Abstract
For this study, an in-depth review of the classification of Lactococcus lactis phages was performed. Reference phages as well as unclassified phages from international collections were analyzed by stringent DNA-DNA hybridization studies, electron microscopy observations, and sequence analyses. A new classification scheme for lactococcal phages is proposed that reduces the current 12 groups to 8. However, two new phages (Q54 and 1706), which are unrelated to known lactococcal phages, may belong to new emerging groups. The multiplex PCR method currently used for the rapid identification of phages from the three main lactococcal groups (936, c2, and P335) was improved and tested against the other groups, none of which gave a PCR product, confirming the specificity of this detection tool. However, this method does not detect all members of the highly diverse P335 group. The lactococcal phages characterized here were deposited in the Félix d'Hérelle Reference Center for Bacterial Viruses and represent a highly diverse viral community from the dairy environment.
Strains of the gram-positive bacterium Lactococcus lactis are used by the dairy industry to acidify milk during the manufacture of fermented products, such as cheese, buttermilk, and sour cream. The use of various L. lactis strains is essential for controlling virulent phages that are responsible for most milk fermentation collapses (40). Lactococcal phages are ubiquitous in the dairy environment, as they are found in raw milk and survive pasteurization (14, 36). Due to their negative effects on fermentation as well as their biodiversity within this ecological niche, numerous lactococcal phages have been isolated and characterized, with the overall aim of improving phage control strategies. Currently, only coliphages have received more attention than lactococcal phages (1, 2). All known L. lactis phages have a double-stranded genome and a noncontractile tail. According to the International Committee on Taxonomy of Viruses, L. lactis phages are members of the Caudovirales order, an extremely large, morphologically and genetically diverse group that encompasses over 95% of all known phages (37). This order contains three families, namely, the Myoviridae (with long, contractile tails), the Siphoviridae (with long, noncontractile tails), and the Podoviridae (with short tails). Lactococcal phages are mainly members of the Siphoviridae family, with a few members from the Podoviridae family.
Over a decade ago, a classification scheme which was mainly based on phage morphology and DNA homology criteria was developed for lactococcal phages (3, 4, 7, 25, 32). It is made up of 12 lactococcal phage groups and has been used successfully for comparing lactococcal phages isolated from around the world. It rapidly became obvious that the vast majority of lactococcal phages belong to one of three main groups, the 936, c2, and P335 groups. Consequently, most studies of lactococcal phages have dealt with these groups. For example, a multiplex PCR method is now available to rapidly assign newly isolated phages to one of these three main groups (29). Moreover, 14 complete lactococcal phage genomic sequences are now available in a public database (www.ncbi.nlm.nih.gov), including 10 from the P335 group, two from the c2 group, and two from the 936 group. The genome of phage asccφ28, from a fourth species (P034), was also reported a few years ago (27, 28), but the sequence is not yet available. Many other lactococcal phage genome projects are also under way.
The cataloging scheme for lactococcal phages has been under scrutiny lately due to the comparative analysis of an increasing number of genomic sequences and the recurring emergence of new virulent phages (13, 42). For instance, comparative genome analysis has led to a proposal to merge the BK5-T species with the P335 species, reducing the number of lactococcal phage species to 11 (30). Based on DNA-DNA hybridization studies, others have also suggested including the 1483 and T187 species in the P335 group (24, 30, 43, 47).
Despite the strategies developed to control phages, this biotechnological problem still remains the most common cause of slow or incomplete milk fermentation. The biodiversity and evolution of phage populations are partly responsible for the difficulty in controlling them. Phage populations in dairy factories are also in a dynamic state, and their composition must be monitored closely to ensure the efficacy of the current control strategies that are based, in part, on the phage species sensitivity of L. lactis strains (40, 42). Consequently, the aim of this study was to reassess the classification system for L. lactis phages.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
The Lactococcus lactis strains and bacteriophages used in this study are listed in Table 1. Strains were grown at 30°C in M17 broth (51) supplemented with 0.5% glucose (GM17) (Quélab) unless otherwise specified. All phages were propagated from a single plaque as described previously (19). High phage titers were obtained by using the method of Jarvis (22). To obtain maximal titers and/or visible plaques, some phages (1483, 949, P087, P369, KSY1, and 1706) and their respective hosts were incubated at 22°C for 24 h. Glycine (0.5%) was also added to the top agar to increase plaque size and facilitate phage enumeration (33). The induction of lactococcal prophages was performed as described previously (41). When needed, phage lysates were concentrated with polyethylene glycol (12, 22, 23) and purified on a discontinuous-step CsCl gradient (49). Ultracentrifugation was performed using a Beckman SW41 Ti rotor at 35,000 rpm for 3 h.
TABLE 1.
Bacteriophages and host strains used in this study
| Family | Phage | Species
|
L. lactis host strain | Reference | Source | |
|---|---|---|---|---|---|---|
| New | Olda | |||||
| Siphoviridae | bIL170 | 936 | 936 | IL1403 | 15 | U. Lavalc |
| c2 | c2 | c2 | LM0231 | 35 | U. Lavalc | |
| CB17 | c2 | Unclassifiedb | SMQ-436 | This study | This study | |
| GR6 | c2 | Unclassifiedb | SMQ-361 | This study | This study | |
| 1483 | P335 | 1483 | 111 | 23, 24 | U. Lavalc | |
| r1t | P335 | P335 | R1K10 | 34, 55 | U. Lavalc | |
| T189 | P335 | T187 | 205.RV | 47 | B. Gellerd | |
| ul36 | P335 | P335 | SMQ-86 | 30 | U. Lavalc | |
| BK5-T | P335 | BK5-T | H2 | 17 | F. K. Vogensene | |
| 949 | 949 | 949 | ML8 | 23 | U. Lavalc | |
| bIL168 | 949 | Unclassifiedb | IL-16 | This study | ||
| P087 | P087 | P087 | C10 | 7, 32 | H. Nevef | |
| 1358 | 1358 | 1358 | 582 | 23 | U. Lavalc | |
| 1706 | 1706 | Unclassifiedb | SMQ-450 | This study | C. Fremauxg | |
| Q54 | Q54 | Unclassifiedb | SMQ-562 | This study | This study | |
| Podoviridae | 1138 | P034 | Unclassifiedb | SMQ-450 | This study | C. Fremauxg |
| P369 | P034 | P034 | F7/2 | 7, 32 | H. Nevef | |
| KSY1 | KSY1 | KSY1 | IE-16 | 50 | U. Lavalc | |
Species from previous classification (25).
Unclassified by multiplex PCR (29).
Félix d'Hérelle Reference Center for Bacterial Viruses, Université Laval, Québec, Canada (www.phage.ulaval.ca).
Department of Microbiology, Oregon State University, Corvallis, Oreg.
Department of Food Science, The Royal Veterinary and Agricultural University (KVL), Frederiksberg, Denmark.
Institute of Microbiology, Federal Dairy Research Center, Kiel, Germany.
Danisco, Dangé Saint-Romain, France.
Phage DNA analysis.
The genomic DNAs of phages ul36, r1t, 1706, Q54, c2, and bIL170 were isolated by using Lambda Maxi DNA purification kits (QIAGEN), with previously described modifications (18). The DNAs of phages P369, 1483, 1358, 949, 1138, P087, and KSY1 were isolated from CsCl-purified phages as reported elsewhere (12). Lastly, the DNAs of phages T189, bIL168, BK5-T, GR6, and CB17 were isolated by using a previously described protocol (41). Restriction endonucleases (Roche Diagnostics) were used as recommended by the manufacturer. After restriction, phage DNA samples were heated for 10 min at 70°C to avoid possible cohesive end ligation. The DNA fragments were separated in 0.8% agarose gels in 1× Tris-acetate-EDTA buffer and visualized by UV photography after being stained with ethidium bromide. The phage DNAs were transferred to positively charged nylon membranes (Roche Diagnostics) by capillary blotting as described by Sambrook and Russell (49). Phage genomic DNAs used as probes were randomly labeled with Dig High-Prime labeling kits (Roche Diagnostics). Prehybridization, hybridization, washes, and detection by chemiluminescence (CDP-star) were performed as suggested by the manufacturer (Roche Diagnostics).
Electron microscopy.
For electron microscopy, 1.5 ml of phage lysate (108 to 109 PFU/ml) was centrifuged for 1 h at 4°C (24,000 × g). The supernatant (approximately 1.4 ml) was gently discarded. The remaining lysate was diluted twice by the addition of 1 ml of ammonium acetate (0.1 M, pH 7.5) and then centrifuged (1 h at 24,000 × g and 4°C). The phage solution (15 μl) was mixed with the stain (15 μl of 2% phosphotungstic acid, pH 7.5) on a nickel Formvar-carbon-coated grid (Pelco International). The liquid was removed after 1 min by touching the edge of the grid with blotting paper. Phage morphology was observed by using a JEOL 1230 transmission electron microscope at 80 kV. Dimensions of the phages are the means of at least 10 specimens.
PCR and DNA sequencing.
For multiplex PCR, the DNA template was made up of a phage lysate treated with DNase (final concentration, 1 μg/ml) for 30 min at 37°C. The conditions and primers used were previously described (29). PCR products corresponding to the genes coding for the major capsid proteins (mcp) of the c2-like phages GR6 and CB17 were sequenced on both strands by using specific oligonucleotides (29) and an ABI Prism 3700 apparatus from the genomic platform at the Centre Hospitalier de l'Université Laval. The 16S rRNA genes were amplified from L. lactis strains SMQ-450 and SMQ-562, using the Bacteria-specific primers SSU27F (5′ AGAGTTTGATCMTGGCTCAG 3′) and SSU1492R (5′ TACGGYTACCTTGTTACGACTT 3′).
Bioinformatic analysis.
Computer-assisted DNA analyses were performed by using version 10.3 of the Genetics Computer Group sequence analysis software package and the ClustalW website (http://www.ebi.ac.uk/clustalw/). PSI-BLAST and Advanced BLAST Search 2.1 were also used for sequence comparisons with databases (5).
Nucleotide sequence accession numbers.
The complete genomic sequences of the P335-like phages analyzed in this study are available under the following GenBank accession numbers (phage name [accession number]): 4268 (AF489524), bIL285 (AF323668), bIL286 (AF323669), bIL309 (AF323670), BK5-T (AF176025), phiLC3 (AF242738), r1t (U38906), TP901-1 (AF304433), Tuc2009 (AF109874), and ul36 (AF349457). For the c2-like phages, in addition to the complete genomic sequences of phages c2 (L48605) and bIL67 (L33769), the nucleotide sequences of the genes coding for the major capsid protein are available for phages eb1 (AF152410), Q44 (AF152412), Q38 (AF152411), CB17 (DQ110947), and GR6 (DQ110948). Complete genomic sequences are available for the 936-like phages sk1 (NC_001835) and bIL170 (NC_001909).
RESULTS
Selection of lactococcal phages.
As a first step, a representative of each of the 12 previously recognized lactococcal phage species was obtained (Table 1). Whenever possible, we selected phages for which the complete genomic sequence was already available. Seven reference phages (949, 1358, 1483, c2, KSY1, bIL170 [936 species], and ul36 [P335 species]) were obtained from the Félix d'Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca) at Université Laval (Quebec City, Canada). Phage T189 was provided by B. Geller (Oregon State University) and was used as a representative of the T187 species since phage T187 is no longer available. Phages P087 and P369 (P034 species) were provided by H. Neve (Institute of Microbiology, Federal Dairy Research Center, Germany), and phage BK5-T was provided by F. K. Vogensen (KVL University, Denmark). Unfortunately, we could not obtain a viable representative of the P107 species, not even from the laboratory that originally isolated this phage (32). All of the samples provided failed to generate the restriction profile for P107 (7), and all of the P107 lysates contained a phage that belonged to the 936 species (data not shown). This lactococcal reference phage thus appears to be extinct. We also included prophage r1t as another representative of the P335 species because it shares only 11% nucleotide sequence identity with the virulent P335-like phage ul36 (30). Lastly, six unclassified lactococcal phages (bIL168, CB17, GR6, Q54, 1138, and 1706) obtained from various collaborators were also included in this study. Whenever possible, the DNA restriction profiles of these phages were compared to published profiles to confirm their identities.
Analysis of genomic DNA by Southern hybridization.
Digoxigenin-labeled genomic probes were produced from each of the previously described lactococcal species and were hybridized with the restricted genomes of the 18 phages studied. To take into account the genetic diversity previously observed within the P335 species (30), a genomic probe was made from each of the two representatives (r1t and ul36) of this group.
Phages bIL170, KSY1, P087, and 1358 only hybridized with themselves (data not shown), which is in agreement with previous reports (7, 23, 25). Phages 1483, T189, and BK5-T (Fig. 1B) shared homology with the P335 phages r1t and ul36. A weak hybridization signal of the r1t probe with the restricted genome of bIL170 (936 species) was also noted (Fig. 1B). Based on the complete genomes of bIL170 (15) and r1t (55), this area was identified as a 950-bp region that shares 91% nucleotide sequence identity and codes for the neck-passage structure protein (9). The significance of this protein remains to be investigated, as not all members of these two phage groups possess this morphological feature. The c2 genomic probe strongly hybridized with the genomes of the unclassified phages CB17 and GR6 (Fig. 1C). The P369 probe strongly hybridized with the genome of phage 1138 (Fig. 1D). Likewise, the genome of phage bIL168 clearly hybridized with the 949 probe (Fig. 1E).
FIG. 1.
(A) Restriction profiles of lactococcal phages analyzed in this study. (B to G) Analysis of phage genomic DNAs by Southern hybridization, using the complete genomes of the following phages as probes: panel B, r1t; panel C, c2; panel D, P369; panel E, 949; panel F, Q54; panel G, 1706. Lanes 1 and 19, 1-kb DNA ladder (Invitrogen); lanes 2, phage bIL170 (genome digested with EcoRV); lanes 3, c2 (EcoRI); lanes 4, Q54 (EcoRI); lanes 5, GR6 (EcoRI); lanes 6, CB17 (EcoRI); lanes 7, ul36 (EcoRV); lanes 8, r1t (EcoRV); lanes 9, 1483 (EcoRV); lanes 10, T189 (EcoRV); lanes 11, P087 (EcoRV); lanes 12, 949 (EcoRV); lanes 13, bIL168 (EcoRV); lanes 14, 1706 (EcoRV); lanes 15, 1358 (EcoRI); lanes 16, KSY1 (AsnI); lanes 17, P369 (EcoRV); lanes 18, 1138 (EcoRV); lanes 20, BK5-T (EcoRV).
None of the probes from the known lactococcal phage species hybridized with the genome of phage Q54 or 1706. A Q54 probe and a 1706 probe also failed to hybridize with the other lactococcal phage genomes (Fig. 1F and G, respectively).
Phage morphology.
The 18 lactococcal phages were observed by transmission electron microscopy to determine their morphology (Table 2). Some of these phages have already been characterized, and our observations were in agreement with published data (3, 7, 23, 25). Fifteen phages had long, noncontractile tails and belonged to the Siphoviridae family, while three phages (P369, KSY1, and 1138) had short tails and belonged to the Podoviridae family. Phages sharing DNA-DNA homology had the same morphotype (1), but some morphological features (tail length and the presence of a collar structure or baseplate) varied slightly. For example, phage bIL170 has a collar structure that is absent from phage sk1 (data not shown). Only one additional gene is necessary for the presence of this structure (11, 15). Moreover, it is known that the tape measure protein determines the tail length and that a mutation in its gene can lead to tail size variations (45, 56).
TABLE 2.
Biodiversity of bacteriophages infecting Lactococcus lactis
Bars, 50 nm.
Phage 1706, which shares no homology with the other phages analyzed in this study, has a morphology not previously reported for lactococcal phages (Table 2). The capsid size of phage 1706 is similar to that of P087, but its noncontractile tail is longer. On the other hand, the morphology of phage Q54 resembles that of phage c2. The host strains for these phages, L. lactis SMQ-450 (host for 1706) and L. lactis SMQ-562 (host for Q54), are used commercially in mesophilic starter cultures to make fermented milk products. The identities of the bacterial species were confirmed by 16S rRNA gene sequencing (data not shown). Moreover, host range analyses revealed that L. lactis SMQ-450 was sensitive to the lactococcal phages 1706 and 1138, while L. lactis SMQ-562 was sensitive to lactococcal phages Q54 and 949.
Based on DNA-DNA hybridization and morphology results, 16 of the phages studied here could be classified into eight groups (Table 1). The two remaining phages (1706 and Q54), which were unrelated to the others, might be members of new emerging groups.
Multiplex PCR test for the identification of phages.
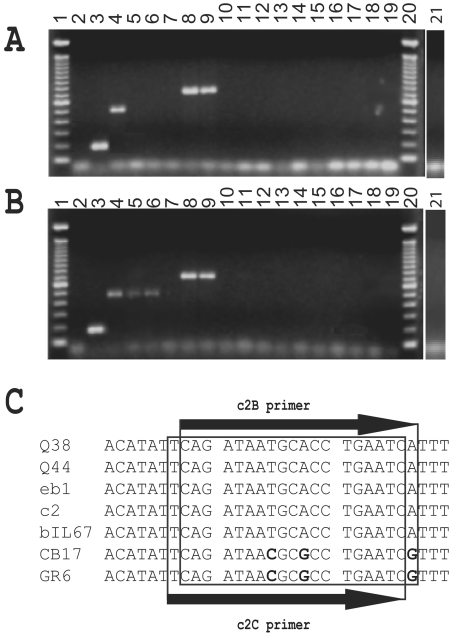
As indicated previously, a multiplex PCR method is available to identify the three predominant lactococcal phage groups for which the sequences of several members are known, namely, 936, c2, and P335-like phages (29). The accuracy of this method was checked with the 18 selected phages (Fig. 2). As expected, PCR amplicons of the predicted sizes were obtained for the reference phages c2, bIL170 (936 species), and r1t (P335 species). Interestingly, phage 1483 gave a PCR product with a size expected for a P335-like phage. This observation, together with DNA-DNA hybridization results (Fig. 1B), suggested that it may belong to the P335 group (Fig. 2). We failed to obtain a PCR product with the other 14 phages. The negative PCR results with phages ul36 and BK5-T (P335 species) have been discussed in the past and are due to the absence of the target region in their genome (29, 30).
FIG. 2.
Identification of lactococcal phage species by multiplex PCR. (A) Three pairs of previously described primers (29) were used for PCR. (B) The same three sets of primers were used, except that the c2B primer was replaced by the c2C primer. Lanes 1 and 20, 100-bp DNA ladder (Invitrogen); lanes 2, negative control; lanes 3, bIL170; lanes 4, c2; lanes 5, GR6; lanes 6, CB17; lanes 7, Q54; lanes 8, r1t; lanes 9, 1483; lanes 10, ul36; lanes 11, T189; lanes 12, P087; lanes 13, 949; lanes 14, bIL168; lanes 15, 1706; lanes 16, 1358; lanes 17, KSY1; lanes 18, P369; lanes 19, 1138; lanes 21, BK5-T. (C) Analysis of the region covered by primers c2B and c2C in c2-like phages for which the gene coding for the major capsid protein is available in GenBank.
Surprisingly, the multiplex PCR method (29) failed to identify phages CB17 and GR6 as members of the c2 species. The two primers (c2A and c2B) used to amplify a 474-bp product specific to c2-like phages are based on two conserved regions (20 nucleotides) in the gene coding for the major capsid protein. The regions covering both primers were sequenced in phages CB17 and GR6 and compared with similar sequences in public databases (Fig. 2C). The c2A primer regions in phages CB17 and GR6 were identical to the same regions for other c2-like phages. However, the c2B primer contained three nucleotide mismatches (Fig. 2C). Of particular interest was a mismatch at the 3′ end that corresponds to the wobble base of the serine codon. A new oligonucleotide (c2C) was designed in which the 20-mer was shifted one nucleotide to avoid the 3′ codon base. The multiplex PCR method was retested using the c2A/c2C primer pair. The three c2-like phages (c2, CB17, and GR6) generated 475-bp PCR products, while the 14 lactococcal phages of the other species did not generate PCR products (Fig. 2B). To confirm the efficacy of this new set of c2-specific primers, 13 other distinct c2-like phages (bIL67, Q44, eb1, ml3, HD1, HD2, HD4, HD7, HD25, HD26, GR3, GR4, and CB27) were tested. PCR products of the expected size were obtained from all of them (data not shown).
DISCUSSION
The lactococcal phage classification system was first set up to characterize these phages and to provide better ways to control phage infections. This pragmatic classification (7, 25) has been applied universally and has led to comparisons of lactococcal phage collections worldwide, providing valuable results. However, more sequence data, together with the isolation of new phages and the absence of an international collection housing reference phages, led us to revise the previous classification and to deposit all the phages used in this study in the Félix d'Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca). In our opinion, this phage set represents the best-characterized bank of viruses that share a well-defined ecological niche and that infect a single bacterial species.
Of the 12 phage groups previously described (7, 25), one appeared extinct (P107), while three others (1483, T187, and BK5-T) were merged with the P335 group, reducing the number of lactococcal phage groups from 12 to 8. Nonetheless, additional lactococcal phage groups may exist. For example, phages 1706 and Q54, which were unrelated to the other phages, could be members of two novel groups. Genomic analysis of these new phages is currently under way and will likely shed light on their origins and relationships with known lactococcal phages.
As indicated above, no member of the P107 species could be retrieved from any collection and thus could not be analyzed in this study. Unfortunately, this is not the first time that a lactococcal phage has been lost. Two previous studies reported the isolation of three phages of the Myoviridae family that infect L. lactis. Phage RZh was described prior to 1970 but has since been lost (53). The second lactococcal myophage was named c10III, but this classification remains uncertain because no contracted sheath was observed (26). In fact, the description and electron micrographs of phage c10III are very similar to those of phage P087 used in the study reported here, and both phages infect the same L. lactis strain (C10). However, since phage c10III is no longer available, the hypothetical relationship with phage P087 could not be investigated. A third putative lactococcal phage from the Myoviridae family was observed by transmission electron microscopy but could not be propagated (52). In the same study, a rare phage with a C1 morphotype (isometric capsids and a short tail) was also observed but was not amplified (52). The loss of these phages points to the need to deposit key phage representatives in public repositories and, ideally, in more than one collection around the world to ensure continued access to them and the reproducibility of future studies.
Three phage groups were merged with P335 due to the high degree of genome mosaicism in this species. Sequencing data revealed that P335 phages have variable genomes that share only 10 to 33% homology (13, 30). This gradient of genetic diversity is likely the consequence of recombination within an infected cell of the genomes of incoming lytic phage and prophage (6, 41). Some members (such as phage ul36) are particularly prone to such recombination (6, 41). This genome plasticity is likely a way to adapt to a new environment including a new host and new phage resistance mechanisms. From a classification point of view, DNA-DNA hybridization (Fig. 1) and a comparative analysis of the 10 P335-like genomes (Fig. 3) showed that the P335-like phages are a perfect example of a polythetic species. Indeed, the P335 species is composed of interconnected isolates with shared properties or modules. However, no single attribute is shared by all known members of this species. Such a mosaic structure makes it unfeasible to detect all the members of this species by using either a single phage genome as a probe for DNA-DNA hybridization or a single pair of primers for PCR assays (30). Previously, it was suggested that a dUTPase gene (Fig. 3) could be used as a target for the PCR detection of P335 phages because, at the time, it was found in all genomes of P335-like phages for which the complete sequence was available (30). However, no dUTPase gene was found in the recently analyzed genome of the virulent phage 4268 (P335 species).
FIG. 3.
Alignment of the genetic maps of 10 completely sequenced P335-like phages (updated from reference 30). Deduced proteins sharing >60% amino acid identity are represented using the same colors and linked with gray shading when possible. Open reading frames with unique sequences are displayed in white. The dUTPase genes are identified by thick lines.
The above results are indicative of the great biodiversity of lactococcal phages. However, phages from genetically different groups do not have the same commercial importance. Three predominant groups (936, c2, and P335) account for 98% of known lactococcal phages and are responsible for most dairy fermentation breakdowns (39, 40). One could speculate on why members of these three lactococcal phage groups predominate in this ecological niche. Clearly, the systematic use of selected L. lactis hosts with specific industrially relevant properties has led to the amplification of these phages in dairy factories. Some of them even survive, to various degrees, the pasteurization process (14, 36). However, one remarkable feature is the relative ease with which phages from these three species can propagate and reach high titers compared to the other lesser-known lactococcal phages. Undoubtedly, this basic fitness parameter explains their widespread distribution and recovery. Other lactococcal phage species are isolated more often from raw milk than from failed fermentations. These phages may not propagate efficiently in a factory environment, which is characterized by higher temperatures, rapid growth of would-be host strains, and mechanical constraints. For some of these species, it has also been difficult to obtain high titers in laboratory conditions, and plaques are usually very small. For example, lactococcal phages of the Podoviridae family (P034 and KSY1) have a long latent period that may explain their low titers (27, 50). Moreover, we observed large numbers of capsids without tails during our electron microscopic observations, indicating that phages with longer tails (949, P087, and 1706) were easily damaged (data not shown). Thus, like other viral communities, rare phages most probably serve as a “bank” and can become abundant in response to very specific environmental conditions (8). Then again, phages Q54 and 1358 are not isolated frequently but were easily amplified by using standard protocols. They may represent emerging groups. It will be interesting to analyze these phages in greater detail to gain a better understanding of their propagation and genetic makeup.
In this study, we used electron microscopic observations, Southern hybridization studies, and comparative sequence analyses (when complete genomes were available) to classify lactococcal phages into 10 groups. Phage classification has always been a subject of controversy (20, 31, 44, 48). For example, it has been suggested that P335 be split into three separate groups, namely, r1t, Sfi11, and Sfi21 (16, 46). This proposal was based on comparative genomics examining a structural gene module (capsid or tail) and on the study of phages active against Streptococcus thermophilus strains. Considering that P335-like phages are prone to genetic recombination through homologous recombination (6, 41) and that comparative complete genome analyses have illustrated the polythetic nature of this species, it may be premature to separate them into different groups. Moreover, despite the fact that L. lactis and S. thermophilus are used in dairy fermentation processes, they are very different bacterial species. S. thermophilus appears to be a species that has only recently emerged that has a clonal structure (21) and is sensitive to only one known polythetic species of temperate/lytic phages (10). In contrast, L. lactis strains are diverse, and as shown in this work, they are sensitive to several genetically unrelated groups of phages. It thus seems a risky exercise to attempt to classify lactococcal phages by singling out one method as a universal taxonomic scheme (44, 54).
The revised classification of lactococcal phages and the improved multiplex PCR test presented in this work provide the basic knowledge and methodology needed to rapidly identify phages responsible for fermentation breakdowns and to follow the evolution of subdominant phages. This report also underscores the need to study the viruses in a given ecological niche as a whole by correlating phenotypic properties with genomic comparisons. Up to now, the only lactococcal phages characterized have been members of the three predominant groups. The characterization of rare phages will likely provide additional information on their biodiversity.
Acknowledgments
We thank Hans-Wolfgang Ackermann, Steve Labrie, Denise Tremblay, and Alain Chopin for helpful discussions, Claudia Bergeron, Geneviève Rousseau, Bruce Geller, Christophe Frémaux, Horst Neve, and Finn K. Vogensen for providing phages and hosts, and Gene Bourgeau for his editorial assistance.
Hélène Deveau is the recipient of a graduate student scholarship from the Fonds Québécois de Recherche sur la Nature et les Technologies (FQRNT). This study was funded by a strategic grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
REFERENCES
- 1.Ackermann, H.-W. 2001. Frequency of morphological phage descriptions in the year 2000. Brief review. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann, H.-W. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245-251. [DOI] [PubMed] [Google Scholar]
- 3.Ackermann, H.-W., E. D. Cantor, A. W. Jarvis, J. Lembke, and J. A. Mayo. 1984. New species definitions in phages of gram-positive cocci. Intervirology 22:181-190. [DOI] [PubMed] [Google Scholar]
- 4.Ackermann, H.-W., M. S. Dubow, A. W. Jarvis, L. A. Jones, V. N. Krylov, J. Maniloff, J. Rocourt, R. S. Safferman, J. Schneider, L. Seldin, T. Sozzi, P. R. Stewart, M. Werquin, and L. Wünsche. 1992. The species concept and its application to tailed phages. Arch. Virol. 124:69-82. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 8.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 9.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 10.Brüssow, H., and F. Desière. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 11.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 12.Chibani Azaiez, S. R., I. Fliss, R. E. Simard, and S. Moineau. 1998. Monoclonal antibodies raised against native major capsid proteins of lactococcal c2-like bacteriophages. Appl. Environ. Microbiol. 64:4255-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chopin, M. C. 1980. Resistance of 17 mesophilic lactic Streptococcus bacteriophages to pasteurization and spray-drying. J. Dairy Res. 47:131-139. [DOI] [PubMed] [Google Scholar]
- 15.Crutz-Le Coq, A.-M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 16.Desière, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brüssow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Leeuwenhoek 82:73-91. [PubMed] [Google Scholar]
- 17.Desière, F., C. Mahanivong, A. J. Hillier, P. S. Chandry, B. E. Davidson, and H. Brüssow. 2001. Comparative genomics of lactococcal phages: insight from the complete genome sequence of Lactococcus lactis phage BK5-T. Virology 283:240-252. [DOI] [PubMed] [Google Scholar]
- 18.Deveau, H., M. R. Van Calsteren, and S. Moineau. 2002. Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl. Environ. Microbiol. 68:4364-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Émond, É., B. J. Holler, I. Boucher, P. A. Vandenbergh, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1997. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. D. Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis, A. W. 1978. Serological studies of a host-range mutant of a lactic streptococcal bacteriophage. Appl. Environ. Microbiol. 36:785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, A. W. 1994. Relationships by DNA-DNA homology between lactococcal phages 7-9, P335 and New Zealand lactococcal phages. Int. Dairy J. 5:355-366. [Google Scholar]
- 25.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 26.Keogh, B. P., and P. D. Shimmin. 1974. Morphology of the bacteriophages of lactic streptococci. Appl. Microbiol. 27:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotsonis, S. E. 1999. Detection and characterization of bacteriophages infecting Lactococcus lactis. Ph.D. thesis. University of Melbourne, Melbourne, Australia.
- 28.Kotsonis, S. E., I. B. Powell, and A. J. Hillier. 1998. Analysis of asccφ28, a Lactococcus lactis phage of a rare morphotype. Aust. J. Dairy Technol. 53:132. [Google Scholar]
- 29.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lembke, J., U. Krisch, A. Lompe, and M. Teuber. 1980. Isolation and ultrastructure of bacteriophages of group N (lactic) streptococci. Zentbl. Bakteriol. Abt. Orig. C 1:79-91. [Google Scholar]
- 33.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 34.Lowrie, R. J. 1974. Lysogenic strains of group N lactic streptococci. Appl. Microbiol. 27:210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madera, C., C. Monjardin, and J. E. Suarez. 2004. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl. Environ. Microbiol. 70:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniloff, J., and H.-W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the other Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Moineau, S. 1999. Applications of phage resistance in lactic acid bacteria. Antonie Leeuwenhoek 76:377-382. [PubMed] [Google Scholar]
- 40.Moineau, S., and C. Lévesque. 2005. Control of bacteriophages in industrial fermentation, p. 286-296. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, Fla.
- 41.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moineau, S., D. Tremblay, and S. Labrie. 2002. Phages of lactic acid bacteria: from genomics to industrial applications. ASM News 68:388-393. [Google Scholar]
- 43.Moineau, S., S. A. Walker, E. R. Vedamuthu, and P. A. Vandenbergh. 1995. Cloning and sequencing of LlaDCHI restriction/modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae DpnII system. Appl. Environ. Microbiol. 61:2193-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson, D. 2004. Phage taxonomy: we agree to disagree. J. Bacteriol. 186:7029-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen, M., S. Ostergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 46.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desière, and H. Brüssow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Relano, P., M. Mata, M. Bonneau, and P. Ritzenthaler. 1987. Molecular characterization and comparison of 38 virulent and temperate bacteriophages of Streptococcus lactis. J. Gen. Microbiol. 133:3053-3063. [DOI] [PubMed] [Google Scholar]
- 48.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Saxelin, M. L., E. L. Nurmiaho, M. P. Korhola, and V. Sundman. 1979. Partial characterization of a new C3-type capsule-dissolving phage of Streptococcus cremoris. Can. J. Microbiol. 25:1182-1187. [DOI] [PubMed] [Google Scholar]
- 51.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teuber, M., and J. Lembke. 1983. The bacteriophages of lactic acid bacteria with emphasis on genetic aspects of group N lactic streptococci. Antonie Leeuwenhoek 49:471-478. [DOI] [PubMed] [Google Scholar]
- 53.Tikhonenko, A. S. 1970. Ultrastructure of bacterial viruses. Plenum Press, New York, N.Y.
- 54.van Regenmortel, M. H., and B. W. Mahy. 2004. Emerging issues in virus taxonomy. Emerg. Infect. Dis. 10:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. Ruiters, G. Venema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 56.Vegge, C. S., L. Brondsted, H. Neve, S. McGrath, D. van Sinderen, and F. K. Vogensen. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187:4187-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]