Abstract
Bacterial diversity is central to ecosystem sustainability and soil biological function, for which the role of roots is important. The high-throughput analysis potential of taxonomic microarray should match the breadth of bacterial diversity. Here, the power of this technology was evidenced through methodological verifications and analysis of maize rhizosphere effect based on a 16S rRNA-based microarray developed from the prototype of H. Sanguin et al. (Environ. Microbiol. 8:289-307, 2006). The current probe set was composed of 170 probes (41 new probes in this work) that targeted essentially the Proteobacteria. Cloning and sequencing of 16S rRNA amplicons were carried out on maize rhizosphere and bulk soil DNA. All tested clones that had a perfect match with corresponding probes were positive in the hybridization experiment. The hierarchically nested probes were reliable, but the level of taxonomic identification was variable, depending on the probe set specificity. The comparison of experimental and theoretical hybridizations revealed 0.91% false positives and 0.81% false negatives. The microarray detection threshold was estimated at 0.03% of a given DNA type based on DNA spiking experiments. A comparison of the maize rhizosphere and bulk soil hybridization results showed a significant rhizosphere effect, with a higher predominance of Agrobacterium spp. in the rhizosphere, as well as a lower prevalence of Acidobacteria, Bacteroidetes, Verrucomicrobia, and Planctomycetes, a new taxon of interest in soil. In addition, well-known taxonomic groups such as Sphingomonas spp., Rhizobiaceae, and Actinobacteria were identified in both microbial habitats with strong hybridization signals. The taxonomic microarray developed in the present study was able to discriminate and characterize bacterial community composition in related biological samples, offering extensive possibilities for systematic exploration of bacterial diversity in ecosystems.
Biodiversity is a major factor influencing ecosystem stability (38, 57). Since bacteria fulfill key ecosystem functions, characterization of their community is important for understanding ecosystem function and for identifying environment quality bioindicators (3, 70, 77). However, bacterial diversity in complex ecosystems remains poorly documented (20, 79). Often, fingerprinting or clone library methods used for diversity and community structure characterizations target the 50 to 200 most dominant populations (25, 46), although the less numerous ones can also be of ecological importance. The main shortcomings of these methods are the difficulties in identifying community members (for fingerprinting methods) and cost and time (for clone library methods). An example of an important complex ecosystem is the rhizosphere.
The rhizosphere has received considerable research attention because many rhizobacteria promote plant growth (41), control soilborne phytoparasites (62, 78), and/or contribute to nutrient cycling and pollutant biodegradation (61). The release of root exudates contributes to higher bacterial population densities and significant community changes compared to bulk soil (54, 55, 69). The rhizosphere is also a hot spot of microbial activity. Culture-dependent methods have shown the prevalence of gram-negative bacteria (Proteobacteria) in the rhizosphere (53, 62). PCR-based methods have extended these findings by showing the occurrence of taxa such as Acidobacteria, Actinobacteria, or Archaea (15, 27, 31, 69). However, much remains to be done to explore rhizobacterial diversity and fully characterize the rhizosphere effect on soil bacteria.
The development of high-throughput tools, such as microarrays, constitutes a new promising step toward monitoring bacterial community composition (24, 28, 40, 48, 49, 59, 71). However, the majority of microarrays developed thus far focus on functional bacterial groups such as diazotrophs (40), methanotrophs (71), ammonia oxidizers (1), sulfate reducers (48) and pollutant biodegraders (9, 42), or on single taxonomic groups, e.g., the Rhodocyclales (49) and Cyanobacteria (14). Little has been done with a microarray approach to explore the entire bacterial diversity (24, 63, 80). With 122 16S rRNA probes covering different taxonomic levels, the taxonomic microarray proposed by Sanguin et al. (63) appeared to be a promising tool for the analysis of Alphaproteobacteria in complex ecosystems.
The objective of the present study was to evaluate the ability of the 16S rRNA microarray approach to assess, under field conditions, the rhizosphere effect of maize on soil bacteria. To this end, the prototype microarray previously developed by Sanguin et al. (63) was improved with additional probes. The data normalization, hybridization specificity, and detection threshold were assessed. Microarray hybridizations of bulk soil and maize rhizosphere were then compared to cloning and sequencing data. The maize rhizosphere effect was investigated by comparing bulk soil and rhizosphere at the levels of (i) the total bacterial community and (ii) a bacterial subcommunity (Agrobacterium genus).
MATERIALS AND METHODS
Environmental samples.
Samples were taken in a field at La Côte Saint-André (France), where the soil (luvisol [32]) was planted with 3-month-old maize (Zea mays cv. PR38a24; Pioneer, Aussonne, France). Three plants (termed 1, 2, and 3), each from a different row and located about 5 m from one another, were used.
The root system of each of the three maize plants was dug up and shaken vigorously to separate soil loosely adhering to the roots. After elimination of the top 5 cm of the root systems (near the crown), each root system with closely adhering soil was transferred to a 1-liter bottle containing a 500-ml sterile distilled water. The bottles were shaken manually for 15 min. The soil fraction was recovered by centrifugation of the extract for 30 min at 6,000 rpm and then stored at −20°C. Bulk soil was taken from the inter-row at three different locations in the same area of the field, at the same depth as for root systems (5 to 20 cm). The three samples were pooled. The composite bulk soil sample was homogenized, and one 15-g subsample was taken for DNA extraction.
Genomic DNA extraction.
DNA from the bacteria Frankia sp. strain ACN14a and Wolbachia pipientis wRi was obtained as described by Simonet et al. (66) and Mavingui et al. (56), respectively. Environmental sample DNA extraction was performed on one bulk soil sample and three rhizosphere samples (equivalent to 1 g of dry weight) using the UltraClean Soil DNA Isolation kit (MO BIO Laboratories, Solana Beach, CA) according to the protocol provided by the manufacturer.
PCR amplification of 16S rRNA genes and labeling.
The 16S rRNA genes from the two reference strains Frankia sp. strain ACN14a and W. pipientis wRi, as well as 20 environmental 16S rRNA clones (3 clones from Sanguin et al. [63] and 17 from the present study; see discussion of cloning and sequencing below) were labeled with Cy3-dCTP (Amersham Biosciences Europe GmbH, Saclay, France) during PCR with universal primers pA and pH' (10) and with primers M13f and M13r (Promega, Madison, WI), respectively, in order to generate internally labeled double-stranded DNA. Two primer pairs were used with environmental DNA samples. First, the universal primers pA and pH′ were used in order to obtain representative 16S rRNA gene amplicons (1.5 kb) of the total community. Second, the reverse primer F2107AgroAT41m (63), which targets the Agrobacterium genus, was used along with pA to obtain Agrobacterium-related 16S rRNA gene amplicons (1 kb).
For the reference strains and the 16S rRNA environmental clones, PCR mixtures (total volume, 50 μl) contained 1× reaction buffer; 1 μM concentrations of each primer; 1.5 mM MgCl2; 50 μM concentrations of dATP, dGTP, and dTTP; 45 μM concentrations of dCTP; 5 μM concentrations of Cy3-dCTP; 50 ng of genomic DNA; and 5 U of Taq DNA polymerase (Invitrogen, Cergy Pontoise, France). For the environmental DNA samples, the PCR mixture (50 μl) contained 1× reaction buffer; each primer at 0.5 μM; 1.5 mM MgCl2; 50 μM concentrations of dATP, dGTP, and dTTP; 45 μM concentrations of dCTP; 5 μM concentrations of Cy3-dCTP; 10 ng of environmental DNA; 0.025 mg of T4 Gene 32 (Roche Applied Science, Meylan, France)/ml; and 1.25 U of Taq Expand High Fidelity (Roche Applied Science). Thermal cycling was carried out with a denaturation step of 94°C for 3 min, 35 cycles with 45 s of denaturation at 94°C, 45 s of annealing at 53 or 55°C (depending on the primer set), 90 s of elongation at 72°C, and a final elongation step for 7 min at 72°C. Labeled amplicons were purified through a QIAquick PCR purification column (QIAGEN, Courtaboeuf, France) according to the manufacturer's instructions. The DNA concentration was determined spectrophotometrically by measuring the optical density at 260 nm.
Cloning and sequencing of environmental 16S rRNA clones.
The total bacterial amplicons from rhizosphere DNA of one maize plant (i.e., plant 2) and from bulk soil DNA were used for cloning into the plasmid vector pGEM-T (pGEM-T Easy Vector System kit; Promega) in order to generate a 16S rRNA clone library. A total of 162 positive clones of 600 were sequenced (Genome Express, Meylan, France), and 17 of them, representative of different major phyla, were used to validate the hybridization reliability of the probe set.
Sequence affiliation was performed by using the two RDP analysis tools Classifier and Sequence Match (http://rdp.cme.msu.edu/index.jsp) at default settings. Chimeric 16S rRNA gene sequences were checked by using two chimera detection programs Pintail (version 0.33; http://www.cardiff.ac.uk/biosi/research/biosoft/Pintail/pintail.html) and Bellerophon (35; http://foo.maths.uq.edu.au/∼huber/bellerophon.pl). All of the sequences described here have been submitted to the EMBL database under accession numbers AJ876433, AJ876449, AJ876455, and AM157230 to AM157351.
Oligonucleotide probe design.
The probe set contained 170 probes, including 129 published probes (120 of them from Sanguin et al. [63]) and 41 new probes (see Table S1 in the supplemental material). The latter were designed by using the phylogenetic software package ARB (52; http://www.arb-home.de). The ARB 16S rRNA database (ssu03jun.arb) was also used to predict probe specificity. The 16S rRNA gene sequences obtained in the present study were aligned by using the alignment tool ARB_EDIT (with the “Integrated Aligners” option) and added to the ARB phylogenetic tree using the “Add Species to Existing Tree” function (with the “Parsimony_Quick Add Marked” option). The ARB database (i.e., PT_SERVER) was updated with the new sequences to allow the evaluation of the probe set on the new sequences by using the Probe Match function. The parameters of the Probe Design function were chosen according to the method of Sanguin et al. (63). The best probes were selected from the Probe Results window. Their predicted melting temperature Tm (according to the nearest-neighbor method) and free energy values (ΔG) of hairpin structures were calculated by using Oligo5 (Molecular Biology Insights, West Cascade, CO). Probes with a Tm of 65 ± 5°C, a %G+C of >50%, no hairpin (or a hairpin with a ΔG > −2 kcal/mol and a Tm < 50°C), and no stable homoduplex were accepted. In some cases, these requirements could not be met, and so probes with suboptimal conditions were accepted in order to provide adequate phylogenetic coverage. Bacteria targeted by each probe, with up to 3.5 weighted mismatches (WMM), were listed with the Probe Match function, and the Probe Match output files were transferred to CalcOligo version 1.07 (7). The CalcOligo output file was used to create an Excel table (available upon request) indicating the predicted Tm (according to the nearest neighbor method), the length, the G+C content of the probes, and the number of WMM for each probe-target combination.
Microarray hybridization.
Probes were custom synthesized (Eurogentec, Seraing, Belgium) with a 5′ C6-NH2 group for covalent attachment onto aldehyde slides AL (Schott Nexterion AG, Mainz, Germany). Microarray manufacturing and processing were performed as described by Sanguin et al. (63). The concentration of probes before printing was 50 μM in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1.5 M betaine. The basic probe pattern on the microarray consisted of two spots for each of the control probes EUB338 and EUB342 (both used as positive controls and landmarks for image analysis) and one spot for each of the other probes. The same basic pattern was replicated six times on the microarray using a MicroGrid II spotter (BioRobotics, Cambridge, United Kingdom).
The labeled DNA (1.5 μg) was dried down in a Speed-Vac (Fisher Bioblock Scientific, Illkirch, France), resuspended in 25 μl of QMT hybridization buffer (Quantifoil, Jena, Germany), and denatured at 95°C for 5 min. The slides were placed in a Genetix hybridization chamber (Proteigene), and the chamber was preheated to 50°C. Labeled DNA was transferred onto the slide, covered with a Hybri-Slip (Grace BioLabs, Bend, OR) (preheated to ∼55°C for 5 min), and transferred back into the hybridization chamber. Hybridization was conducted overnight at 50°C. Slide-washing steps were performed as described by Sanguin et al. (63).
Scanning and data collection.
The slides were scanned at 10-μm resolution with a GeneTac LS IV scanner (GenomicSolutions, Huntingdon, United Kingdom) at 532 nm for Cy3, and the images were analyzed by using GenePix 4.01 software (Axon, Union City, CA). The spot quality was visually checked, and the spots of poor quality (i.e., the presence of dust, hybridization signal saturation, etc.) were excluded from further analyzes, as was done by Sanguin et al. (63).
Filtration, normalization, and statistics.
Data filtration was conducted with the R statistical computing environment (http://www.r-project.org). Individual spots were considered positive if 80% of the spot pixels had an intensity higher than the median local background pixel intensity plus twice the standard deviation of the local background. This procedure decreases the probability of false-positive signals to <2.5% (59). To further decrease the risk of false-positives, a given feature probe was considered truly hybridized when at least three of six replicate spots provided a significantly positive hybridization.
Two types of normalization were evaluated in order to reduce the intraslide variability in microarray experiments. A transformation was first applied on the intensity signals for each spot. The intensity signals (median of signal minus background) were replaced by their square root to decrease the impact on the analysis of the few probes displaying particularly intense signals. The first type of normalization (63) was based on the positive control EUB342, which targets the whole Bacteria domain amplified with the primer set used. For the second one, the intensity of each spot was expressed as the percentage of the total intensity signal of its basic pattern. Microarray hybridization variability according to the type of normalization was analyzed by comparing the distribution of probe variation coefficients (CVs) computed by using each spot from two slides (n = 12).
Comparison of microarray results for environmental samples was carried out on the normalized intensity signals by principal component analysis (PCA) using the package ade4 (73; http://pbil.univ-lyon1.fr/ADE-4/) for R or by hierarchical cluster analysis and the Kmeans test using R. The PCA was performed by using the covariance matrix, and the hierarchical cluster analysis was based on Ward's method. The probes EUB338, EUB342, and AntiPA (positive controls) and CF319, CF319m, Irog2-mrc, GAMrc, and Verru1 (nonspecific hybridizations) were removed for PCA (and for probe validation analysis) because they do not provide any biological information for the comparison of samples.
Evaluation of the detection threshold.
Spiking experiments were performed by adding 16S rRNA PCR products from either of two strains (Frankia sp. strain ACN14a or W. pipientis wRi) to the PCR product obtained from the maize rhizosphere. The two strains were chosen because the probes targeting them did not produce any signal when hybridized with maize rhizosphere DNA. Furthermore, Wolbachia is a common cytoplasmic insect symbiont, and the absence of actinorhizal symbiotic hosts for Frankia in this area is well documented. 16S rRNA genes were then amplified, labeled, mixed, and hybridized. The 16S rRNA amplicons were used at 0.3, 0.1, 0.03, and 0.01% of the total amount of DNA. Two replicates were carried out for each mixture.
RESULTS
Development of the probe set.
The current probe set was composed of 170 probes, which targeted essentially the Proteobacteria. A total of 41 new probes were designed here, and nine others were obtained from the literature (2, 34, 45, 51) (see Table S1 in the supplemental material). A total of 62 probes targeted the Alphaproteobacteria, 32 targeted the Betaproteobacteria, 26 targeted the Gammaproteobacteria, 4 targeted the Deltaproteobacteria, and 1 targeted the Epsilonproteobacteria. The other principal phyla also represented on the microarray were the Firmicutes (eight probes), Bacteroidetes (seven probes), Actinobacteria (five probes), Planctomycetes (four probes), Acidobacteria (three probes), Verrucomicrobia (two probes), Chloroflexi (two probes), Cyanobacteria (two probes), Fibrobacteres (one probe), and Fusobacteria (one probe).
Data normalization.
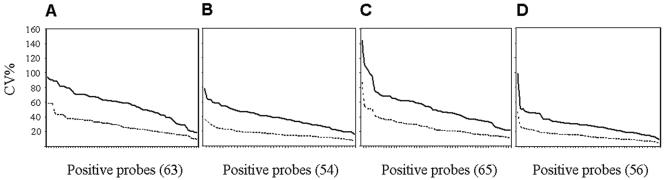
The comparison of the CV of hybridization data from rhizosphere and bulk soil obtained with the two types of data normalization (Fig. 1) indicated that the CV was considerably reduced when the hybridization intensities were replaced by their square root and then divided by the total intensity signal of their basic pattern. Four classes of intensity—i.e., less than 9 × 10−3 relative fluorescent units (RFU), 9 × 10−3 to 1.5 × 10−2 RFU, 1.5 × 10−2 to 3 × 10−2 RFU, and more than 3 × 10−2 RFU—were defined based on the quartiles of the normalized hybridization intensities from all environmental samples. These four classes were subsequently used to contrast the various hybridization intensity levels recorded in the present study and especially to define strong intensity as 3 × 10−2 RFU or more.
FIG. 1.
Effect of microarray data normalization on CVs for positive probes for four environmental samples: bulk soil (A), rhizosphere plant 1 (B), rhizosphere plant 2 (C), and rhizosphere plant 3 (D). Normalization was based on either the control probe EUB342 (solid line) or the total intensity signal of the basic pattern (dotted line). The positive probes have been ordered by decreasing CV values, and the number in parentheses indicates the number of positive probes.
Probe set validation.
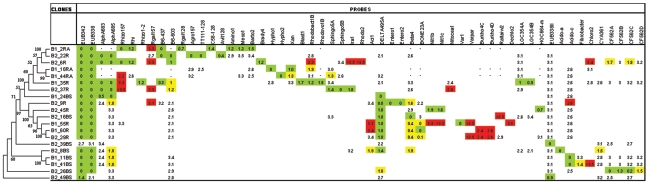
For the 20 16S rRNA clones studied, all probe-target combinations that displayed a perfect match yielded a hybridization signal (except for BONE23A with the clone B1_55R) (Fig. 2). Box plot analysis showed that normalized hybridization intensities depended on WMM values (data not shown), thereby strengthening and expanding preliminary results (63). Indeed, hybridization intensities were higher in cases with perfect matches (median of the normalized intensities = 9 × 10−2 RFU) compared to nonperfect matches (WMM = 0.1 to 1.9; 3 × 10−2 RFU). These perfect matches involved 22 probes; 12 of which were checked in the present study and targeted various taxa, i.e., Betaproteobacteria, Acidobacteria, Planctomycetes, Bacteroidetes, Enterobacteriaceae, Hyphomicrobium, and Variovorax.
FIG. 2.
Comparison of experimental and expected hybridization results for 20 16S rRNA clones from bulk soil and maize rhizosphere. Green shading indicates expected positive results, red shading indicates unexpected positive results, a white background indicates expected negative results (without showing WMM), and yellow shading indicates unexpected negative results. The number in the cells represents the WMM value. The 144 probes situated between positions 114 and 1291 (E. coli numbering), which corresponded to sequences checked for both strands, were analyzed. Concerning the clones B1_2RA, B1_16RA, and B1_44RA (Sanguin et al. [63]), the probes targeting the 16S rRNA gene at a position >600 were excluded from the analysis (shown as a dash). Probes yielding only expected negative results with all 16S rRNA clones tested are not shown (87 probes). The 16S rRNA neighbor-joining phylogenetic tree was constructed by using the Kimura two-parameter correction of evolutionary distances. Bootstrap values greater than 50 are shown.
The comparison of experimental and theoretical hybridizations (2,847 individual probe-target hybridizations), based on the hybridization threshold (WMM = 2) defined by Sanguin et al. (63), revealed 0.91% false positives and 0.81% false negatives (Fig. 2). Consequently and according to previous observations in Sanguin et al. (63), a few probes were characterized either for their nonspecific hybridizations (Irog2-mrc, GAMrc, Verru1, Rgal157, and Halo2) or for their extended target patterns compared to the perfect match target pattern (CF319, CF319m, and DELTA495a). Most importantly, the hierarchic taxonomic approach followed in the present study proved functional. For instance, hybridization pattern analysis of the clone B2_22R, which was affiliated with the Agrobacterium genomic species G1 (Table S2 in the supplementary material), allowed the affiliation of the clone to phylum (Alpha683 and Alpha685), genus (Agro157), and genomic species (TT111-128).
Microarray analysis of the total rhizosphere bacterial community.
With the maize rhizosphere, probes targeting Alphaproteobacteria, Deltaproteobacteria, Actinobacteria, Acidobacteria, Planctomycetes, and Bacteroidetes yielded strong hybridization signals (>3 × 10−2 RFU) in most cases, whereas the levels for Firmicutes were only ca. 1.4 × 10−2 RFU (data not shown). For taxonomic levels below that of the phylum, strong hybridization signals were observed for several Alphaproteobacteria probes, such as those targeting the three families Rhizobiaceae (except Agrobacterium), Bradyrhizobiaceae, and Brucellaceae (i.e., with probes Rhizo157 and Rhi) and Sphingomonas spp. (Sphingo5B). For the Agrobacterium genus, 10 probes yielded positive signals of various intensity levels, among which the three specific probes targeting the genomic species of Agrobacterium biovar 1 (probes B6-128, C58-128, and TT111-128) yielded strong hybridization signals. In addition, the Mesorhizobium probe also yielded significant hybridization intensities.
Cloning and sequencing analysis of rhizosphere and bulk soil bacterial communities.
Sequencing analysis of 101 rhizosphere clones and 61 bulk soil clones was performed with the two RDP analysis tools Classifier and Sequence Match to determine the taxonomic assignment of clones. 16S rRNA gene sequences were then analyzed with Pintail and Bellerophon programs, revealing the presence of 25 and 15 potential chimeric sequences for rhizosphere and bulk soil samples, respectively (which were then discarded).
Results from maize rhizosphere revealed a high dominance of Proteobacteria (97%), mainly Betaproteobacteria (64%) and Alphaproteobacteria (28%), whereas only two clones (i.e., 3%) (B1_21R and B1_36R) were affiliated with Acidobacteria (Table 1 and Table S2 in the supplemental material). Proteobacteria (57%) and Acidobacteria (17%) dominated bulk soil, but several major phyla were also observed, i.e., Planctomycetes, Bacteroidetes, Gemmatimonadetes, Chloroflexi/Thermomicrobia, Verrucomicrobia, Actinobacteria, and Firmicutes, as well as the TM7 division.
TABLE 1.
Simplified taxonomic affiliation of 16S rRNA clones from bulk soil and maize rhizosphere (plant 2)
| Taxonomic affiliation | No. of clones
|
|
|---|---|---|
| Maize rhizosphere | Bulk soil | |
| Proteobacteria | 72 | 26 |
| Alphaproteobacteria | 24 | 12 |
| Rhizobiales | 3 | 7 |
| Sphingomonadales | 19 | 1 |
| Caulobacterales | 2 | |
| Rhodospirillales | 2 | |
| Betaproteobacteria | 44 | 9 |
| Burkholderiales | 42 | 4 |
| Gammaproteobacteria | 5 | 1 |
| Xanthomonadales | 4 | 1 |
| Enterobacteriales | 1 | |
| Deltaproteobacteria | 1 | 2 |
| Myxococcales | 1 | 1 |
| Acidobacteria | 2 | 8 |
| Actinobacteria | 1 | |
| Firmicutes | 1 | |
| Bacilli | 1 | |
| Bacillales | 1 | |
| Bacteroidetes | 2 | |
| Sphingobacteria | 1 | |
| Sphingobacteriales | 1 | |
| Gemmatimonadetes | 2 | |
| Thermomicrobia/Chloroflexi | 2 | |
| Planctomycetes | 2 | |
| Planctomycetacia | 1 | |
| Planctomycetales | 1 | |
| Verrucomicrobia | 1 | |
| Verrucomicrobiae | 1 | |
| Verrucomicrobiales | 1 | |
| TM7 | 1 | |
Comparison of microarray and cloning-sequencing methods.
First, sequence comparison of the probes and clones (positions 114 to 1291, Escherichia coli numbering) revealed that all clones that had perfect matches with corresponding probes were positive in the hybridization experiment (except for BONE23A and Beta4) for rhizosphere and bulk soil samples (data not shown). Strong hybridization signals (>3 × 10−2 RFU) were observed for 64 and 58% of the probes displaying a perfect match with at least one clone for rhizosphere and bulk soil, respectively. Among the probes yielding strong hybridization signals, 22 and 8% displayed a WMM of >2 (the threshold used by Sanguin et al. [63]) for the rhizosphere and bulk soil, respectively.
Second, microarray and cloning-sequencing results were compared based on taxonomic implications. For the rhizosphere, probes targeting phyla such as Actinobacteria (HGC236-m), Planctomycetes (Plancto4-mA and Plancto-mB), and Bacteroidetes (CFB562A) yielded strong hybridization signals, but no sequence affiliated to these phyla was obtained by cloning-sequencing. Likewise, only one clone affiliated with Agrobacterium genomic species 1 was retrieved by cloning-sequencing, whereas the majority of probes targeting the different genomic species within Agrobacterium biovar 1 yielded strong hybridization signals. Nevertheless, the majority of taxa recovered by cloning-sequencing (based on Sequence Match scores of >0.800) corresponded to at least one positive probe (e.g., Sphingomonas, Agrobacterium, Bradyrhizobium, Variovorax, and Enterobacter spp.). In several cases, strong hybridization signals corresponded to a high number of clones with perfect matches (e.g., probe Sphingo5B). The probe set was not adapted for identification at genus level within certain taxonomic groups retrieved by cloning-sequencing (such as Aquabacterium, Caulobacter, Pseudoxanthomonas, Stenotrophomonas, and Rhodanobacter spp.).
For bulk soil, the clone sequence match scores with RDP-II data were often not high enough to enable reliable comparisons between microarray and cloning-sequencing results at the genus level, but such comparisons could be done for higher taxonomic levels. All of the phyla identified by cloning-sequencing were also found by hybridization, except the two that lacked microarray probes (TM7 division and Gemmatimonadetes).
Maize rhizosphere effect on bacterial community structure.
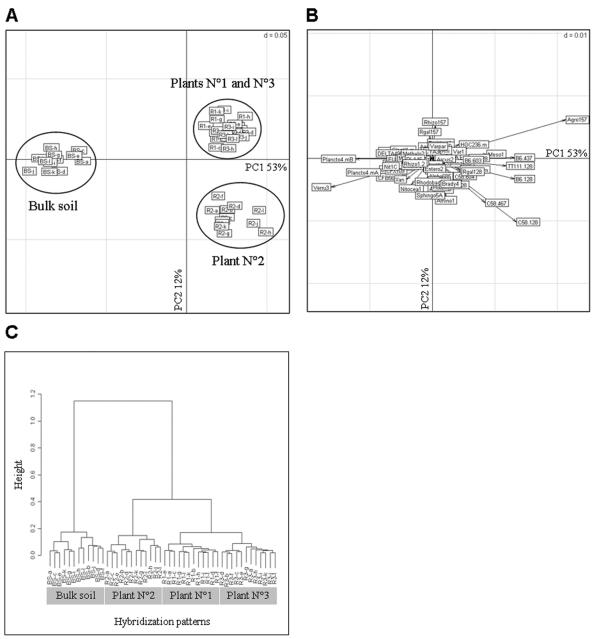
Hybridization data from bulk soil and the rhizosphere of three maize plants were analyzed by PCA (Fig. 3). Bulk soil and rhizosphere samples were well separated along the first PCA axis (as much as 53% of the total variability). The 53% inertia of this axis indicates that the maize rhizosphere effect is a strong structuring agent of the bacterial community in this soil. The probes targeting different genomic species of Agrobacterium biovar 1 and Mesorhizobium contributed significantly to rhizosphere hybridization patterns, whereas for bulk soil it was the probes targeting Planctomycetes and Verrucomicrobia. For most probes, the differences between rhizosphere and bulk soil consisted in differences in intensity levels, but several probes yielded hybridization signals only in bulk soil (the Planctomycetes probes PLA46 and EUB338II) or in the rhizosphere (probes C58-128 and AF310-128, which target certain Agrobacterium species). Two Planctomycetes probes (PLA46 and Plancto4-mA) hybridized specifically with the Planctomyces clone B2_39BS (confidence threshold of 100%). The two other Planctomycetes probes also hybridized with 16S rRNA clones affiliated to other taxa, i.e., (i) Verrucomicrobia (clone B2_49BS) and Bacteroidetes (B2_26BS) for probe Plancto4-mB and (ii) Bacteroidetes (B2_26BS) for EUB338II.
FIG. 3.
Hybridization pattern analysis of bacterial communities from bulk soil and maize rhizosphere performed by PCA (A and B) and hierarchical cluster analysis (C). BS indicates bulk soil, and R1, R2, and R3 indicate the rhizospheres of plants 1, 2, and 3, respectively. The six replicates on the first slide are indicated by the letters a to f, and those on the second slide are indicated by the letters g to l. For PCA, samples are shown in panel A, and positive probes are shown in panel B. The hierarchical cluster analysis in panel C was done by using Ward's method.
Interestingly, plant-to-plant variability of bacterial community composition was found when we compared plant 2 with plants 1 and 3 along the first axis of the PCA carried out on rhizosphere samples (data not shown). Four probes explained most of this variability. The probes Agro157 (Agrobacterium spp.) and Rhizo157 (Rhizobiaceae [except Agrobacterium]/Bradyrhizobiaceae/Brucellaceae) were associated with plants 1 and 3, and probe C58-128 (Agrobacterium genomic species G6/G8 and A. rubi) was associated with plant 2. The robustness of the results was also tested by hierarchical cluster analysis (Fig. 3C) and the Kmeans method (data not shown), confirming the two major effects observed with PCA, i.e., the differentiation between bulk soil and maize rhizosphere and plant-to-plant variability. However, these two methods also separated plant 1 from plant 3.
Maize rhizosphere effect on a bacterial subcommunity.
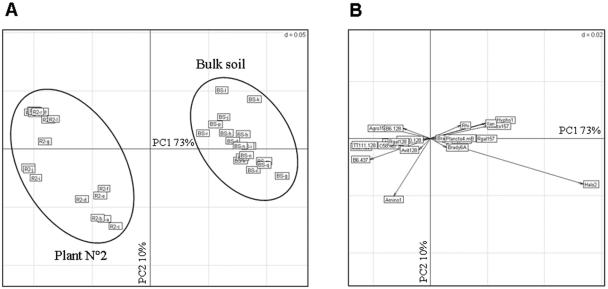
Since Agrobacterium appeared as a major taxonomic group in the maize rhizosphere, Agrobacterium-related PCR amplicons (63) were obtained and hybridization patterns for bulk soil and the rhizosphere of maize plant 2 were compared by PCA (Fig. 4). The results confirmed those obtained with the total community, i.e., all Agrobacterium probes (except Arub128) that were positive in the subcommunity approach were positive in the total community analysis. The abundance of Agrobacterium populations (biovar 1) in this soil is in the order of 103 CFU g−1 in bulk soil (75) and 105 CFU g−1 in the maize rhizosphere (unpublished data). Here, bulk soil and rhizosphere of maize plant 2 were clearly separated along the first axis (73% of the total variability). Probes Agro157, B6-437, TT111-128, C58-128, and Amino1, which target mostly Agrobacterium genomic species (Table S1 in the supplemental material), contributed significantly to the rhizosphere patterns. Previous results indicated that Amino1 was likely hybridized with Agrobacterium strains (63). The probes C58-128 and AF310-128 yielded hybridization signals only with the rhizosphere. The probes Rhizo157 (targeting Rhizobiaceae [excepted Agrobacterium]/Bradyrhizobiaceae/Brucellaceae), Xan (Xanthobacter), and Hypho1 (Hyphomicrobium) contributed to bulk soil patterns, hybridization signals for the last two probes being almost nonexistent in the rhizosphere.
FIG. 4.
Hybridization pattern analysis of Agrobacterium subcommunity from bulk soil and maize rhizosphere (plant 2) performed by PCA. BS indicates bulk soil, and R2 indicates the rhizosphere of plant 2. The six replicates on the first slide are indicated by the letters a to f, those on the second slide are indicated by the letters g to l, and those on the third slide are indicated by the letters m to r. The samples are shown in panel A, and the positive probes are shown in panel B.
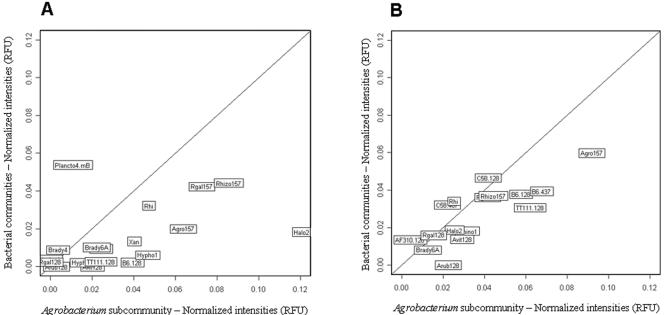
When we compared the results for the total bacterial community and the Agrobacterium subcommunity, the probes targeting Agrobacterium spp. gave higher hybridization signals with the Agrobacterium-related amplicon than the total bacterial amplicon, except for the two probes targeting the genomic species G6/G8 (i.e., C58-128 and C58-467) (Fig. 5).
FIG. 5.
Comparison of hybridization patterns between total bacterial communities and the Agrobacterium subcommunity for bulk soil (A) and for the rhizosphere of maize plant 2 (B). Only the probes that yielded hybridization signals with the Agrobacterium subcommunity are shown.
Detection threshold of DNA from a complex sample.
The detection threshold was studied by using a 16S rRNA gene amplicon from maize rhizosphere mixed with different quantities of 16S rRNA gene amplicons from W. pipientis wRi or Frankia sp. strain ACN14a, whose specific probes (Wol3/Wol4 and Actino2/Actino3, respectively) were validated experimentally in the present study (data not shown). Hybridization signals of the four genus-specific probes were detected down to 0.03% (i.e., 0.45 ng) of spiked DNA in the mixture (Table 2). Considering that W. pipientis and Frankia sp. strain ACN14a contain, respectively, one (6) and two (unpublished result) 16S rRNA gene copies and that one 16S rRNA gene molecule weighs 1.64 × 10−9 ng, this level of 0.03% would correspond to a number of amplified sequences on the order of 108.
TABLE 2.
Spiking experiments for evaluation of the microarray detection threshold in a complex DNA mixturea
| Relative amt of spiked DNA (%) |
Frankia sp. strain ACN14a
|
W. pipientis wRi
|
||
|---|---|---|---|---|
| Actino2 | Actino3 | Wol3 | Wol4 | |
| 0.3 | + | + | ND | ND |
| 0.1 | + | + | ND | ND |
| 0.03 | + | + | + | + |
| 0.01 | − | − | − | − |
| 0 | − | − | − | − |
ND, not done. Two slides were studied per sample. The symbol “+” indicates that all 12 replicates of two slides were positive, and “−” indicates that all 12 replicates were negative.
DISCUSSION
Scope.
Bacterial diversity is central to ecosystem sustainability, and its assessment is essential for understanding soil biological function, for which the role of roots is important (67). Taxonomic microarrays have become a part of the technological response to this need in microbial ecology. Here, the 16S rRNA-based microarray developed from the prototype of Sanguin et al. (63) was applied to the study of maize rhizosphere microbial communities.
Methodological evaluation: probe set and detection limit.
Basic verifications, e.g., the evaluation of false positives (0.91%) and false negatives (0.81%) and detection limits were consistent with previous estimates (0.7 to 9%) in taxonomic microarray development (7, 47, 49). Furthermore, this supports the use of the hybridization threshold (WMM = 2) defined by Sanguin et al. (63) to predict probe-target hybridization.
The detection limit has been generally estimated to 1 to 5% of community members in the environment (7, 17, 23, 28, 74) or less (1%) when tyramide signal amplification with a 70mer microarray is used (23). Wu et al. (81) claimed a detection limit of 5 × 106 cells. Nevertheless, these estimates are highly dependent on the probes tested (28). In the present study, the detection limit was estimated to be 0.03% of a DNA type in a complex sample, i.e., 0.45 ng of DNA or (at least) 108 amplicon copies. This detection limit is an intrinsic feature of the microarray technology used, and it should be extrapolated cautiously to estimate the detection threshold of community members, since DNA extraction and PCR from complex samples were not taken into account. The evaluation of a true detection limit would require extensive additional experiments based on numerous probes. The probes for Agrobacterium biovar 1 hybridized in the case of bulk soil, in which this taxon amounts to 103 CFU g−1 (75), and this level is among the lowest detection levels published thus far in microarray studies.
Microarray reliability for community analysis.
Effective data treatment is essential for reliable results and for comparing different experiments (8, 37). Without transformation of the hybridization intensities, strong hybridization signals have an unacceptably large effect on the outcome of the analysis. The log transformation is extensively used with microarray data (17, 50, 72) since it enables data normalization and provides variation stabilization of strong hybridization levels (http://www.stat.berkeley.edu/users/terry/zarray/Html/log.html). However, the log transformation may lead to certain bias (26, 65). First, it is not adapted to low hybridization signals, as it maximizes their weight in comparison experiments and inflates the variance of near-background observations. Second, it drastically reduces the contribution of strong hybridization signals. Here, the square root was used as a compromise between no transformation and the log transformation of data, and this was validated by the relationship between perfect match and high intensity. In addition, the variability inherent to microarray processing, labeling, and hybridization methods (19, 43, 68) needs to be limited. Local normalization using the total intensity signal of basic patterns appeared to be a better reference for data normalization than the positive control (probe EUB342) as used previously (63), which is subjected to spot-to-spot variability between replicates on a given slide and between slides. In addition, using EUB342 as a reference for comparison experiments may induce an important bias because it cannot detect all environmental bacteria, as indicated by (i) fluorescence in situ hybridization analyses with probe EUB338 (very similar to EUB342) (21, 58) and (ii) in silico analysis (available upon request).
Comparison of cloning-sequencing and microarray results.
The cloning-sequencing was carried out in parallel to hybridizations to evaluate the efficiency of the microarray to detect sequences in the environment. The concordance between the two methods was not perfect, since cloning is limited by the number of clones sequenced and the dependence of cloning efficiency on GC content and sequence length, whereas the microarray approach is limited by the number and taxonomic coverage of the probes. However, two essential criteria on which the taxonomic microarray technology is based were satisfied. First, the existence of a relationship between WMM and the intensity level (63) was confirmed, since high-intensity levels (i.e., above 3 × 10−2 RFU) corresponded in most cases to perfect match. This confirms the reliability of the microarray technology to analyze communities. This reliability is important since not all probes have been verified individually. Second, based on strong hybridization signals, false positives amounted to 8 and 22% for bulk soil and rhizosphere samples, respectively. These results are even more encouraging if one considers that these numbers include true false positives plus potential hybridizations with unknown sequences in the complex samples. Increasing the number of sequenced clones would thus decrease the number of false positives. The taxonomic significance of the rhizosphere results demonstrates a good agreement between clones and the microarray for several Proteobacteria taxa (e.g., Sphingomonas, Agrobacterium, Bradyrhizobium, Variovorax, and Enterobacter spp.). The comparison also exposed the bias of the diversity coverage by the probe set. This bias led to the underestimation of certain taxa such as Aquabacterium spp. (no probe targeting this taxonomic group on the microarray), which appeared as an important constituent of the maize bacterial community based on our cloning-sequencing data and the results described by Schmalenberger and Tebbe (64). On the other hand, however, microarray results revealed the presence of major phylogenetic groups such as Actinobacteria, Bacteroidetes, and Planctomycetes, which were not found by cloning-sequencing. The bulk soil results showed a better agreement between the two methods at the phylum level than the rhizosphere results. However, identification of 16S rRNA clones at lower taxonomic levels was not possible because both the confidence threshold of clone identification was too low and the probe set still was insufficient for several phyla.
Microarray characterization of the maize rhizosphere effect.
Microarray data revealed a clear maize rhizosphere effect on bacterial community composition. This is in agreement with the impact of maize artificial root exudates observed on the bacterial community structure (5). Our microarray results also indicated a plant-to-plant variation, which may be important to take into account when functional implications of the rhizosphere are assessed.
Overall, the maize rhizosphere was characterized by a higher proportion of Agrobacterium (16, 60) and Mesorhizobium spp. and a lower prevalence of Verrucomicrobia, Planctomycetes, Acidobacteria, and Bacteroidetes compared to bulk soil. When the microarray approach was narrowed to a subcommunity, using Agrobacterium primers, the same maize rhizosphere effect on Agrobacterium diversity patterns was observed. Thus, while Agrobacterium DNA was only one component of the total DNA, reliable information on the bacterial subcommunity structure was obtained from a total community analysis. Nevertheless, the accuracy of the Agrobacterium subcommunity analysis was improved by using the Agrobacterium amplicon and thus enabled the detection of A. rubi.
The hybridization signal for Planctomycetes was strong in bulk soil. Since the Planctomycetes probes have been validated by 16S rRNA clones, this substantiates the wide occurrence of this taxon in soil (12, 22, 36), although this group is mainly documented in aquatic systems (11, 18, 30, 76). Furthermore, their differential distribution between rhizosphere and bulk soil is an interesting new result. The higher hybridization level of Planctomycetes in bulk soil compared to the rhizosphere is in agreement with the fact that certain members of this group have been described as being oligotrophs (29), which corresponds to the prevailing conditions in bulk soil. For Verrucomicrobia, which is a widespread but poorly characterized taxonomic group in soil (12, 36), the corresponding probe was not verified, but data pointed to a lower abundance of this group in the rhizosphere, similar to that found by Buckley et al. (13) when cultivated and noncultivated soil were compared.
Acidobacteria and Bacteroidetes also showed a lower contribution to the hybridization patterns of the rhizosphere versus bulk soil. The presence of these two taxa in soil has been established (22, 44) but, like Planctomycetes and Verrucomicrobia, the difficulty in isolating representatives has hampered the assessment of their ecological role (39). The importance of Actinobacteria in bulk soil and/or the rhizosphere has been shown before (60, 69), but most data originate from polluted soils or extreme environments (4, 31, 33).
Conclusion.
In our study, microarray analyzes demonstrated changes in community structure due to the maize rhizosphere effect. These changes included poorly documented taxa. In addition, each plant's rhizosphere may harbor a specific community structure. Probe set validation, detection limit, and the discrimination between related environmental samples confirm the potential of the 16S rRNA microarray for a systematic exploration of bacterial diversity. This potential will be further improved with the extension of the current probe set to a wider taxonomic range.
Supplementary Material
Acknowledgments
This study was supported in part by the ACI Ecologie Quantitative program (“Analyse Spatiale de la Diversité Bactérienne dans le Sol: Développement d'une Puce à ADN pour l'Analyze de la Colonization des Micro-agrégats par une Bactérie Tellurique, le Modèle Agrobacterium” [Ministère de la Recherche]), the Genome program from CNRS (“Réalisation de Réseau d'Oligonucléotides pour le Séquençage d'ADN′” [ROSA]), the Bio-ingéniérie pour la Santé, Génomique, et Innovations Médicales 2002 program (Génotube [02 L 0263; Ministère de la Recherche and Ministère des Finances]), the Thématique Prioritaire program (“Les Communautés Microbiennes: Outils d'Analyze du Potentiel Biologique des Sols” [Région Rhônes-Alpes]), and COST Action 853 Agricultural Biomarkers for Array Technology.
We are grateful to P. Simonet, P. Mavingui and P. Normand (UMR CNRS 5557) for strains and/or useful discussions. We thank L. Bodrossy (Department of Biotechnology, Division of Life and Environmental Sciences, ARC Seibersdorf Research GmbH [Austria]) and J. Bernillon and C. Oger (DTAMB) for help and discussions. The present study made use of the DTAMB/Genopôle Rhône-Alpes gene array platform located at IFR41 in University Lyon 1.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adamczyk, J., M. Hesselsoe, N. Iversen, M. Horn, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barea, J. M., M. J. Pozo, R. Azcon, and C. Azcon-Aguilar. 2005. Microbial co-operation in the rhizosphere. J. Exp. Bot. 56:1761-1778. [DOI] [PubMed] [Google Scholar]
- 4.Basil, A. J., J. L. Strap, H. M. Knotek-Smith, and D. L. Crawford. 2004. Studies on the microbial populations of the rhizosphere of big sagebrush (Artemisia tridentata). J. Ind. Microbiol. Biotechnol. 31:278-288. [DOI] [PubMed] [Google Scholar]
- 5.Baudoin, E., E. Benizri, and A. Guckert. 2003. Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil. Biol. Biochem. 35:1183-1192. [Google Scholar]
- 6.Bensaadi-Merchermek, N., J. C. Salvado, C. Cagnon, S. Karama, and C. Mouches. 1995. Characterization of the unlinked 16S rDNA and 23S-5S rRNA operon of Wolbachia pipientis, a prokaryotic parasite of insect gonads. Gene 165:81-86. [DOI] [PubMed] [Google Scholar]
- 7.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 8.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 9.Bonch-Osmolovskaya, E. A., M. L. Miroshnichenko, A. V. Lebedinsky, N. A. Chernyh, T. N. Nazina, V. S. Ivoilov, S. S. Belyaev, E. S. Boulygina, Y. P. Lysov, A. N. Perov, A. D. Mirzabekov, H. Hippe, E. Stackebrandt, S. l'Haridon, and C. Jeanthon. 2003. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl. Environ. Microbiol. 69:6143-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce, K. D., W. D. Hiorns, J. L. Hobman, A. M. Osborn, P. Strike, and D. A. Ritchie. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummer, I. H., A. D. Felske, and I. Wagner-Dobler. 2004. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl. Environ. Microbiol. 70:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 13.Buckley, D. H., and T. M. Schmidt. 2001. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol. Ecol. 35:105-112. [DOI] [PubMed] [Google Scholar]
- 14.Castiglioni, B., E. Rizzi, A. Frosini, K. Sivonen, P. Rajaniemi, A. Rantala, M. A. Mugnai, S. Ventura, A. Wilmotte, C. Boutte, S. Grubisic, P. Balthasart, C. Consolandi, R. Bordoni, A. Mezzelani, C. Battaglia, and G. De Bellis. 2004. Development of a universal microarray based on the ligation detection reaction and 16S rRNA gene polymorphism to target diversity of Cyanobacteria. Appl. Environ. Microbiol. 70:7161-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelius, M. K., and E. W. Triplett. 2001. The Diversity of Archaea and Bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 16.Cheneby, D., S. Perrez, C. Devroe, S. Hallet, Y. Couton, F. Bizouard, G. Iuretig, J. C. Germon, and L. Philippot. 2004. Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N2O-reducing abilities. Can. J. Microbiol. 50:469-474. [DOI] [PubMed] [Google Scholar]
- 17.Cho, J. C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2003. Molecular evidence for novel Planctomycete diversity in a municipal wastewater treatment plant. Appl. Environ. Microbiol. 69:7354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook, K. L., and G. S. Sayler. 2003. Environmental application of array technology: promise, problems and practicalities. Curr. Opin. Biotechnol. 14:311-318. [DOI] [PubMed] [Google Scholar]
- 20.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 21.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 22.Davis, K. E., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denef, V. J., J. Park, J. L. M. Rodrigues, T. V. Tsoi, S. A. Hashsham, and J. M. Tiedje. 2003. Validation of a more sensitive method for using spotted oligonucleotide DNA microarrays for functional genomics studies on bacterial communities. Environ. Microbiol. 5:933-943. [DOI] [PubMed] [Google Scholar]
- 24.DeSantis, T. Z., C. E. Stone, S. R. Murray, J. P. Moberg, and G. L. Andersen. 2005. Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol. Lett. 245:271-278. [DOI] [PubMed] [Google Scholar]
- 25.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durbin, B. P., J. S. Hardin, D. M. Hawkins, and D. M. Rocke. 2002. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18(Suppl. 1):S105-S110. [DOI] [PubMed] [Google Scholar]
- 27.Filion, M., R. C. Hamelin, L. Bernier, and M. St-Arnaud. 2004. Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl. Environ. Microbiol. 70:3541-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke-Whittle, I. H., S. H. Klammer, and H. Insam. 2005. Design and application of an oligonucleotide microarray for the investigation of compost microbial communities. J. Microbiol. Methods 62:37-56. [DOI] [PubMed] [Google Scholar]
- 29.Fuerst, J. A. 1995. The Planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141(Pt. 7):1493-1506. [DOI] [PubMed] [Google Scholar]
- 30.Gade, D., H. Schlesner, F. O. Glockner, R. Amann, S. Pfeiffer, and M. Thomm. 2004. Identification of Planctomycetes with order-, genus-, and strain-specific 16S rRNA-targeted probes. Microb. Ecol. 47:243-251. [DOI] [PubMed] [Google Scholar]
- 31.Gremion, F., A. Chatzinotas, and H. Harms. 2003. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal contaminated bulk and rhizosphere soil. Environ. Microbiol. 5:896-907. [DOI] [PubMed] [Google Scholar]
- 32.Grundmann, G. L., A. Dechesne, F. Bartoli, J. L. Chassé, J. P. Flandrois, and R. Kizungu. 2001. Simulation of the spatial distribution of micro-habitat of NH4+- and NO2−-oxidizing bacteria in soil. Soil Sci. Soc. Am. J. 65:1709-1716. [Google Scholar]
- 33.Hery, M., A. Herrera, T. M. Vogel, P. Normand, and E. Navarro. 2005. Effect of carbon and nitrogen input on the bacterial community structure of Neocaledonian nickel mine spoils. FEMS Microbiol. Ecol. 51:333-340. [DOI] [PubMed] [Google Scholar]
- 34.Hiraishi, A., M. Iwasaki, and H. Shinjo. 2000. Terminal restriction pattern analysis of 16S rRNA genes for the characterization of bacterial communities of activated sludge. J. Biosci. Bioeng. 90:148-156. [DOI] [PubMed] [Google Scholar]
- 35.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 36.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 38.Ives, A. R., K. Gross, and J. L. Klug. 1999. Stability and variability in competitive communities. Science 286:542-544. [DOI] [PubMed] [Google Scholar]
- 39.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins, B. D., G. F. Steward, S. M. Short, B. B. Ward, and J. P. Zehr. 2004. Fingerprinting diazotroph communities in the Chesapeake Bay by using a DNA macroarray. Appl. Environ. Microbiol. 70:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloepper, J. W., R. M. Zablotowicz, E. M. Tipping, and R. Lifshitz. 1991. Plant growth promotion mediated by bacterial rhizosphere colonizers, p. 315-326. In D. L. Keister and P. B. Cregan (ed.), The rhizosphere and plant growth. Kluwer, Dordrecht, The Netherlands.
- 42.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, M. L., F. C. Kuo, G. A. Whitmore, and J. Sklar. 2000. Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 97:9834-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipski, A., U. Friedrich, and K. Altendorf. 2001. Application of rRNA-targeted oligonucleotide probes in biotechnology. Appl. Microbiol. Biotechnol. 56:40-57. [DOI] [PubMed] [Google Scholar]
- 46.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loy, A., K. Kusel, A. Lehner, H. L. Drake, and M. Wagner. 2004. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal co-occurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loy, A., C. Schulz, S. Lucker, A. Schopfer-Wendels, K. Stoecker, C. Baranyi, A. Lehner, and M. Wagner. 2005. 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order “Rhodocyclales.” Appl. Environ. Microbiol. 71:1373-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu, C. 2004. Improving the scaling normalization for high-density oligonucleotide GeneChip expression microarrays. BMC Bioinformatics 5:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 52.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lubmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahaffee, W. F., and J. W. Kloepper. 1997. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.). Microb. Ecol. 34:210-223. [DOI] [PubMed] [Google Scholar]
- 54.Maloney, P. E., A. H. C. van Bruggen, and S. Hu. 1997. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb. Ecol. 34:109-117. [DOI] [PubMed] [Google Scholar]
- 55.Marschner, P., W. Marino, and R. Lieberei. 2002. Seasonal effects on microorganisms in the rhizosphere of two tropical plants in a polyculture agroforestry system in Central Amazonia, Brazil. Biol. Fertil. Soils 35:68-71. [Google Scholar]
- 56.Mavingui, P., V. Van Tran, E. Labeyrie, E. Rancès, F. Vavre, and P. Simonet. 2005. Efficient procedure for purification of obligate intracellular Wolbachia pipientis and representative amplification of its genome by multiple-displacement amplification. Appl. Environ. Microbiol. 71:6910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCann, K. S. 2000. The diversity-stability debate. Nature 405:228-233. [DOI] [PubMed] [Google Scholar]
- 58.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of Planctomycetes with 16S rRNA-targeted probes. Microbiology 144(Pt. 12):3257-3266. [DOI] [PubMed] [Google Scholar]
- 59.Peplies, J., S. C. Lau, J. Pernthaler, R. Amann, and F. O. Glockner. 2004. Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ. Microbiol. 6:638-645. [DOI] [PubMed] [Google Scholar]
- 60.Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, and J. C. Germon. 2002. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl. Environ. Microbiol. 68:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pilon-Smits, E. 2005. Phytoremediation. Annu. Rev. Plant Biol. 56:15-39. [DOI] [PubMed] [Google Scholar]
- 62.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2003. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 44:35-43. [DOI] [PubMed] [Google Scholar]
- 63.Sanguin, H., A. Herrera, C. Oger-Desfeux, A. Dechesne, P. Simonet, E. Navarro, T. M. Vogel, Y. Moënne-Loccoz, X. Nesme, and G. L. Grundmann. 2006. Development and validation of a prototype 16S rRNA-based taxonomic microarray for Alphaproteobacteria. Environ. Microbiol. 8:289-307. [DOI] [PubMed] [Google Scholar]
- 64.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-262. [DOI] [PubMed] [Google Scholar]
- 65.Sharov, V., K. Y. Kwong, B. Frank, E. Chen, J. Hasseman, R. Gaspard, Y. Yu, I. Yang, and J. Quackenbush. 2004. The limits of log-ratios. BMC Biotechnol. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simonet, P., P. Normand, A. Moiroud, and M. Lalonde. 1985. Restriction enzyme digestion patterns of Frankia plasmids. Plant Soil. 87:49-60. [Google Scholar]
- 67.Singh, B. K., P. Millard, A. S. Whiteley, and J. C. Murrell. 2004. Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends. Microbiol. 12:386-393. [DOI] [PubMed] [Google Scholar]
- 68.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stralis-Pavese, N., A. Sessitsch, A. Weilharter, T. Reichenauer, J. Riesing, J. Csontos, J. C. Murrell, and L. Bodrossy. 2004. Optimization of diagnostic microarray for application in analysing landfill methanotroph communities under different plant covers. Environ. Microbiol. 6:347-363. [DOI] [PubMed] [Google Scholar]
- 72.Taboada, E. N., R. R. Acedillo, C. C. Luebbert, W. A. Findlay, and J. H. Nash. 2005. A new approach for the analysis of bacterial microarray-based comparative genomic hybridization: insights from an empirical study. BMC Genomics 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thioulouse, J., D. Chessel, S. Dolédec, and J. M. Olivier. 1997. ADE-4: a multivariate analysis and graphical display software. Stat. Comput. 7:75-83. [Google Scholar]
- 74.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-675. [DOI] [PubMed] [Google Scholar]
- 75.Vogel, J., P. Normand, J. Thioulouse, X. Nesme, and G. L. Grundmann. 2003. Relationship between spatial and genetic distance in Agrobacterium spp. in 1 cubic centimeter of soil. Appl. Environ. Microbiol. 69:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ward, N., F. A. Rainey, E. Stackebrandt, and H. Schlesner. 1995. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 61:2270-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Webster, N. S., R. I. Webb, M. J. Ridd, R. T. Hill, and A. P. Negri. 2001. The effects of copper on the microbial community of a coral reef sponge. Environ. Microbiol. 3:19-31. [DOI] [PubMed] [Google Scholar]
- 78.Weller, D. M., J. M. Raaijmakers, B. B. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 79.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.