity of detection, the one-step format is recommended for less time-consuming and more convenient manipulation, thus reducing cross-contamination risks.
Best standard molecules for accurate quantification.
Another set of controls are required for reliable quantifications of the samples. These controls, added at known concentrations to the samples, and so-called internal controls should be able to trace both the efficiency of the nucleic acid extraction and the efficiency of the RT-PCR. The ssRNA molecule may be a good candidate for the last step, provided that an RNase-free nucleic acid suspension is achieved. The actual virus, although by itself the best control for the extraction efficiency, is hardly useful since it is indistinguishable from the potential naturally occurring HAV present in the same samples. Alternatively, a model virus should be used for this purpose, such as the mengovirus.
A one-step TaqMan real-time RT-PCR based on the amplification of a related 5′NCR fragment from the mengovirus genome was developed and optimized for the working conditions of the HAV assay, in order that it could be quantified in the same plate. The sensitivity of this technique was 100 physical particles/reaction, with a linear range of from 2.0 × 102 to 2.0 × 109 particles/ml.
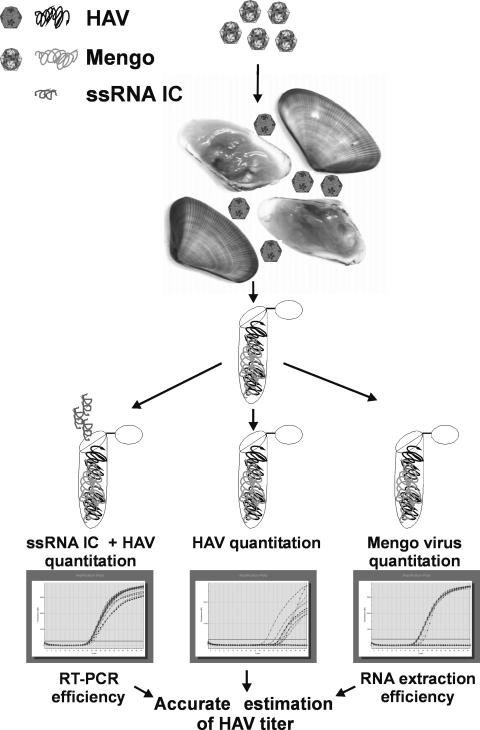
A schematic representation of the whole proposed control process is shown in Fig. 3. This approach consists of the addition of a known concentration of mengovirus to the sample to be analyzed, before performing the virus nucleic acids extraction. Once the extraction is performed, three separate subsamples of the nucleic acid suspension are analyzed. To one subsample a known concentration of the ssRNA control is added. With two of the subsamples, including the one with the ssRNA control, quantification of HAV is performed. With the third subsample, quantification of mengovirus is assessed. Two values will be obtained for HAV, corresponding to the level of virus present in the sample on the one hand and to the addition of the HAV genomes plus the ssRNA molecules on the other. At this point the efficiency of the RT-PCR may be estimated by comparing the number of ssRNA molecules detected (by subtracting the number of HAV genomes quantified in one subsample from the sum of HAV plus ssRNA in the other subsample) with the number of ssRNA molecules added. Finally, from the third subsample, the efficiency of the nucleic acid extraction may be calculated by comparing the numbers of detected and added mengovirus genomes.
FIG. 3.
Proposed standardized procedure for an accurate estimation of HAV genome copies in food or clinical samples. IC, internal control.
Altogether, an accurate estimation of the numbers of HAV genomes present in a given sample may be achieved. However, an important issue must be solved before conducting the whole test: the determination of the optimal amount of mengovirus to be added. Three different concentrations of mengovirus were tested in serum, stool suspensions and shellfish digestive tissues (Table 1), ranging from 8.6 × 106 to 8.6 × 102 infectious units/ml in the two former cases and from 1.0 × 106 to 1.0 × 102 infectious units/g of digestive tissues in the third type of sample. Each spiked sample was extracted twice, and each nucleic acid suspension was titrated by real-time TaqMan RT-PCR. Although a certain level of variability was observed in all types of samples, this variability was more related to the nature of the particular samples rather than to the concentration of viruses added. In view of the low viral nucleic acid recovery efficiencies (Table 1), mostly in shellfish samples, concentrations of mengovirus of around 105 infectious units/ml were selected for clinical samples (serum and stool), while 106 infectious units/g were employed for shellfish samples. In an attempt to eliminate the possibility of different behaviors of mengovirus and HAV during the extraction procedures, a comparative experiment was performed in which the pHM175 43c strain of HAV was added at different concentrations to the different types of samples (Table 2). The extraction efficiency was similar for both viruses, thus validating the use of the mengovirus control.
TABLE 1.
Validation of mengovirus and ssRNA internal control as standards for determination of nucleic acid extraction and RT-PCR efficiencies, respectively
| Sample | Nucleic acid extraction
|
RT-PCR
|
||||
|---|---|---|---|---|---|---|
| Spiked mengovirusa | Recovered mengovirusb (mean ± SD) | Efficiency (%, mean ± SD) | Spiked ssRNAc | Recovered ssRNAd (mean ± SD) | Efficiency (%, mean ± SD) | |
| Serum | ||||||
| 1 | 8.6 × 106 | (1.1 ± 1.4) × 107 | 100.0 ± 0.0 | 1.2 × 108 | (2.5 ± 1.0) × 108 | 100.0 ± 0.0 |
| 8.6 × 104 | (3.6 ± 1.0) × 105 | 100.0 ± 0.0 | 1.2 × 108 | (2.0 ± 0.6) × 108 | 100.0 ± 0.0 | |
| 8.6 × 102 | (3.6 ± 2.3) × 103 | 100.0 ± 0.0 | 1.2 × 108 | (2.1 ± 1.3) × 108 | 100.0 ± 0.0 | |
| 2 | 8.6 × 106 | (1.1 ± 3.7) × 106 | 12.3 ± 4.4 | 1.6 × 108 | (1.9 ± 0.5) × 108 | 100.0 ± 0.0 |
| 8.6 × 104 | (2.5 ± 1.6) × 104 | 28.8 ± 19.0 | 1.6 × 108 | (2.8 ± 0.4) × 108 | 100.0 ± 0.0 | |
| 8.6 × 102 | (2.6 ± 1.9) × 102 | 30.7 ± 21.7 | 1.6 × 108 | (1.6 ± 0.3) × 108 | 93.5 ± 9.2 | |
| Stool | ||||||
| 1 | 8.6 × 106 | (3.4 ± 0.4) × 106 | 39.5 ± 4.9 | 1.2 × 108 | (2.2 ± 1.7) × 107 | 18.1 ± 14.3 |
| 8.6 × 104 | (5.1 ± 2.2) × 104 | 59.0 ± 25.4 | 1.2 × 108 | (9.6 ± 0.2) × 106 | 8.3 ± 1.8 | |
| 8.6 × 102 | (4.3 ± 2.3) × 102 | 49.3 ± 25.9 | 1.2 × 108 | (1.5 ± 0.8) × 107 | 12.3 ± 6.6 | |
| 2 | 8.6 × 106 | (3.0 ± 1.3) × 106 | 34.6 ± 15.3 | 1.6 × 108 | (3.1 ± 0.3) × 108 | 100.0 ± 0.0 |
| 8.6 × 104 | (3.5 ± 6.6) × 103 | 40.0 ± 7.6 | 1.6 × 108 | (3.4 ± 0.8) × 108 | 100.0 ± 0.0 | |
| 8.6 × 102 | (9.3 ± 10.0) × 102 | 65.7 ± 48.4 | 1.6 × 108 | (1.6 ± 0.6) × 108 | 100.0 ± 0.0 | |
| Shellfish | 1.0 × 106 | (9.1 ± 9.2) × 102 | 0.1 ± 0.1 | 1.5 × 108 | 1.3 ± 0.4 × 108 | 84.3 ± 21.4 |
| 1.0 × 104 | (2.2 ± 0.7) × 101 | 0.2 ± 0.1 | 1.5 × 108 | (1.2 ± 0.3) × 108 | 79.9 ± 17.5 | |
| 1.0 × 102 | NDe | 1.5 × 108 | (1.6 ± 0.4) × 108 | 93.2 ± 6.7 | ||
The inocula added are expressed as TCID50/ml for sera and stool suspensions and as TCID50/g of hepatopancreas for shellfish.
Each spiked sample was extracted twice, and each nucleic acid suspension was titrated by real-time TaqMan RT-PCR with a standard curve made by means of mengovirus infectious units.
The inocula added are expressed as the number of molecules of the ssRNA internal control per ml of nucleic acid suspension.
Each spiked nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of HAV-derived ssRNA molecules estimated from the OD260 readings.
ND, not detected.
TABLE 2.
Comparison of infectious mengovirus and infectious HAV in nucleic acid extraction
| Sample | Mengovirus
|
HAV
|
||||
|---|---|---|---|---|---|---|
| Spikeda | Recoveredb | Nucleic acid extraction efficiency (%) | Spikedc | Recoveredd | Nucleic acid extraction efficiency (%) | |
| Serum | 8.6 × 106 | 1.3 × 105 | 1.5 | 2.0 × 106 | 1.1 × 105 | 5.5 |
| 8.6 × 104 | 1.0 × 104 | 11.6 | 2.0 × 104 | 6.7 × 102 | 3.3 | |
| Stool | 8.6 × 106 | 2.1 × 106 | 23.8 | 2.0 × 106 | 2.8 × 105 | 14.0 |
| 8.6 × 104 | 3.0 × 104 | 34.6 | 2.0 × 104 | 1.4 × 103 | 7.0 | |
| Shellfish | 1.0 × 106 | 1.6 × 103 | 0.2 | 2.0 × 106 | 4.0 × 103 | 0.2 |
| 1.0 × 104 | 2.7 × 101 | 0.3 | 2.0 × 104 | 3.3 × 101 | 0.2 | |
The inocula added are expressed as TCID50/ml for sera and stool suspensions and as TCID50/g of hepatopancreas for shellfish.
Each spiked sample was extracted, and the nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of mengovirus infectious units.
The pHM175 43c strain of HAV was added, and virus titers are expressed as TCID50/ml for sera and stool suspensions and as TCID50/g for shellfish.
Each spiked sample was extracted, and the nucleic acid suspension was titrated by real-timeTaqMan RT-PCR using a standard curve made by means of HAV infectious units.
Regarding the number of ssRNA molecules, it was decided to add a rather high concentration (around 108 molecules/ml) in order to minimize potential competition in case of an HAV-positive sample.
This complete approach was applied to the quantification of experimentally contaminated clinical and shellfish samples as well as to naturally contaminated clinical and shellfish samples.
Quantification of HAV in clinical samples.
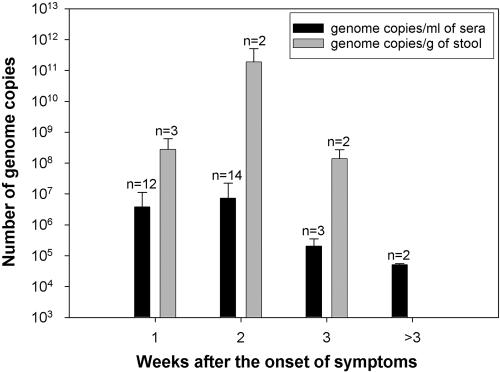
This complete approach was applied for the quantification of HAV in clinical samples from patients suffering from acute hepatitis and diagnosed positive for hepatitis A by means of immunoglobulin M positivity. Only samples belonging to genotypes IA (sporadic cases) and IB (shellfish-borne outbreak) were available. In the latter case, the quantitative results (Table 3) revealed a high and long lasting viremia, with the peak occurring at two weeks after the onset of symptoms (Fig. 4). Although for genotype IA samples no information was available about the time that the sera was obtained, if it is assumed that the most probable time is around 1 to 2 weeks, then no significant differences in the number of genome copies per ml was observed with regard to genotype IB, with the exception of sample 2. This sample showed a significantly higher number of HAV genomes, clearly exceeding even the average detected at 2 weeks postsymptoms. However, it should be mentioned that this sample was from a human immunodeficiency virus-positive patient.
TABLE 3.
Quantification of HAV in serum samples from patients infected with either genotype IA or IB
| Genotypea | Sample (wkb) | Nucleic acid extraction
|
RT-PCR
|
HAV titerg
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Spiked mengovirusc | Recovered mengovirusd | Efficiency (%) | Spiked ssRNAe | Recovered ssRNAf | Efficiency (%) | Crude sample | Final estimate | ||
| IA | 1 | 1.3 × 105 | 3.2 × 104 | 24.7 | 1.5 × 108 | 1.6 × 108 | 100.0 | 1.6 × 105 | 6.4 × 105 |
| 2 | 1.3 × 105 | 5.4 × 104 | 41.9 | 1.5 × 108 | 1.4 × 108 | 94.1 | 3.2 × 108 | 7.6 × 108 | |
| 3 | 1.3 × 105 | 5.7 × 104 | 43.5 | 1.5 × 108 | 1.5 × 108 | 98.2 | 3.2 × 107 | 7.3 × 107 | |
| IB | 1 (1) | 1.3 × 105 | 6.6 × 104 | 50.0 | 1.5 × 108 | 2.3 × 108 | 100.0 | 4.8 × 105 | 9.6 × 105 |
| 2 (1) | 1.3 × 105 | 1.7 × 104 | 13.0 | 1.5 × 108 | 2.6 × 108 | 100.0 | 1.4 × 105 | 1.1 × 106 | |
| 3 (2) | 1.3 × 105 | 6.6 × 103 | 5.0 | 1.5 × 108 | 1.9 × 108 | 100.0 | 1.9 × 106 | 3.8 × 107 | |
| 4 (2) | 1.3 × 105 | 1.5 × 104 | 11.5 | 1.5 × 108 | 1.2 × 108 | 78.4 | 9.2 × 105 | 8.0 × 106 | |
| 5 (3) | 1.3 × 105 | 1.7 × 104 | 13.0 | 1.5 × 108 | 1.4 × 108 | 94.1 | 7.4 × 104 | 5.6 × 105 | |
| 6 (3) | 1.3 × 105 | 3.8 × 104 | 28.5 | 1.5 × 108 | 1.8 × 108 | 100.0 | 6.8 × 104 | 2.4 × 105 | |
| 7 (6) | 1.3 × 105 | 8.6 × 103 | 6.5 | 1.5 × 108 | 1.4 × 108 | 94.1 | 3.1 × 103 | 4.8 × 104 | |
| 8 (6) | 1.3 × 105 | 6.3 × 104 | 48.5 | 1.5 × 108 | 2.1 × 108 | 100.0 | 2.6 × 104 | 5.4 × 104 | |
IB samples are from different patients from an outbreak.
Week after the onset of symptoms at which the samples were taken.
The inocula added are expressed as TCID50/ml of serum.
Each spiked sample was extracted, and the nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of mengovirus infectious units.
The inocula added are expressed as the number of molecules of the ssRNA internal control per ml of nucleic acid suspension.
Each spiked nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of HAV-derived ssRNA molecules estimated from the OD260 readings.
The titer is expressed as the number of genome copies per ml of serum.
FIG. 4.
Time course for HAV genome copies in sera and feces from patients of a shellfish-borne outbreak.
Regarding the titers detected in feces, the results revealed numbers higher than expected (Table 4), with a peak again at around 2 weeks after the onset of symptoms (Fig. 4).
TABLE 4.
Quantification of HAV in fecal samples from patients with acute hepatitis A from a genotype IB outbreak
| Sample (wka) | Nucleic acid extraction
|
RT-PCR
|
HAV titerf
|
|||||
|---|---|---|---|---|---|---|---|---|
| Spiked mengovirusb | Recovered mengovirusc | Efficiency (%) | Spiked ssRNAd | Recovered ssRNAe | Efficiency (%) | Crude sample | Final estimate | |
| 1 (1) | 1.3 × 105 | 2.8 × 104 | 21.0 | 1.5 × 108 | 9.6 × 107 | 64.9 | 1.1 × 107 | 5.1 × 108 |
| 2 (1) | 1.3 × 105 | 2.7 × 103 | 2.1 | 1.5 × 108 | 5.2 × 107 | 35.1 | 9.2 × 105 | 4.4 × 107 |
| 3 (1) | 1.3 × 105 | 3.5 × 104 | 26.5 | 1.5 × 108 | 1.3 × 107 | 8.6 | 3.6 × 105 | 4.2 × 106 |
| 4 (2) | 1.3 × 105 | 8.0 × 103 | 6.0 | 1.5 × 108 | 1.0 × 106 | 1.0 | 2.2 × 108 | 2.2 × 1010 |
| 5 (2) | 1.3 × 105 | 1.3 × 103 | 1.0 | 1.5 × 108 | 1.1 × 108 | 70.3 | 5.6 × 109 | 5.6 × 1011 |
| 6 (3) | 1.3 × 105 | 2.8 × 103 | 2.1 | 1.5 × 108 | 3.0 × 107 | 21.6 | 5.6 × 106 | 2.7 × 108 |
| 7 (3) | 1.3 × 105 | 5.1 × 104 | 38.0 | 1.5 × 108 | 9.6 × 106 | 6.5 | 4.6 × 105 | 7.1 × 106 |
Week after the onset of symptoms at which the samples were taken.
The inocula added are expressed as TCID50/ml of fecal suspension.
Each spiked sample was extracted, and the nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of mengovirus infectious units.
The inocula added are expressed as the number of molecules of the ssRNA internal control per ml of nucleic acid suspension.
Each spiked nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of HAV-derived ssRNA molecules estimated from the OD260 readings.
The titer is expressed as the number of genome copies per g of feces.
Quantification of HAV in shellfish samples.
The same approach was applied for the quantification of the HAV present in the shellfish samples associated with the above-mentioned outbreak (Table 5). Three out of seven tested samples could be quantified, showing estimated titers ranging from 7.5 × 103 to 7.3 × 105 HAV genomes per gram of digestive tissues, or around 1 × 103 to 1 × 105 per g of clam. The rest of the samples were either negative for viruses or contained virus levels below the detection limit, which strongly depends on the efficiency of the extraction procedure.
TABLE 5.
Quantification of HAV in shellfish samples associated with a genotype IB outbreak
| Sample | Nucleic acid extraction
|
RT-PCR
|
HAV titere
|
|||||
|---|---|---|---|---|---|---|---|---|
| Spiked mengovirusa | Recovered mengovirusb | Efficiency (%) | Spiked ssRNAc | Recovered ssRNAd | Efficiency (%) | Crude sample | Final estimate | |
| 1 | 8.6 × 105 | 8.6 × 102 | 0.10 | 1.5 × 108 | 1.4 × 108 | 91.9 | NDf | |
| 2 | 8.6 × 105 | 6.4 × 102 | 0.07 | 1.5 × 108 | 1.1 × 108 | 75.7 | 3.5 × 101 | 6.5 × 104 |
| 3 | 8.6 × 105 | 1.1 × 103 | 0.13 | 1.5 × 108 | 1.2 × 108 | 83.8 | ND | |
| 4 | 8.6 × 105 | 1.3 × 103 | 0.15 | 1.5 × 108 | 1.3 × 108 | 89.2 | 1.1 × 103 | 7.9 × 105 |
| 5 | 8.6 × 105 | 8.0 × 102 | 0.09 | 1.5 × 108 | 9.6 × 107 | 64.8 | 4.4 × 100 | 7.5 × 103 |
| 6 | 8.6 × 105 | 5.9 × 102 | 0.07 | 1.5 × 108 | 1.2 × 108 | 78.4 | ND | |
| 7 | 8.6 × 105 | 1.2 × 103 | 0.14 | 1.5 × 108 | 1.0 × 108 | 70.3 | ND | |
The inocula added are expressed as TCID50/g of hepatopancreas.
Each spiked sample was extracted, and the nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of mengovirus infectious units.
The inocula added are expressed as the number of molecules of the ssRNA internal control per ml of nucleic acid suspension.
Each spiked nucleic acid suspension was titrated by real-time TaqMan RT-PCR using a standard curve made by means of HAV-derived ssRNA molecules estimated from the OD260 readings.
The titer is expressed as the number of genome copies per g of hepatopancreas.
ND, not detected (<6.66 copies per g of hepatopancreas, assuming a 100% efficiency for both extraction and RT-PCR).
DISCUSSION
In the present work a standardized methodology for an accurate quantification of HAV in clinical and food samples is presented. The general approach is based on the use of several controls to measure the efficiency of the critical steps for quantification: the nucleic acid extraction and the RT-PCRs. The first involves the use of a nonpathogenic virus with structural characteristics similar to those of the target virus. Since HAV belongs to the Picornaviridae family, another member of the same family may be used to validate the behavior of HAV during the nucleic acid extraction procedures. Encephalomyocarditis virus has been proposed as a model for HAV in validation studies of HAV removal in blood product manufacturing by several agencies, such as the European Agency for the Evaluation of Medicinal Products (http://www.emea.eu.int/pdfs/human/bwp/026995en.pdf) and the U.S. Food and Drug Administration (http://www.fda.gov/cber/sba/igivbax042705S.pdf). However, the use of this virus as is hampered by its potential pathogenicity in several animals, including primates (4) and even humans (15). Mengovirus is serologically indistinguishable from encephalomyocarditis virus and is nonpathogenic for humans, although it may infect several animals. It has been previously demonstrated that the removal of the poly(C) tract from the 5′NCR of the wild-type mengovirus gives rise to a mutant strain, i.e., mengovirus vMC0, with the same growth properties but with no pathogenic capacity (17). The use of mengovirus strain vMC0 as a control for HAV is proposed, since it represents a phenotypic variant of mengovirus which is avirulent in all animal species (murine and nonmurine) so far tested and is used as a vaccine for a wide variety of hosts, including baboons, macaques, and domestic pigs (21). In addition to its nonpathogenic phenotype, another important issue for the choice of this virus is the structure of the 5′NCR of its genomic RNA. The real-time TaqMan RT-PCR developed in the present work for the quantification of HAV is based on the amplification of a fragment of the 5′NCR. This region is the most conserved of the HAV genome, and of any other picornavirus, and consequently is a good choice for the development of quantification techniques. The RNA sequences that constitute the 5′NCR fold into complex multidomain structures, which are involved in both translation (9) and replication (23). The high conservation of the 5′NCR in all picornaviruses thus relies on its structural function. In fact, phylogenetic analyses of the 5′NCR of picornavirus reveal extensive structure-conserving substitutions within predicted stems, a high degree of sequence conservation in predicted loops, and clustering of regions with sequence divergence in spacer regions between domains (24). In particular, the chosen target region for HAV spans stem-loop domains II and III of the 5′NCR and thus contains an upstream internal ribosome entry site (IRES) region and the 5′ boundary of the IRES. Three different types of IRES have been defined among picornaviruses: type I in entero- and rhinoviruses; type II in cardio-, aphtho-, and paraechoviruses; and type III in hepatoviruses (9). Mengovirus is a cardiovirus, and thus belongs to type II, which appears to be more closely related to type III IRES than type I is. In this context, and having in mind that the quantification of the control virus and HAV should be performed in the same plate and obviously under the same program conditions, it appears again reasonable to use mengovirus as a control. The target region selected for the quantification of mengovirus was related as much as possible to that of HAV in terms of structure, length, and base composition. However, since the IRES types are indeed different, the sensitivity achieved for the mengovirus quantification was significantly lower than that for HAV. The importance of this lower sensitivity is, however, negligible due to the fact that only a comparison between the number of viruses added and recovered is sought. To further confirm the validity of the mengovirus vMC0 strain as a control, a comparison analysis with respect to the behavior of the pHM175 43c HAV strain was performed, and the results demonstrated similar patterns for the two viruses in serum, fecal, and shellfish samples.
A requirement for addition of some kind of internal controls for the follow-up of the molecular reactions has been suggested by many authors (12, 22). The control for an RT-PCR should rely on an ssRNA molecule that corresponds to the viral target and is thus amplifiable with the same pair of primers under exactly the same conditions and with the same efficiency. Cloning the pHM175 43c amplimer under the control of a strong promoter, i.e., the SP6 promoter, allows the synthesis of huge amounts of RNA transcripts, which can be comparatively quantified before and after being added to the sample under analysis and thus can act as an internal control for the RT-PCR. Finally, a third step that should be carefully controlled is the choice of the best standard molecule for the generation of the standard curve for the target quantification. After testing of the above-mentioned ssRNA internal control molecule, a dsDNA molecule corresponding to the pHM175 43c amplimer, and the actual pHM175 43c RNA genome, the use of the dsDNA molecule is recommended. The alternative of using the actual virus may involve two inherent difficulties. On the one hand, due to the encapsidation of the viral genome, its quantification intrinsically comprises the extraction efficiency, and since it is an RNA molecule, it inherently takes into account both the RT and the PCR efficiencies. In the approach here presented, all these efficiencies are validated externally, and thus the use of the virus as a standard may lead to an overestimation. With the dsDNA molecules, only the efficiency of the PCR would be twice quantified; it is, however, well known that the most critical step in an RT-PCR is the RT reaction rather than the PCR, and some kind of molecule must nevertheless be used. On the other hand, this choice avoids an extra manipulation of a human pathogen.
In summary, the whole test involves the generation of three different standard curves: one for the quantification of mengovirus (made with mengovirus titrated by infectivity), another for the quantification of the ssRNA molecule internal control (made with the ssRNA molecule titrated by means of the OD260), and a third for the quantification of HAV (made with the dsDNA molecule titrated again by means of the OD260). All these standard curves are obtained in a single plate, which additionally contains wells devoted to the calculation of extraction and enzyme efficiencies, as well as the samples to be tested, all under exactly the same conditions.
Another important issue for any diagnostic technique, and particularly when a real-time TaqMan RT-PCR is evaluated, is its specificity, breadth, and sensitivity. All these points are interconnected and depend mostly on the target sequences for primers and probe. The selected targets must guarantee an absolute specificity and must reach equilibrium between high sensitivity, broad reactivity, and reliability of quantification. The specificity of the proposed primer-probe sequences has been shown to be excellent against a broad panel of enteric viruses, including the other enteric hepatitis virus (i.e., hepatitis E virus), many gastroenteritis viruses, and many enteroviruses. Mentioning sensitivity and broad reactivity makes inevitable the comparison with other primer-probe sets. Several TaqMan assays based on the use of primer-probe sets of the 5′NCR (5, 14), which makes all of them potentially suitable for broad reactivity, have been previously described. Three genotypes (two of them subdivided into two subtypes) have been described for human HAV, based on a variable region of the genome around the VP1X2A junction. However, in spite of this variability, all genotypes are well conserved in the 5′NCR for the reasons discussed above. In one of these studies (14), a broad reactivity was demonstrated through the quantification of five different strains representing all the human types. This should be, however, the rule for all those techniques based on highly conserved sequences in the 5′NCR. In fact, it is quite surprising that in work of Jothikumar et al. (14) the efficiencies of amplification for genotypes IIA, IIB, and, to a lesser extent, III are lower than those for of genotypes IA and IB, since they have only one and two mismatches, respectively, in the probe sequence. In our case, the only available genotypes are IA and IB, since the others do not occur in our geographical area. However, through the alignment of the available GenBank sequences it can be concluded that all genotypes should be equally well quantified. Regarding the reverse primer (HAV240), only a point insertion is detected in genotype III; for the forward primer (HAV68), the homology of the available sequences is 100%; and finally, for the probe sequence (HAV150), only one point mutation is shown, affecting particularly genotype IB, which is the one corresponding to the pHM175 43c strain and which it is clearly well quantified. In addition, the demonstration of the detection of one particular strain of a given genotype does not guarantee the detection of all the strains belonging to this genotype due to the quasispecies dynamics of replication of RNA viruses and particularly of HAV (28). In this context, an alternative for the verification of the robustness of the target sequences of primers and probe may be the analysis of the mutant spectra. In our case, this has shown that the chosen target regions are well preserved, presenting only very few mutations. Moreover, this type of analysis has confirmed the coincidence of the master or dominant sequence with the consensus or average sequence, an important requirement for a reliable, reproducible, and comparable quantification. In fact, this is again an expected result due to the above-mentioned function of the 5′NCR, and consequently, although not proven for the assay described by Jothikumar et al. (14), the same conclusions might be assumed for its primer-probe set. A different situation is that of the assay developed by Costa-Mattioli et al. (5), in which the reverse primer is located in a spacer region that accumulates many mutations. However, it should be pointed out that the objective of their work was not the development of a broadly reactive assay but to study the viremic phase of the HAV infection.
The most limiting step regarding the sensitivity of an RT-PCR is the RT rather than the PCR, and the secondary structure of RNA is accordingly critical. In this context, high ratios of G · C to A · U plus G · U pairs in the stem structures may imply very high temperatures for RNA secondary structure denaturation. Bearing this consideration in mind, it is clearly seen that the reverse primer target of the assay described by Costa-Mattioli et al. (5) is advantageous, since it is from an nonstructured spacer, while the target regions for the assays described by Jothikumar et al. (14) and the one presented here are more structured, with ratios of G · C to A · U plus G · U of 1.8 and 0.5, respectively. This difference could contribute to the slightly lower sensitivity showed by the first of these assays. Yet, in some circumstances, such as food samples harboring very low loads of viruses (see sample 5 of the shellfish involved in a hepatitis A outbreak shown in Table 5) or plasma pools, exquisite levels of sensitivity can make a difference.
The procedure developed here has been tested with different one-step RT-PCR kits from Applied Biosystems, Invitrogen, QIAGEN, and Stratagene. All these kits worked similarly well in terms of sensitivity, with the exception of that from Applied Biosystems, which offered a sensitivity two log units lower and which in some conditions, such as with plasma samples, did not enable virus detection, presumably due to the inhibition of the Taq Gold polymerase by anticoagulant factors (data not shown).
From the titers of HAV found in serum samples, it may be concluded that viremia is higher and longer than previously thought, as other authors have recently pointed out (5, 20). An increased and longer pattern of excretion in feces is also observed. These data not only change the whole picture of the infection cycle but also reinforce the need for the quantification of HAV in those samples that are susceptible to contamination, such as some types of food, water, and plasma. Additionally an association between low or very low levels of viremia with fulminant hepatitis has recently been described (26), which justifies the need for an extremely sensitive assay.
It was the purpose of this work not to validate nucleic acid extraction procedures but to provide tools for their validation. In this context, it is noteworthy that the extraction step has been shown to be the most critical point in viral quantification assays. Consequently, validation studies of existing or new methods, mostly for shellfish, are urgently required. Standardized methodologies and standard reagents, such as those developed in this study, should be employed for this purpose.
Quality control and quality assurance procedures must be implemented through the use of automated standardized molecular procedures, such as the one described here, that may enable inclusion in regulatory standards for viruses in food or blood products.
Acknowledgments
We thank Ann C. Palmenberg, University of Wisconsin, for advice and useful discussion. We acknowledge the assistance of the Genomics Unit of the Serveis Científic-Tècnics of the University of Barcelona.
M. I. Costafreda is the recipient of a fellowship in the European Union project SEAFOODplus, Food-CT-2004-506359. This study was supported in part by grants Food-CT-2004-506359 and SP22-CT-2004-502571 from the European Union, BIO2002-02625 and BIO2005-05022 from the Spanish Ministry of Education and Science, and 2005SGR00966 from the Generalitat de Catalunya and by the Centre de Referència de Biotecnologia de Catalunya (CeRBa), Generalitat de Catalunya.
REFERENCES
- 1.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch, A., J. F. Gonzalez-Dankaart, I. Haro, R. Gajardo, J. A. Pérez, and R. M. Pintó. 1998. A new continuous epitope of hepatitis A virus. J. Med. Virol. 54:95-102. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, A., F. Lucena, J. M. Díez, R. Gajardo, M. Blasi, and J. Jofre. 1991. Human enteric viruses and indicator microorganisms in a water supply associated with an outbreak of infectious hepatitis. J. Am. Water Works Assoc. 83:80-83. [Google Scholar]
- 4.Citino, S. B., B. L. Homer, J. H. Gaskin, and D. J. Wickham. 1988. Fatal encephalomyocarditis virus infection in a Sumatran orangutan (Pongo pygmaeus abelii). J. Zool. Wildl. Med. 19:214-218. [Google Scholar]
- 5.Costa-Mattioli, M., S. Monpoeho, E. Nicand, M. H. Aleman, S. Billaudel, and V. Ferre. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Viral Hep. 9:101-106. [DOI] [PubMed] [Google Scholar]
- 6.Costa-Mattioli, M., A. D. Napoli, V. Ferre, S. Billaudel, R. Perez-Bercoff, and J. Cristina. 2003. Genetic variability of hepatitis A virus. J. Gen. Virol. 84:3191-3201. [DOI] [PubMed] [Google Scholar]
- 7.Deng, M. Y., S. P. Day, and D. O. Cliver. 1994. Detection of hepatitis A virus in environmental samples by antigen-capture PCR. Appl. Environ. Microbiol. 60:1927-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dentinger, C., W. Bower, O. Nainan, M. Cotter, G. Myers, L. Dubusky, S. Fowler, E. Salehi, and B. Bell. 2001. An outbreak of hepatitis A associated with green onions. J. Infect. Dis. 183:1273-1276. [DOI] [PubMed] [Google Scholar]
- 9.Ehrenfeld, E., and N. L. Teterina. 2002. Initiation of translation of picornavirus RNAs: structure and function of the internal ribosome entry site, p. 159-169. In B. L. W. E. Semler (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 10.Halliday, M. L., L.-Y. Kang, T.-Z. Zhou, M.-D. Hu, Q.-C. Pan, T.-Y. Fu, Y. S. Huang, and S.-L. Hu. 1991. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. J. Infect. Dis. 164:852-859. [DOI] [PubMed] [Google Scholar]
- 11.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-840. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 12.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansen, R. W., J. E. Newbold, and S. M. Lemon. 1988. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology 163:299-307. [DOI] [PubMed] [Google Scholar]
- 14.Jothikumar, N., T. L. Cromeans, M. D. Sobsey, and B. H. Robertson. 2005. Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 71:3359-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkland, P. D., R. A. Hawkes, H. M. Naim, and C. R. Boughton. 1989. Human infection with encephalomyocarditis virus in New South Wales. Med. J. Aust. 151:176. [DOI] [PubMed] [Google Scholar]
- 16.Lemon, S. M., and L. N. Binn. 1983. Antigenic relatedness of two strains of hepatitis A virus determined by cross neutralization. Infect. Immun. 42:418-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, L. R., G. M. Duke, J. E. Osorio, D. J. Hall, and A. C. Palmenberg. 1996. Mutational analysis of the mengovirus poly(C) tract and surrounding heteropolymeric sequences. J. Virol. 70:2027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mast, E. E., and M. J. Alter. 1993. Epidemiology of viral hepatitis: an overview. Semin. Virol. 4:273-283. [Google Scholar]
- 19.Noble, R. C., E. M. Kane, S. A. Reeves, and I. Roeckle. 1984. Posttransfusional hepatitis A in a neonatal intensive care unit. JAMA 252:2711-2715. [PubMed] [Google Scholar]
- 20.Normann, A., C. Jung, A. Vallbracht, and B. Flehmig 2004. Time course of hepatitis A viremia and viral load in the blood of human hepatitis A patients. J. Med. Virol. 72:10-16. [DOI] [PubMed] [Google Scholar]
- 21.Osorio, J. E., G. B. Hubbard, K. F. Soike, M. Girard, S. van der Werf, J. C. Moulin, and A. C. Palmenberg. 1996. Protection of non-murine mammals against encephalomyocarditis virus using a genetically engineered Mengo virus. Vaccine 14:155-161. [DOI] [PubMed] [Google Scholar]
- 22.Parshionikar, S. U., J. Cashdollar, and G. Shay Fout. 2004. Development of homologous viral internal controls for use in RT-PCR assays of waterborne enteric viruses. J. Virol. Methods 121:39-48. [DOI] [PubMed] [Google Scholar]
- 23.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. W. E. Semler (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 24.Poyry, T., L. Kinnunen, and T. Hovi. 1992. Genetic variation in vivo and proposed functional domains of the 5′ noncoding region of poliovirus RNA. J. Virol. 66:5313-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid, T. M. S., and H. G. Robinson. 1987. Frozen raspberries and hepatitis A. Epidemiol. Infect. 98:109-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezende, G., A. M. Roque-Afonso, D. Samuel, M. Gigou, E. Nicand, V. Ferre, E. Dussaix, H. Bismuth, and C. Féray. 2003. Viral and clinical factors associated with the fulminant course of hepatitis A virus. Hepatology 38:613-618. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, K. Ito, T. Ishizu, Y. Moritsugu, and S. M. Lemon. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 73:1365-1377. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, G., A. Bosch, G. Gomez-Mariano, E. Domingo, and R. M. Pinto. 2003. Evidence for quasispecies distributions in the human hepatitis A virus genome. Virology 315:34-42. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez, G., R. M. Pinto, H. Vanaclocha, and A. Bosch. 2002. Molecular characterization of hepatitis A virus isolates from a transcontinental shellfish-borne outbreak. J. Clin. Microbiol. 40:4148-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheretz, R. J., B. A. Russell, and P. D. Reunman. 2005. Transmission of hepatitis A by transfusion of blood products. Arch. Intern. Med. 1441:1579-1580. [PubMed] [Google Scholar]