Abstract
The degradation of polycyclic aromatic hydrocarbons (PAHs) by bacteria has been widely studied. While many pure cultures have been isolated and characterized for their ability to grow on PAHs, limited information is available on the diversity of microbes involved in PAH degradation in the environment. We have designed generic PCR primers targeting the gene fragment encoding the Rieske iron sulfur center common to all PAH dioxygenase enzymes. These Rieske primers were employed to track dioxygenase gene population shifts in soil enrichment cultures following exposure to naphthalene, phenanthrene, or pyrene. PAH degradation was monitored by gas chromatograph with flame ionization detection. DNA was extracted from the enrichment cultures following PAH degradation. 16S rRNA and Rieske gene fragments were PCR amplified from DNA extracted from each enrichment culture and an unamended treatment. The PCR products were cloned and sequenced. Molecular monitoring of the enrichment cultures before and after PAH degradation using denaturing gradient gel electrophoresis and 16S rRNA gene libraries suggests that specific phylotypes of bacteria were associated with the degradation of each PAH. Sequencing of the cloned Rieske gene fragments showed that different suites of genes were present in soil microbe populations under each enrichment culture condition. Many of the Rieske gene fragment sequences fell into clades which are distinct from the reference dioxygenase gene sequences used to design the PCR primers. The ability to profile not only the bacterial community but also the dioxygenases which they encode provides a powerful tool for both assessing bioremediation potential in the environment and for the discovery of novel dioxygenase genes.
Polycyclic aromatic hydrocarbons (PAHs) are widespread in nature due to both their natural production in the environment and input from anthropogenic activities, such as the burning of fossil fuels, the use of wood preservatives such as creosote, and the generation of wastes from coal gasification plants. Some PAHs, especially the higher molecular weight compounds, are toxic to living organisms, and this toxicity is enhanced by their intrinsic chemical stability and resistance to many forms of degradation (13, 16). Therefore, there is great interest in developing strategies to remove PAHs from contaminated sites. Many of these remediation strategies employ microorganisms which can degrade PAHs. Bioremediation of contaminated sites relies on either the activity of microorganisms already present at the site or the addition of selected microorganisms with desired catabolic traits in bioaugmentation techniques (9, 48). Molecular ecological approaches, combined with traditional laboratory enrichments, are often utilized to identify bacterial populations that are functionally important in the biodegradation of organic pollutants. For example, a comprehensive phylogenetic study of PAH-degrading bacteria from geographically diverse soils suggests that PAH degradation is associated with distinct genera independent of geographic location (38). Similar research demonstrated a shift in microbial community structure from alpha- and betaproteobacteria to gammaproteobacteria when a microbial community was exposed to a mixture of aromatic hydrocarbons (53). Molecular-based techniques have also been used to follow the establishment of a soil-derived consortium capable of mineralizing benzo[a]pyrene during the biodegradation of a complex hydrocarbon mixture (26). While these studies suggest that diverse microorganisms are present in polluted environments, a greater understanding of the diversity, ecology, and biochemistry of these PAH-degrading microorganisms is necessary in order to effectively remediate contaminated environments.
Aerobic degradation of lower molecular weight PAHs such as naphthalene (NP) by cultured microorganisms has been studied extensively (9). Bacterial degradation of PAHs under aerobic conditions begins with the addition of both molecules of molecular oxygen to the aromatic ring by a dioxygenase system. Aromatic ring dioxygenases are multicomponent enzymes which consist of an electron transport chain containing a ferredoxin and a reductase and a terminal dioxygenase (21). The dioxygenase is composed of two subunits. The alpha subunit is the catalytic component and contains two conserved regions: the [Fe2-S2] Rieske center and the mononuclear iron-containing catalytic domain. The Rieske cluster accepts electrons from the ferredoxin and passes them on to the mononuclear iron for catalysis (19, 42). The majority of information on PAH degradation pathways has come from studies on gram-negative bacteria, particularly the pseudomonads (7, 49). The best studied PAH dioxygenase is naphthalene dioxygenase from Pseudomonas putida NCIB 9816-4 (28, 35, 43), encoded by the NAH plasmid pDTG1 (14). These nah genes have been found in a wide variety of bacteria and geographic locations (1, 20, 36, 54, 55). Other more distantly related PAH degradation genes have also been described. Burkholderia sp. strain RP007, which was isolated from a PAH-contaminated site in New Zealand based on its ability to degrade phenanthrene (PH), contains a suite of PAH catabolic genes, the phn genes, which, while possessing activity similar to that of the nah genes, are only distantly related on the DNA and amino acid level (33). Competitive PCR studies by Laurie and Lloyd-Jones also showed that the Pseudomonas-type nah genes are not always dominant in the environment and that the phn-type genes can have a greater ecological significance than the nah-like genotype (34). Less information is available on PAH degradation by gram-positive bacteria, although recent reports have documented genetic and biochemical analysis of PAH degradation by Rhodococcus, Mycobacterium, Terrabacter, and Nocardioides (2, 8, 29, 41). Indeed, as more PAH-degrading bacteria have been isolated and characterized, it has become apparent that pseudomonads and the nah-like genes represent only a fraction of the PAH degradation picture. However, surveys of PAH degradation potential frequently rely on nah-based primers or probes to assess biodegradation potential in the environment (1, 52, 56).
Recently there have been a number of reports that describe primers or probes that can be used to measure PAH degradation potential in the environment (4, 17, 50, 55). Widada et al. (55) used a combination of isolation of PAH-degrading bacteria and PCR amplification of dioxygenase genes with a suite of degenerate primers to assess the diversity of PAH-degrading bacteria from different geographic locations. Even using a variety of primers, they failed to identify PAH dioxygenases genes from 7 of the 19 PAH-degrading isolates tested. Baldwin et al. (4) described a suite of aromatic oxygenase PCR primer pairs which amplify naphthalene, toluene, and biphenyl dioxygenase genes and their use in real-time PCR assessment of PAH biodegradation. While their study had the advantage of bypassing the culturing and isolation of the PAH degraders, which probably misses many of the important players in PAH biodegradation, they require the use of multiple primer sets to assess bioremediation at a site. Here we describe primers which target the conserved Rieske center of PAH dioxygenases and their use in monitoring PAH dioxygenase population shifts during degradation of naphthalene, phenanthrene, and pyrene (PY) in a series of enrichment experiments. The Rieske primers allow us to target only the dioxygenases which oxidize neutral aromatic hydrocarbons and represent a useful tool for assessing biodegradation potential in contaminated environments without cultivation and isolation of bacteria.
MATERIALS AND METHODS
Soil.
Medium loam soil was collected from a PAH-impacted site in Somerset County, New Jersey. Soil was sieved to a particle size of ≤2 mm and consisted of 37% sand, 43% silt, 20% clay. It contained 2.77% organic carbon and had a pH of 4.95 in water. PAH residue analysis of background soil performed by Accutest Laboratories (Dayton, NJ) showed the following PAH content: 220 mg/kg phenanthrene; 457 mg/kg pyrene; 7 mg/kg acenaphthylene; 8 mg/kg dibenzo[a, h]anthracene; 11 mg/kg acenaphthene; 17 mg/kg benzo[g, h, i]perylene; 20 mg/kg indeno[1,2,3-cd]pyrene; 21 mg/kg fluorene; 31 mg/kg anthracene; 45 mg/kg benzo[a]pyrene; 68 mg/kg benzo[k]fluoranthene; 83 mg/kg benzo[b]fluoranthene; 108 mg/kg benzo[a]anthracene; 166 mg/kg chrysene; and 513 mg/kg fluoranthene.
Enrichment cultures.
Triplicate cultures were initiated in 150-ml serum bottles using 1 g of PAH-impacted soil and 25 ml of minimal medium (51) spiked with 200 mg/liter of NP, PH, or PY. Bottles were sealed using Teflon caps and shaken at 200 rpm for up to 250 h at 30°C. At each time point, samples were removed and separate triplicate cultures were used for cell enumeration and DNA extraction or PAH analysis.
Cell enumeration.
One milliliter of the enrichment culture was removed and fixed overnight at 4°C with 100 μl of 37% formaldehyde. Samples were vortexed to ensure even sample distribution, diluted 1:200 using sterile minimal medium, and placed with ice in an ultrasonic water bath for 5 min. Cells were stained with 5 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI [Sigma Aldrich, St. Louis, MO]) for 1 h in the dark at 4°C. Resulting solutions were filtered through a Millipore 0.2-μm prestained black 22-mm diameter filter, and a total of 9 grid fields per slide were counted using a Zeiss Axiovert 200 M epifluorescent microscope (45).
PAH analysis.
Dichloromethane (25 ml) was added to the enrichment cultures, and PAHs were extracted overnight at 200 rpm. Following extraction, the solvent layers were removed and dried with 4 g of anhydrous sodium sulfate. Extracts (1 ml) were analyzed on a Varian CP-3800 gas chromatograph with flame ionization detection using an RTX-5 column. The gas chromatograph program consisted of 6 min at 40°C followed by a 10°C/min increase to 300°C. The concentration of each PAH was calculated by comparison against individual PAH standard curves.
DNA extraction, PCR, and DGGE analysis.
At each time point, enrichment cultures were transferred to 50-ml Teflon tubes and centrifuged for 10 min at 27,000 × g. Total community DNA was extracted from the soil pellet using the UltraClean Soil DNA kit (MoBio Laboratories, Solana Beach, CA) and further purified using the Geneclean Spin kit (Q-BIOgene, Irvine, CA). For denaturing gradient gel electrophoresis (DGGE) analysis, a 193-bp sequence of the V3 region of the 16S rRNA genes was amplified using the primer set 341FGC and 534R as described by Muyzer et al. (39). PCR products were purified using QIAquick PCR purification columns (QIAGEN, Valencia, CA) and analyzed by denaturing gradient gel electrophoresis using a Dcode Universal Mutation Detection system (Bio-Rad Laboratories, Hercules, CA). Briefly, samples were run on 10% polyacrylamide gels with a denaturant gradient from 40% to 60%. Electrophoresis was carried out for 16 h at 70 V and 60°C. The gels were stained for 1 h with SYBR Green I and imaged using a gel documentation station (Kodak, Rochester, NY). Variability in the DGGE profiles for each treatment was determined by principal component analysis (PCA) based on the number of bands shared between profiles.
ARDRA.
For amplified ribosomal DNA restriction analysis (ARDRA), a 585-bp sequence of the 16S rRNA genes was amplified using the primer set 341F and 907R as described by Muyzer et al. (39). PCR products were gel purified, ligated into the pCR4-TOPO vector, and transformed into One Shot TOP10 chemically competent Escherichia coli following the manufacturer's protocol. One hundred randomly picked colonies per sample were each grown overnight at 37°C in Luria broth containing antibiotics. Plasmid DNA extraction was performed using the Qiaprep Spin Miniprep kit (QIAGEN, Valencia, CA), and the insert was amplified using primers 341F and 907R. Ten microliters of each amplicon was digested for 3 h at 37°C with 2.5 U of HaeIII, 2.5 U of RsaI, and 2.5 U of HinfI in 15-μl reactions. Restriction patterns were separated on 2% Metaphor agarose gels and imaged with SYBR Green I DNA stain. Each different restriction pattern was defined as an operational taxonomical unit (OTU). Amplicons from unique OTUs were sequenced, edited, and aligned using Lasergene sequence analysis software (DNAstar, Madison, WI). The distribution of OTUs in each treatment was determined and used to calculate the Shannon-Weaver index of diversity {H = −Σ[ni · log(ni)], where ni represents the relative contribution of each OTU to the entire library}. Amplicons from unique OTUs were sequenced, confirmed, and hand aligned using Lasergene sequence analysis software (DNAStar, Madison, WI). Corrected sequences were screened against those in the GenBank database using Blastn.
Rieske primer design.
Amino acid sequences of the large subunit of dioxygenases targeting aromatic hydrocarbons were aligned using the ClustalW function in MegAlign (DNAStar, Madison, WI). The ProSite motif database describes the bacterial ring hydroxylating dioxygenase alpha-subunit signature as C-x-H-R-[GAR]-x (7, 8)-[GEKVI]-[NERAQ]-x (4, 5)-C-x-[FY]-H (40). However, close inspection of the aligned aromatic hydrocarbon dioxygenase amino acid sequences revealed differences in the Rieske sequences of the dioxygenases which attack nonpolar aromatic compounds, such as PAHs, polychlorinated biphenyls, benzene, toluene, and xylene, and those which attack polar aromatic compounds, such as benzoate, toluate, and phthalate. Primers were then designed based on this Rieske motif (Rieske_f, CRHRG; Rieske_r, CSYHGW) which allowed us to distinguish between dioxygenases targeting polar and nonpolar aromatic hydrocarbons. The sequences of the primers are the following: Rieske_f, TGYMGNCAYMGNGG; Rieske_r, CCANCCRTGRTANSWRCA. A second set of primers was designed substituting inosine residues for “N” residues to reduce the degeneracy of the primers. The Rieske primers are similar to those described by Cigolini et al. (12) and Kasuga et al. (27) but incorporate more degeneracies in order to amplify a wider diversity of PAH dioxygenase genes. The primers amplify a 78-bp PCR product. PCRs were prepared in either 25 or 50 μl containing 2 ng μl−1 DNA, 1× PCR buffer, 2.5 mM MgCl2, 1 μM each forward and reverse primers, and 1 U Taq polymerase (Sigma, St. Louis, MO) and amplified with the following program: an initial denaturation step of 94°C for 5 min; 35 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 30 s; and a final extension of 72°C for 5 min. The primers were tested on a series of bacterial strains which grow on benzene, toluene, naphthalene, phenanthrene, and biphenyl with successful results.
Amplification of dioxygenase genes from enrichment cultures.
DNA isolated from various time points of the PAH enrichments was screened for PAH dioxygenase genes using the Rieske primers. PAH dioxygenase genes were amplified as described above and cloned into pCR2.1 TOPO (Invitrogen, Carlsbad, CA). Colonies were picked, and plasmid DNA was isolated using the Concert 96 kit (Invitrogen). Rieske clones were sequenced with an ABI Prism 3100 Genetic Analyzer using Big Dye v3.1 chemistry (Applied Biosystems, Foster City, CA).
Phylogenetic analysis.
The nucleotide sequences were assembled, and the primer sequence was removed using SeqMan (DNAStar, Madison, WI). Rieske sequences which were 95% identical were assigned to the same clone family and aligned with reference sequences from GenBank using the ClustalW function in MegAlign (DNAStar). The phylogenetic tree was constructed by neighbor-joining analysis using Molecular Evolutionary Genetics Analysis software (32) with 1,000 bootstrap replicates.
Rieske clone library diversity analysis.
Accumulation curves were plotted for each library. The data were rarefied using the internet-based Rarefaction Calculator of Krebs and Brzustowski (31). The nonparametric richness estimates Chao1 and ACE (10, 11) and Shannon-Weaver diversity indices (H′) (44) were calculated for each clone library.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study were deposited in the GenBank database with the following accession numbers: DQ270422 to DQ270455 (16S rRNA clones) and DQ325359 to DQ325435 (Rieske clones).
RESULTS
Bacterial growth and PAH biodegradation.
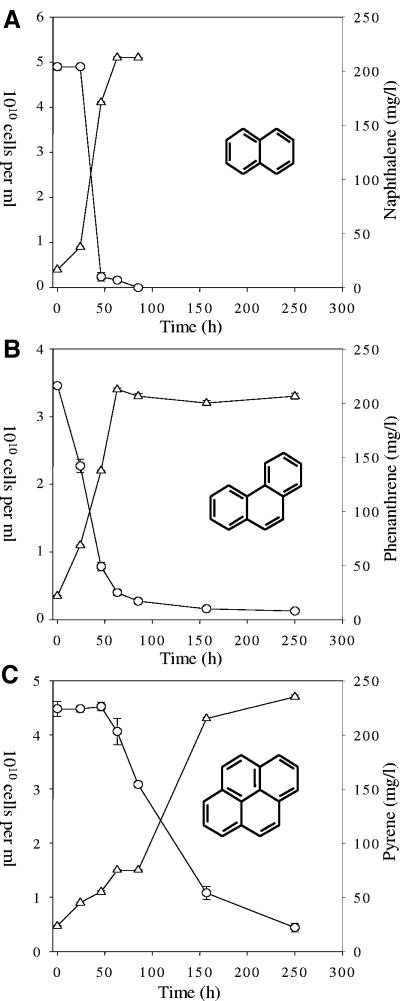
Enrichment cultures were monitored over time to examine the growth and biodegradation potential of soil bacterial communities enriched in the presence of different PAHs. Direct counting of DAPI-stained cells showed that the initial bacterial density in all enrichment cultures was approximately 4 × 109 cells/ml (Fig. 1). NP degradation by the enrichment cultures occurred after a 24-h lag and showed greater than 95% reduction of NP by 46 h (Fig. 1A). At the onset of NP degradation, the enrichment cultures demonstrated exponential-phase growth and reached a maximum cell density of approximately 5 × 1010 cells/ml by 63 h (Fig. 1A). At 80 h, the soil microbial community was in stationary-phase growth and NP concentration was below the level of detection and was not examined further. On the other hand, the PH enrichment cultures showed no lag time before the onset of growth and PH degradation and an overall slower degradation rate, with greater than 90% degradation occurring by 85 h (Fig. 1B). Initiation of PY degradation by the enrichment cultures occurred after a 46-h lag time with approximately 90% PY degradation occurring by 250 h (Fig. 1C). No lag time was observed for the growth of the PY enrichment cultures with cultures reaching stationary-phase growth by 250 h (Fig. 1C).
FIG. 1.
Microbial growth (Δ) and biodegradation of PAHs (O) over time. (A) Enrichment cultures with NP; (B) enrichment cultures with PH; (C) enrichment cultures with PY. Monitoring of NP cultures ended at 84 h due to rapid degradation of the compound in comparison to PH and PY. Data represent the averages and standard errors of triplicate data.
Bacterial community structure.
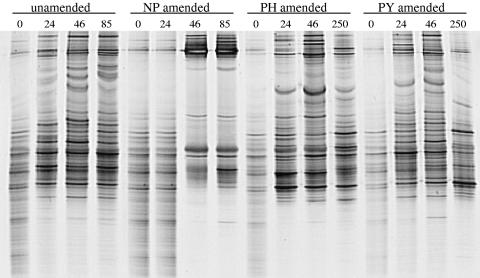
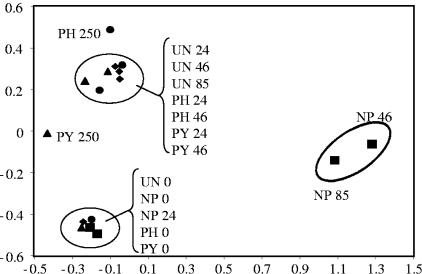
The effect of individual PAHs on the bacterial community structure was determined throughout the time course using 16S rRNA gene-based PCR-DGGE for each treatment. As expected, all treatments showed identical banding patterns at time zero, indicating that each enrichment culture was initiated with the same bacterial community (Fig. 2). A decrease in the number of observed bands in the unamended cultures throughout the time course suggests an inherent decrease in species richness due to laboratory conditions and utilization of the background PAH contaminants in the soil (Fig. 2). However, DGGE analysis of cultures enriched on individual PAHs resulted in unique banding profiles, suggesting the selection of PAH-specific bacterial communities (Fig. 2). In order to determine relationships between the bacterial community fingerprints shown in Fig. 2, principal component analysis (PCA) was performed based on the presence or absence of each DGGE band under the different conditions. PCA shows that time-zero samples from all treatments as well as the 24-h sample from the NP treatment form a distinct cluster, suggesting similar bacterial communities (Fig. 3). However, after the onset of PAH degradation, each treatment formed a distinct cluster, suggesting the formation of PAH-specific bacterial communities (Fig. 3). Also, PCA suggests that the PH- and PY-degrading communities change in a fashion similar to that of the unamended cultures, while the NP-degrading community is unique, forming a cluster separated from the other treatments.
FIG. 2.
Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene fragments from PAH-amended enrichment cultures over time. The time of sampling (hours) is listed above the lanes. DGGE lanes are representative of triplicate data.
FIG. 3.
Ordination plots, by treatments and time, of bacterial communities generated by principle component analysis of bacterial species occurrence from 16S rRNA gene DGGE profiles. ⧫, unamended (UN); ▪, naphthalene amended (NP); •, phenanthrene amended (PH); ▴, pyrene amended (PY).
To further examine the effect of individual PAHs on the bacterial community composition, phylogenetic analysis of cloned 16S rRNA genes was performed. To ensure comparison of active PAH-degrading communities, clone libraries were constructed from DNA extracted from enrichment cultures demonstrating approximately 90% degradation of respective PAHs. For further comparison, a clone library was also constructed for the unamended enrichment cultures at 85 h. One hundred clones from each clone library were screened by ARDRA, and the unique OTUs were sequenced. Phylogenetic analysis of representative OTUs from 16S rRNA gene libraries show that enrichment culture clones are distributed throughout the alpha-, beta-, and gamma-proteobacteria, the acidobacteria, and the actinobacteria. The relative contribution of each OTU type to the overall bacterial diversity among the various treatments was examined (Table 1). While the clone library constructed from unamended soil enrichments showed a slight predominance of OTUs 1 and 4, it contained the greatest number of OTUs (28) of the enrichments and had the highest H′ value (1.25), suggesting a high level of bacterial diversity. In contrast, cultures enriched on NP showed greatly reduced diversity (H′ = 0.09), with only five OTUs and a strong selection for a Pseudomonas fluorescens-like phylotype (OTU 29). While PH-amended cultures demonstrated the greatest diversity (H′ = 0.90) among the PAH treatments with 12 OTUs and selection towards OTUs 1, 2, 22, 31, and 32, an overall decrease in diversity was observed compared to that of the unamended cultures. Similarly, cultures enriched on PY also demonstrated decreased diversity (H′ = 0.61) compared to unamended and PH-amended cultures, with 10 OTUs and a strong selection for a Stenotrophomonas maltophilia-like phylotype (OTU 31). Furthermore, the composition of enrichment culture diversity with respect to bacterial taxa showed similar distribution of the alphaproteobacteria (27%), the betaproteobacteria (34%), and the acidobacteria (22%) in the unamended cultures (Table 2). With the exception of the NP-amended cultures, the distribution of alpha- and betaproteobacteria in PAH-amended cultures remained most similar to the unamended cultures. However, in all PAH-amended cultures, an increase in gammaproteobacteria and a decrease in actinobacteria and acidobacteria was observed.
TABLE 1.
GenBank database sequences with the highest identity match to dominant OTUs and distribution of OTUs in PAH enrichment cultures
| OTU | BLAST match | Accession no. | Identity (%) | No. of clones
|
|||
|---|---|---|---|---|---|---|---|
| UNa | NP | PH | PY | ||||
| 1 | Burkholderia sp. strain Dint1 | AM062710 | 100 | 20 | 1 | 19 | 9 |
| 2 | Sphingomonas elodea | AF503278 | 98 | 9 | 18 | 10 | |
| 4 | Acidobacterium Ellin7137 | AY673303 | 95 | 12 | 2 | ||
| 5 | Uncultured alphaproteobacterium clone EB1127 | AY395446 | 98 | 5 | |||
| 7 | Burkholderia sp. | AB191222 | 99 | 7 | |||
| 12 | Acidobacterium group bacterium clone FTL227 | AF529104 | 96 | 6 | |||
| 22 | Gammaproteobacterium SA29-B | AB174845 | 99 | 3 | 13 | 3 | |
| 29 | Pseudomonas fluorescens A1XB1-4 | AY512614 | 99 | 96 | 4 | ||
| 31 | Stenotrophomonas maltophilia ZJUB-041 | DQ223428 | 99 | 1 | 22 | 61 | |
| 32 | Massilia sp. | AY177372 | 98 | 11 | |||
| 33 | Sphingomonas agrestis | AY506539 | 99 | 1 | 6 | ||
UN, unamended.
TABLE 2.
Number of clones from unamended and PAH-amended enrichment culture 16S rRNA gene libraries representing different bacterial taxaa
| Enrichment culture | Proteobacteria
|
Actinobacteria | Acidobacteria | ||
|---|---|---|---|---|---|
| Alpha | Beta | Gamma | |||
| Unamended | 27 | 34 | 6 | 11 | 22 |
| NP amended | 1 | 1 | 97 | 0 | 1 |
| PH amended | 24 | 34 | 40 | 0 | 2 |
| PY amended | 24 | 10 | 64 | 0 | 2 |
The total number of clones sampled for each library was 100.
Rieske primer design and verification.
The Rieske primers were tested against a variety of bacteria known to degrade PAHs. The primers successfully amplified 78-bp DNA fragments from Burkholderia xenovorans LB400 (biphenyl dioxygenase), Comamonas testosteroni GZ39 (PAH dioxygenase), Pseudomonas putida F1 (toluene dioxygenase), and Pseudomonas putida NCIB 9816-4 (naphthalene dioxygenase). In each case, a single 78-bp PCR product was obtained whose sequence was identical to the published dioxygenase sequences from these strains. These strains also contain dioxygenase genes encoding enzymes which oxidize polar compounds, but in all cases our primers amplified only the gene encoding the neutral dioxygenase (data not shown). In organisms containing more than one PAH dioxygenase (Mycobacterium sp. strains PYO1 and PYR1), multiple dioxygenase sequences were obtained after cloning the PCR product and sequencing 20 random clones (data not shown).
Amplification of Rieske gene fragments from environmental samples.
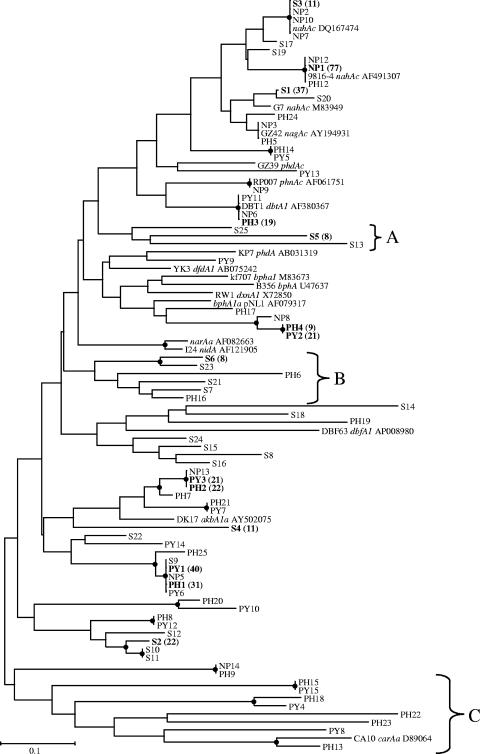
The Rieske primers were utilized to assay the PAH dioxygenase gene complement from creosote-contaminated soil. Using total community DNA as the template, the Rieske primers amplified a single 78-bp PCR product (data not shown). The unamended soil library contained 25 Rieske clone families and was dominated by one family, S1, which represented 30% of the clones sequenced (Fig. 4). The closest match for this family is naphthalene dioxygenase from Comamonas testosteroni GZ42 (Blastn and BlastP identity of 100%) (24). Three other clone families, S2, S3, and S4, represented 36% of the clone library. Clone family S2 is most closely related to DxnA1, a dioxin dioxygenase from Sphingomonas sp. strain RW1 (3) (Table 3). Clone family S3 is most closely related to naphthalene dioxygenase from Polaromonas naphthalenivorans and P. putida G7 (Table 3) (49). Clone family S4, however, which represents 9% of the soil Rieske library, does not exhibit significant similarity to any dioxygenase sequences currently in the GenBank database. Indeed, 18% of the soil clone library sequences do not exhibit significant similarity to any dioxygenase sequences currently in GenBank, and a further 13% have only low matches (<50% identity) to uncharacterized dioxygenase sequences from GenBank. Some of these Rieske sequences form clusters with clone sequences from the enrichment libraries, while others are unique to the soil library (Fig. 4). Analysis of the relationship of the Rieske sequences to one another shows that clone families S5, S13, and S25 form a unique group (labeled A) and comprise 8% of the soil library. A second cluster of sequences with no known relatives in GenBank (labeled B) includes S6, S7, S21, and S23 in addition to PH6 and PH16. The remaining soil library sequences are distributed among 14 clone families and represent a wide diversity of Rieske sequences (Fig. 4).
FIG. 4.
Phylogenetic distribution of the Rieske gene fragment families from the soil (S) and enrichment libraries. The dendrogram was constructed from a ClustalW alignment of the Rieske sequences by neighbor-joining analysis using Mega 3.0. Nodes supported by bootstrap values greater than 50 are indicated with a filled black circle. Major clone families are in boldface and have the number of clones observed indicated in parentheses. Reference sequences from GenBank include the accession number. The scale bar represents substitutions per nucleotide.
TABLE 3.
Best matches in the GenBank database for the dominant Rieske clone families in the naphthalene, phenanthrene, and pyrene enrichments and the unamended creosote-contaminated soil libraries
| Clone family | BlastN result
|
BlastP result
|
||||
|---|---|---|---|---|---|---|
| Accession no. | % Identity | Name | Accession no. | % Identity | Name | |
| S1 | AY194931 | 98 | nagAc | AAP30901 | 100 | NagAc |
| S2 | X72850 | 65 | dxnA1 | CAA51365 | 60 | DxnA1 |
| S3 | DQ167474 | 100 | nahAc | AAA25902 | 100 | G7 NahAc |
| NP1 | AF491307 | 100 | NCIB9816-4 nahAc | AAO64274 | 100 | NCIB9816-4 NahAc |
| NP2 | DQ167474 | 100 | nahAc | AAA25902 | 100 | G7 NahAc |
| NP3 | AY194931 | 100 | nagAc | AAP30901 | 100 | NagAc |
| PH1 | AY502075 | 63 | akbA1a | AAR90131 | 66 | DK17 AkbA1a |
| PH2 | AY502075 | 74 | akbA1a | AAR90131 | 86 | DK17 AkbA1a |
| PH3 | AF380367 | 100 | dbtAc | AAK62353 | 100 | DbtAc |
| PH4 | AF079317 | 74 | bphA1a | AAD03980 | 80 | BphA1a |
| PH5 | AY194931 | 100 | nagAc | AAP30901 | 100 | NagAc |
| PY1 | AY502075 | 63 | akbA1a | AAR90131 | 66 | DK17 AkbA1a |
| PY2 | AF079317 | 74 | bphA1a | AAD03980 | 73 | BphA1a |
| PY3 | AY502075 | 74 | akbA1a | AAR90131 | 80 | DK17 AkbA1a |
The Rieske gene fragment library from the NP enrichment was much less diverse than the library constructed from the contaminated soil. Only 12 clone families were identified in the NP enrichment library (Table 4). One family (NP1) contained 78% of the clones sequenced (Fig. 4). NP1 is 100% identical to the naphthalene dioxygenase of Pseudomonas putida NCIB9816-4 at both the nucleotide and amino acid levels (Table 3) (14). The next most dominant clone families, NP2 and NP3, each account for 4% of the library and are most closely related to naphthalene dioxygenases from P. putida G7 (100%) and Comamonas testosteroni GZ42 (100%), respectively (Table 3). It is interesting that these families account for such a small fraction of the NP enrichment, when sequences similar to nagAc from C. testosteroni GZ42 accounted for 30% of the clones in the soil used to establish the NP enrichment culture and sequences similar to P. putida G7 nahAc accounted for 9% of the soil library. 16S-DGGE and 16S rRNA clone library profiling revealed that the NP enrichment culture is dominated by one phylotype which is most closely related to Pseudomonas fluorescens (Table 1). An NP-degrading isolate was obtained from the enrichment. The 16S rRNA and Rieske gene fragment from this isolate were sequenced and are identical to those obtained from the enrichment culture (data not shown). The P. fluorescens pure culture thus links the observed 16S and Rieske gene fragment data.
TABLE 4.
Diversity analysis of the Rieske gene fragment librariesa
| Library | N | n | H′ | Chao1 |
|---|---|---|---|---|
| Soil | 122 | 25 | 2.41 | 62 |
| Naphthalene | 99 | 12 | 1.14 | 21 |
| Phenanthrene | 113 | 23 | 2.30 | 37 |
| Pyrene | 106 | 15 | 1.83 | 47 |
N, the number of clones sequenced; n, the number of clone families; H′, the Shannon-Weaver diversity index; and Chao1, the Chao1 nonparametric richness estimate.
In contrast to the Rieske sequences seen in the NP enrichment, the Rieske sequences seen in the PH enrichment library are more diverse. The PH enrichment culture library yielded 23 clone families from 113 clones sequenced (Table 4). The PH enrichment culture clone library contained three dominant families, PH1, PH2, and PH3, which represented 27, 19, and 17% of the clones sequenced, respectively. BLAST analysis revealed that PH1 and PH2 are most closely related to alkylbenzene dioxygenase from Rhodococcus sp. strain DK17 (30) (Table 3). Clone families PH1 and PH2 share 67% nucleotide identity. Clone family PH3 is most closely related to dibenzothiophene dioxygenase from Burkholderia sp. strain DBT1 (100% identity) (15). A fourth family, PH4, accounts for 8% of the library and is most closely related to an uncharacterized aromatic dioxygenase from Sphingomonas aromaticovorans (46) (Table 3). The PH library also contained a number of Rieske families for which no significant similarities were found in GenBank. These include PH18, which clusters with PY4 and PH15, and PH22 and PH23, which cluster together with PY8 and PY15 to form clade C, which groups with carbazole dioxygenase from Pseudomonas sp. strain CA10 (47) (Fig. 4).
The PY enrichment culture clone library contained 15 clone families. The library was dominated by clone family PY1, accounting for 38% of the clones sequenced (Fig. 4). Clone family PY1 is most closely related to an alkylbenzene dioxygenase from Rhodococcus sp. strain DK17 (Table 3) (30). A second clone family, PY3, which accounts for 22% of the enrichment library, is also most closely related to alkylbenzene dioxygenase from Rhodococcus sp. strain DK17. Clone family PY2 is most closely related to an uncharacterized aromatic dioxygenase from Sphingomonas aromaticovorans (Table 3). The remaining 12 PY enrichment Rieske families occurred rarely (Fig. 4). There was significant overlap between the PH and PY libraries, with the dominant families in both libraries being most closely related to alkylbenzene dioxygenase from Rhodococcus sp. strain DK17. Indeed, clone families PY1 and PH1 have identical nucleotide sequences, as do PY2 and PH4 as well as PY3 and PH2.
Diversity and species richness of the soil and enrichment libraries.
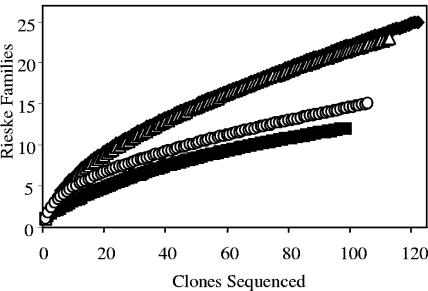
The Shannon-Weaver diversity index and the Chao1 nonparametric species richness estimators were calculated for each Rieske library. The Shannon-Weaver index indicates that the soil enrichment library was the most diverse, followed, in order, by the PH, PY, and NP libraries (Table 4). This order is in agreement with the 16S-based bacterial community diversity reported above. The Chao1 estimate of species richness also indicates a difference in the estimated species richness of the soil and enrichment libraries. Enrichment on the different model PAHs led to selection of different dioxygenase gene populations. Species accumulation curves were plotted for each library using the internet-based rarefaction calculator (31) in order to assess how well our sequencing effort had sampled our libraries. Even though more than 100 clones were sequenced from each library, none of the accumulation curves reached an asymptote (Fig. 5). This is an indication that further sequencing would likely yield new Rieske gene fragment families. However, even our limited sampling effort indicates clear differences in the richness of the four libraries, with the NP and PY libraries being significantly less diverse than the soil and PH libraries. Furthermore, our sampling clearly identified dominant Rieske gene fragment families in each enrichment culture.
FIG. 5.
Rieske clone family accumulation curves showing the number of clones sequenced versus the number of Rieske families observed. Soil (⧫), naphthalene (▪), phenanthrene (▵), and pyrene (○) samples are shown.
DISCUSSION
The ability of bacteria to utilize individual PAHs as carbon and energy sources has been extensively documented over the last few decades (1, 9, 38). While these studies have been essential for establishing metabolic pathways for the biodegradation of individual PAHs, there is increasing interest in understanding how organic pollutants affect the overall structure of microbial communities. In this study, we assessed the influence of individual PAHs on bacterial community structure by monitoring PAH biodegradation and changes in diversity of both PAH dioxygenase genes and bacterial species over time for cultures enriched from a single PAH-impacted soil. Previous studies assessing the genetic potential for PAH degradation in the environment have been limited by the narrow scope of the PCR primers used to screen for PAH dioxygenases which detect only a fraction of the PAH degraders in the environment (37, 55). PCR primers based on the nahAc gene frequently detect only dioxygenases from pseudomonads and fail to detect dioxygenases from other genera known to degrade PAHs, such as Rhodococcus, Terrabacter, and Nocardioides (1, 17, 34). One approach to this problem has been to design PCR primers which target specific subsets of PAH dioxygenase genes. Baldwin et al. (4) employed this approach in the design of a set of real-time PCR primers which target naphthalene, toluene, and biphenyl dioxygenase as well as toluene and phenol monooxygenase genes. Stach and Burns (50) took a similar approach in utilizing three different primer sets in assessing the diversity of dioxygenase genes in enrichment versus biofilm cultures. While these different primer sets are useful in assessing the PAH biodegradation potential of specific gene families in an environment, they are less useful in assessing the overall diversity of PAH degradation genes in a sample, as they are rather narrowly targeted and may fail to amplify previously uncharacterized dioxygenase genes. We have designed PCR primers that allow us to detect those dioxygenases oxidizing PAHs and other neutral aromatic hydrocarbons. The Rieske primers have the advantage of amplifying a wide variety of dioxygenase genes, ranging from naphthalene and biphenyl dioxygenases to pyrene and dioxin dioxygenases, and provide sequence information that can be used at a later date to interrogate metagenomic libraries and identify full-length novel dioxygenase genes.
In this study, we isolated DNA from a PAH-contaminated site and used the Rieske PCR primers to screen for the presence of PAH dioxygenase genes. The site was contaminated with a variety of high-molecular-weight PAHs. Enrichment of the bacterial population in the presence of model two, three, and four ring PAHs led to changes in both the microbial community structure and the dioxygenase gene profile. Our Rieske gene fragment survey revealed the presence of a wide diversity of dioxygenase genes, ranging from the well characterized naphthalene dioxygenase family to less well studied genes such as those of the dioxin dioxygenase family. A significant number of the clones sequenced had no relatives in the GenBank database.
While the unamended soil library was quite diverse, enrichment on NP led to the domination of the Rieske library by one nahAc-like Rieske sequence. It is interesting that the dominant nahAc clone from the soil library is not the dominant nucleotide sequence observed in the NP enrichment library. Sequences identical to P. putida G7 nahAc and C. testosteroni nagAc accounted for 41% of the soil library but were barely detected in the NP enrichment library. Instead, the NP enrichment library was dominated by a Rieske fragment most closely related to P. putida NCIB 9816-4 nahAc. The NP-enriched cultures exhibited a 24-h lag time before the onset of NP degradation followed by near-complete degradation by 46 h. Analysis of the initial PAH levels in the contaminated soil used in our study did not show the presence of NP, so the lag time was likely due to the absence of chronic exposure to NP, thus requiring time for an increase in the number or activity of NP-degrading populations. This is supported by molecular analysis of the NP-amended cultures showing that the microbial community remained similar to unamended cultures until after 24 h, at which point there was a shift in community structure corresponding to increased NP degradation and the development of an almost monoculture-like microbial community consisting predominantly of a single Pseudomonas fluorescens-like phylotype. It is interesting that the NP enrichment was dominated so quickly by one organism and one dioxygenase gene. Stach and Burns (50) reported a similar decrease in diversity in batch cultures compared to biofilm cultures in a study that enriched with NP and PH.
A frequent criticism of enrichment cultures is that they select only for the fastest growing organisms under the conditions utilized and do not reflect what occurs in nature (18, 23). While enrichment on NP led to a profound decrease in diversity, the PH enrichment library was more complex and more closely resembled the unamended soil library, possibly because the original soil contained relatively high PH concentrations. The prior adaptation of the microbial community to the presence of PH is supported by molecular analysis of the community structure. DGGE fingerprints for PH-amended cultures did show a slight reduction in band number; however, the overall community fingerprint changed in a way similar to that of the unamended cultures. This is not surprising, given that the three ring and higher PAHs in the original soil materials most likely caused a shift in the unamended samples. Clone libraries and phylogenetic analysis show similar enrichment for OTU 1 and OTU 2 (phylotypes closely affiliated with Burkholderia and Sphingomonas) in the PH-amended and unamended cultures. Moreover, while small numbers of OTUs 22, 31, and 32 (phylotypes affiliated with Nitrosococcus, Stenotrophomonas, and Massilia) were observed in the unamended cultures, they were enriched to high numbers in PH-amended cultures. Members of these genera have been demonstrated to degrade a range of PAHs, suggesting they may play a role in PH degradation in our cultures (5, 6, 22, 55, 57).
nahAc-like genes were rare in the PH enrichment. In contrast to the NP enrichment library, which was dominated by one Rieske sequence, the PH enrichment library contained a number of equally dominant Rieske sequences and remained quite diverse. This contrasts with the findings of Stach and Burns, who reported a decrease in dioxygenase gene diversity in both PH and NP enrichment and biofilm communities (50). The Rieske data also contrast with the microbial community profiling results in that the unamended and PH enrichments share some dominant bacterial phylotypes but have no dominant Rieske families in common. It is possible that the bacterial community analysis detects not only bacteria growing on the added PH but also on by-products from PH degradation. The use of the Rieske primers has the advantage of allowing us to target bacteria possessing the ability to catalyze the first step in PH biodegradation.
Molecular analysis of the adaptation of the microbial community to growth on PY shows that after PY degradation, the community was enriched in three different phylotypes. As in the unamended and PH-amended cultures, OTUs 1 and 2 (Burkholderia and Sphingomonas) were observed in PY-enriched cultures. However, we observed a strong selection for OTU 31 (Stenotrophomonas), resulting in a low Shannon-Weaver diversity index. Previous studies have documented the presence of members of the genus Stenotrophomonas in PAH-contaminated soils, and individual bacteria isolated from these sites have been shown to mineralize PY (6, 25, 58). The PH and PY enrichment libraries had a number of Rieske clone families in common, suggesting that these enzymes may be involved in both PH and PY degradation. Both the PH and PY libraries contained alkylbenzene dioxygenase-like sequences and an uncharacterized aromatic dioxygenase-like sequence as two of their most dominant clone families. Both libraries also contained a number of sequences that have no close relatives in the GenBank database, indicating that there remains a wealth of untapped dioxygenase sequence diversity and potentially functional diversity in contaminated environments.
The goal of our current studies is to design a better method of predicting the overall biodegradative and bioremediative potential of contaminated sites. Our approach used traditional laboratory biodegradation studies combined with molecular ecological techniques to characterize shifts in both microbial community and functional gene profiles during the biodegradation of a range of model PAHs. Overall, our study suggests that contaminated environments can harbor a wide diversity of dioxygenase genes, allowing for succession of different dioxygenase gene populations in response to exposure to different PAHs. The combination of the Rieske primers and bacterial community profiling represents a powerful tool for both assessing bioremediation potential in the environment and for the discovery of novel dioxygenase genes.
Acknowledgments
This work was supported by NIH grant ES012824 to G.J.Z. and J.J.K.
We thank Laurie Seliger for excellent DNA sequencing assistance.
REFERENCES
- 1.Ahn, Y., J. Sanseverino, and G. S. Sayler. 1999. Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegradation 10:149-157. [DOI] [PubMed] [Google Scholar]
- 2.Allen, C. C. R., D. R. Boyd, M. J. Larkin, K. A. Reid, N. D. Sharma, and K. Wilson. 1997. Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038. Appl. Environ. Microbiol. 63:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodour, A. A., J. M. Wang, M. L. Brusseau, and R. M. Maier. 2003. Temporal change in culturable phenanthrene degraders in response to long-term exposure to phenanthrene in a soil column system. Environ. Microbiol. 5:888-895. [DOI] [PubMed] [Google Scholar]
- 6.Boonchan, S., M. L. Britz, and G. A. Stanley. 1998. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol. Bioeng. 59:482-494. [DOI] [PubMed] [Google Scholar]
- 7.Bosch, R., E. Garcia-Valdes, and E. R. Moore. 1999. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149-157. [DOI] [PubMed] [Google Scholar]
- 8.Brezna, B., A. A. Khan, and C. E. Cerniglia. 2003. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol. Lett. 223:177-183. [DOI] [PubMed] [Google Scholar]
- 9.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 10.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 11.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 12.Cigolini, J. F., A. K. Goyal, and G. J. Zylstra. 1997. Universal PCR primers for detection, identification, and cloning of genes for dioxygenase enzymes involved in ring oxidation of aromatic compounds. Abstr. 97th Gen. Meet. Am. Soc. Microbiol. 1997, abstr. Q-49, p. 463.
- 13.Collins, J. F., J. B. Brown, G. V. Alexeeff, and A. G. Salmon. 1998. Potency equivalency factors for some polycyclic aromatic hydrocarbons and polycyclic aromatic hydrocarbon derivatives. Regul. Toxicol. Pharmacol. 28:45-54. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, J. J., and G. J. Zylstra. 2004. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341:753-768. [DOI] [PubMed] [Google Scholar]
- 15.Di Gregorio, S., C. Zocca, S. Sidler, A. Toffanin, D. Lizzari, and G. Vallini. 2004. Identification of two new sets of genes for dibenzothiophene transformation in Burkholderia sp. DBT1. Biodegradation 15:111-123. [DOI] [PubMed] [Google Scholar]
- 16.Dibble, A. S., C. Cheng, and C. A. H. Biggar. 1990. Polycyclic aromatic hydrocarbon carcinogens, p. 109-127. In M. W. Pariza, H. U. Aeschbacher, J. S. Fenton, and S. Sato (ed.), Mutagens and carcinogens in the diet. Wiley-Liss, New York, N.Y.
- 17.Dionisi, H., C. Chewning, K. Morgan, F. Menn, J. Easter, and G. Sayler. 2004. Abundance of dioxygenase genes similar to Ralstonia sp. strain U2 nagAc is correlated with naphthalene concentrations in coal tar-contaminated freshwater sediments. Appl. Environ. Microbiol. 70:3988-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraro, D. J., L. Gakhar, and S. Ramaswamy. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175-190. [DOI] [PubMed] [Google Scholar]
- 20.Ferrero, M., E. Llobet-Brossa, J. Lalucat, E. Garcia-Valdes, R. Rossello-Mora, and R. Bosch. 2002. Coexistence of two distinct copies of naphthalene degradation genes in Pseudomonas strains isolated from the western Mediterranean region. Appl. Environ. Microbiol. 68:957-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 22.Greene, E., J. Kay, K. Jaber, L. Stehmeier, and G. Voordouw. 2000. Composition of soil microbial communities enriched on a mixture of aromatic hydrocarbons. Appl. Environ. Microbiol. 66:5282-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder, W., and L. Dijkhuizen. 1982. Strategies of mixed substrate utilization in microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297:459-480. [DOI] [PubMed] [Google Scholar]
- 24.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juhasz, A. L., and R. Naidu. 2000. Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a]pyrene. Int. Biodeterior. Biodegrad. 45:57-88. [Google Scholar]
- 26.Kanaly, R. A., R. Bartha, K. Watanabe, and S. Harayama. 2000. Rapid mineralization of benzo[a]pyrene by a microbial consortium growing on diesel fuel. Appl. Environ. Microbiol. 66:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 28.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Struct. Fold. Des. 6:571-586. [DOI] [PubMed] [Google Scholar]
- 29.Khan, A. A., R. F. Wang, W. W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, D. Y., J. C. Chae, G. J. Zylstra, Y. S. Kim, S. K. Kim, M. H. Nam, Y. M. Kim, and E. B. Kim. 2004. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 70:7086-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krebs, C. J., and J. Brzustowski. The rarefaction calculator. [Online.] http://www2.biology.ualberta.ca/jbrzusto/rarefact.php.
- 32.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 33.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, K. U., B. Kauppi, R. E. Parales, D. T. Gibson, and S. Ramaswamy. 1997. Purification and crystallization of the oxygenase component of naphthalene dioxygenase in native and selenomethionine-derivatized forms. Biochem. Biophys. Res. Commun. 241:553-557. [DOI] [PubMed] [Google Scholar]
- 36.Lloyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 37.Meyer, S., R. Moser, A. Neef, U. Stahl, and P. Kampfer. 1999. Differential detection of key enzymes of polyaromatic-hydrocarbon-degrading bacteria using PCR and gene probes. Microbiology 145:1731-1741. [DOI] [PubMed] [Google Scholar]
- 38.Mueller, J. G., R. Devereux, D. L. Santavy, S. E. Lantz, S. G. Willis, and P. H. Pritchard. 1997. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek 71:329-343. [DOI] [PubMed] [Google Scholar]
- 39.Muyzer, G., S. Hottentrager, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR amplified 16S rDNA: a new molecular approach to analyze the genetic diversity of mixed microbial communities. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishing, Dordrecht, The Netherlands.
- 40.Nam, J. W., H. Nojiri, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254-263. [DOI] [PubMed] [Google Scholar]
- 41.Nojiri, H., H. Habe, and T. Omori. 2001. Bacterial degradation of aromatic compounds via angular dioxygenation. J. Gen. Appl. Microbiol. 47:279-305. [DOI] [PubMed] [Google Scholar]
- 42.Parales, R. E. 2003. The role of active-site residues in naphthalene dioxygenase. J. Ind. Microbiol. Biotechnol. 30:271-278. [DOI] [PubMed] [Google Scholar]
- 43.Parales, R. E., K. Lee, S. M. Resnick, H. Y. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pielou, E. C. 1977. Mathematical ecology, 2nd ed. Wiley, New York, N.Y.
- 45.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 46.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato, S., N. Ouchiyama, T. Kimura, H. Nojiri, H. Yamane, and T. Omori. 1997. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J. Bacteriol. 179:4841-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shuttleworth, K. L., and C. E. Cerniglia. 1995. Environmental aspects of PAH biodegradation. Appl. Biochem. Biotechnol. 54:291-302. [DOI] [PubMed] [Google Scholar]
- 49.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 50.Stach, J. E. M., and R. G. Burns. 2002. Enrichment versus biofilm culture: a functional and phylogenetic comparison of polycyclic aromatic hydrocarbon-degrading microbial communities. Environ. Microbiol. 4:169-182. [DOI] [PubMed] [Google Scholar]
- 51.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Appl. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 52.Stapleton, R., and G. Sayler. 1998. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb. Ecol. 36:349-361. [DOI] [PubMed] [Google Scholar]
- 53.Stoffels, M., R. Amann, W. Ludwig, D. Hekmat, and K. H. Schleifer. 1998. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl. Environ. Microbiol. 64:930-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuomi, P. M., J. M. Salminen, and K. S. Jorgensen. 2004. The abundance of nahAc genes correlates with the C-14-naphthalene mineralization potential in petroleum hydrocarbon-contaminated oxic soil layers. FEMS Microbiol. Ecol. 51:99-107. [DOI] [PubMed] [Google Scholar]
- 55.Widada, J., H. Nojiri, K. Kasuga, T. Yoshida, H. Habe, and T. Omori. 2002. Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl. Microbiol. Biotechnol. 58:202-209. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye, D. Y., M. A. Siddiqi, A. E. Maccubbin, S. Kumar, and H. C. Sikka. 1996. Degradation of polynuclear aromatic hydrocarbons by Sphingomonas paucimobilis. Environ. Sci. Technol. 30:136-142. [Google Scholar]
- 58.Zocca, C., S. Di Gregorio, F. Visentini, and G. Vallini. 2004. Biodiversity amongst cultivable polycyclic aromatic hydrocarbon-transforming bacteria isolated from an abandoned industrial site. FEMS Microbiol. Lett. 238:375-382. [DOI] [PubMed] [Google Scholar]