Abstract
Fuel oxygenates such as methyl and ethyl tert-butyl ether (MTBE and ETBE, respectively) are degraded only by a limited number of bacterial strains. The aerobic pathway is generally thought to run via tert-butyl alcohol (TBA) and 2-hydroxyisobutyrate (2-HIBA), whereas further steps are unclear. We have now demonstrated for the newly isolated β-proteobacterial strains L108 and L10, as well as for the closely related strain CIP I-2052, that 2-HIBA was degraded by a cobalamin-dependent enzymatic step. In these strains, growth on substrates containing the tert-butyl moiety, such as MTBE, TBA, and 2-HIBA, was strictly dependent on cobalt, which could be replaced by cobalamin. Tandem mass spectrometry identified a 2-HIBA-induced protein with high similarity to a peptide whose gene sequence was found in the finished genome of the MTBE-degrading strain Methylibium petroleiphilum PM1. Alignment analysis identified it as the small subunit of isobutyryl-coenzyme A (CoA) mutase (ICM; EC 5.4.99.13), which is a cobalamin-containing carbon skeleton-rearranging enzyme, originally described only in Streptomyces spp. Sequencing of the genes of both ICM subunits from strain L108 revealed nearly 100% identity with the corresponding peptide sequences from M. petroleiphilum PM1, suggesting a horizontal gene transfer event to have occurred between these strains. Enzyme activity was demonstrated in crude extracts of induced cells of strains L108 and L10, transforming 2-HIBA into 3-hydroxybutyrate in the presence of CoA and ATP. The physiological and evolutionary aspects of this novel pathway involved in MTBE and ETBE metabolism are discussed.
Methyl tert-butyl ether (MTBE) and the related ethyl tert-butyl ether (ETBE) are branched alkyl ethers that have been used as oxygenating compounds in gasoline since the 1980s (45, 47). The worldwide production of the leading fuel oxygenate MTBE currently amounts to 20 Mt per year. Due to this extensive use, freshwater resources have been widely impacted by MTBE (1, 34, 45), e.g., through accidental spills and leaking storage tanks. This contamination collides with human drinking water demands, since odor and taste thresholds for MTBE are low and because of its classification as a possible human carcinogen (49). In the future, ETBE will replace MTBE in some countries, as legislation promotes the use of biofuels to reduce carbon dioxide emissions (13). In contrast to MTBE, ETBE can be easily produced in sufficient amounts from isobutene and ethanol that has been won through biotechnological processes from renewable biomass (38).
Both MTBE and ETBE have been demonstrated to be degradable by bacteria and fungi (12, 16). However, strains using these oxygenates as the sole source of carbon and energy are rarely found. Bacterial isolates capable of aerobic growth on MTBE include the β-proteobacterial strains Methylibium petroleiphilum PM1 (20, 35) and Hydrogenophaga flava ENV 735 (21), as well as the gram-positive Mycobacterium austroafricanum IFP 2012 (18). An ETBE-growing strain is Rhodococcus ruber IFP 2001 (15, 22), showing an incomplete degradation. Generally, degradation rates are slow, and growth is inefficient on these substrates.
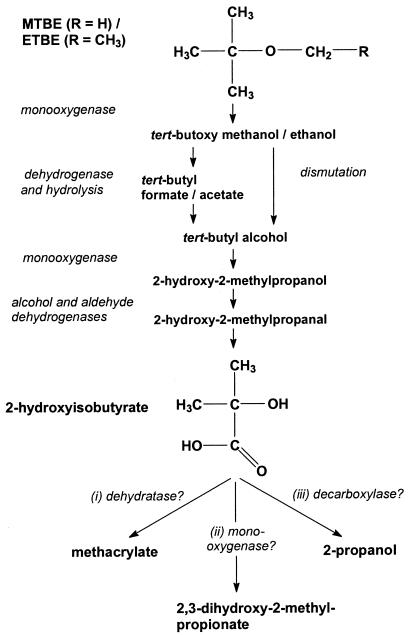
Until now, the aerobic degradation pathways of MTBE and ETBE have not been fully elucidated. Particularly, biochemical and genetic data are missing. Nevertheless, there is agreement on the first steps of oxidation (Fig. 1) (16, 44, 46). Initially, the methyl and ethyl groups, respectively, are attacked by monooxygenase systems, resulting in unstable hemiacetals. These compounds can spontaneously decompose to tert-butyl alcohol (TBA) and an aldehyde, i.e., formaldehyde or acetaldehyde. On the other hand, it was demonstrated that the hemiacetal derived from MTBE is oxidized to tert-butylformate (TBF), which is further hydrolyzed to TBA and formate (44). In the case of ETBE, analogous reactions would yield TBA and acetate. The intermediate one- and two-carbon aldehydes and acids are expected to be oxidized by conventional dehydrogenase systems. TBA, however, is thought to be hydroxylated, again by the ether monooxygenase or different enzymes, to 2-hydroxy-2-methylpropanol, which is further oxidized by dehydrogenases to 2-hydroxyisobutyric acid (2-HIBA). Until 2-HIBA, the tert-butyl moiety is maintained (Fig. 1), but it can be speculated that this bulky structure is destroyed in the next steps to connect the special fuel oxygenate reactions with the general metabolism and, thus, enable complete oxidation. Unfortunately, the reaction sequence of this central part of the alkyl tert-butyl ether pathways is unclear. Since the work of Steffan et al. (46), three possible reactions have been discussed: (i) dehydration to methacrylate, (ii) hydroxylation to 2,3-dihydroxy-2-methylpropanoate, and (iii) decarboxylation to isopropanol with further oxidation to acetone. The latter proposal is favored by several authors (12, 14, 18, 41), although supporting experimental data are limited. In most studies, it was only demonstrated that isopropanol and acetone are degraded by the strains investigated (11, 18, 27, 46) or that an acetone-oxidizing activity is induced (14, 19). In the case of Mycobacterium vaccae Job5, an intermediate of acetone metabolism, hydroxyacetone, was detected during MTBE degradation (27). However, the enzymes involved in 2-HIBA metabolism or a specific pathway have not been identified thus far.
FIG. 1.
Proposed pathways for the aerobic degradation of the fuel oxygenates MTBE and ETBE (16, 44, 46).
In the present study, we investigated the degradation pathway of 2-HIBA in the MTBE-degrading β-proteobacterium strain L108, a novel isolate capable of growing on fuel oxygenate ethers as the sole source of carbon and energy, as well as in two TBA-degrading strains, a mutant of L108, strain L10, and strain CIP I-2052, previously isolated by Piveteau et al. (37). Results are presented that support an alternative route to the above-mentioned proposals consisting of a cobalamin-depending mutase reaction and transforming 2-HIBA in one step into 3-hydroxybutyrate. Our results also show that use of the mutase reaction results in an exceptional nutritional demand for cobalt during growth on substrates possessing the tert-butyl moiety. Furthermore, the necessity of cobalamin biosynthesis is rate limiting for the growth on TBA and 2-HIBA.
MATERIALS AND METHODS
Sources of bacterial strains and chemicals.
Strain L108 was isolated in the present study from MTBE-contaminated groundwater, which was sampled in 2003 at the industrial park of Leuna, Germany, after enrichment on liquid MTBE mineral salt medium and streaking on R2A agar plates. Strain L10 was obtained by incubating an MTBE-grown culture of strain L108 on R2A agar plates and searching for colonies incapable of MTBE degradation. Strains L108 and L10 were cultivated on MTBE and TBA mineral salt medium, respectively, supplemented with 50 μg of cobalt ions per liter. The isolates were added to the strain collection of the Department of Environmental Microbiology of the UFZ. Strain CIP I-2052 (37) was provided by F. Fayolle (IFP [France]) and cultured further on TBA in our laboratories. All chemicals used in the present study were of the highest purity available and purchased from either Sigma-Aldrich Chemie (Taufkirchen, Germany) or Merck (Darmstadt, Germany).
Media and cultivation techniques.
For testing and growing bacteria on definite substrates as sole source of carbon and energy, the following mineral salt medium was used (in mg/liter): NH4Cl, 761; KH2PO4, 340; K2HPO4, 436; CaCl2 · 6H2O, 5.5; MgSO4 · 7H2O, 71.2; ZnSO4 · 7H2O, 0.44; MnSO4 · H2O, 0.615; CuSO4 · 5H2O, 0.785; Na2MoO4 · 2H2O, 0.252; and FeSO4 · 7H2O, 4.98, which was adjusted to pH 7 and supplemented after autoclaving with the following vitamins (in μg/liter): biotin, 20; folic acid, 20; pyridoxine-HCl, 100; thiamine-HCl, 50; riboflavin, 50; nicotinic acid, 50; Ca-pantothenate, 50; p-aminobenzoic acid, 50; and lipoic acid, 50. To the mineral salt medium, cobalt ions were usually added at 50 μg of cobalt per liter or cobalt was replaced by cyanocobalamin as individually indicated. Substrates were added from aqueous stock solutions. For isolation of strains and purity tests, bacterial cultures were streaked on R2A agar (Merck no. 100416) plates. Generally, liquid cultures were incubated at 30°C on rotary shakers. Appropriate substrate concentrations of up to 1 g/liter and volumes of up to 500 ml of culture medium were applied. In the case of volatile compounds such as ethers and alcohols, close systems (glass serum bottles sealed with butyl rubber stoppers) with sufficient headspace volume to guarantee aerobic conditions were used; in other cases, conic flasks with cellulose stoppers were used. Cobalt- and cobalamin-deficient cultures were obtained by incubation on mineral salt medium for two passages (3% [vol/vol] inoculum) omitting supplementation with cobalt or cobalamin.
Short-term degradation tests.
Strains were grown on TBA, 2-HIBA, 3-hydroxybutyrate, isobutyrate, butyrate, methacrylate, methylmalonate, acetate, or glucose until late exponential growth phase. Cells were then harvested by centrifugation at 6,000 × g and 4°C for 10 min and washed twice with mineral salt solution. To the final cell suspension 50 μg of cyanocobalamin per liter was added. Degradation tests were performed with 25 ml of a cell suspension of 1 to 2 g of biomass (dry weight) per liter in 240-ml serum bottles sealed with butyl rubber stoppers and incubated on a rotary shaker at 30°C. Initial 2-HIBA values were 200 or 500 mg/liter, in the case of low or high activity, respectively. The concentration of 2-HIBA was monitored by sampling at 20-min intervals for a total period of 2 h. Activity values were determined by using linear regression analysis on the decrease of 2-HIBA within this time period.
Preparation of cell extract and enzyme assay.
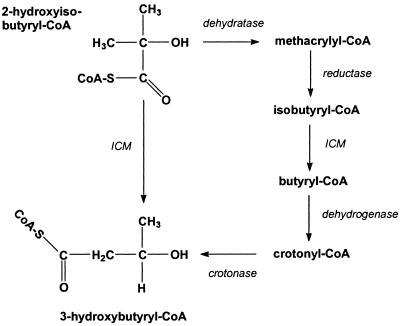
Biomass was obtained as described for the short-term degradation tests from 2-HIBA and acetate cultures. Cells were suspended in potassium phosphate buffer (50 mM [pH 7.2], supplemented with 10 μM adenosylcobalamin) at about 25 mg of protein per ml and disrupted under nitrogen atmosphere in the dark using stirring glass beads as previously described (40). Then, intact cells and cell debris were removed by centrifugation (20 min, 16,000 × g, twice). The supernatant, hereafter referred to as the cell extract, usually contained 2 to 4 mg of protein per ml and was stored under nitrogen atmosphere at −25°C. Assays were performed within the next few days since storage beyond 3 weeks resulted in nearly complete loss of activity. For measuring the enzymatic transformation activity of 2-HIBA into 3-hydroxybutyrate, cell extracts were mixed with the above-mentioned phosphate buffer, adjusting protein concentrations to 1 to 2 mg of protein per ml. After supplementation with magnesium ions (MgCl2 · 6H2O), ATP (disodium salt, Merck no. 1.01432), and coenzyme A (CoA; trilithium salt, Sigma no. C3019) to final concentrations of 1 mM each, the assays were started by adding 2 g of 2-HIBA per liter. All assays were incubated under nitrogen atmosphere in the dark at 30°C on a rotary shaker. Samples were taken at appropriate time intervals and analyzed for 3-hydroxybutyrate and other possible carbonic acid intermediates and products (Fig. 6).
FIG. 6.
Possible pathways for 2-HIBA degradation using a cobalamin-dependent carbon skeleton-rearranging step catalyzed by ICM. Initially, 2-HIBA may be activated to 2-hydroxyisobutyryl-CoA by acyl-CoA synthetases or CoA transferases.
Proteome analysis.
Total protein of acetate-, MTBE-, TBA-, and 2-HIBA-grown cells of strain L108 were compared by two-dimensional electrophoresis as previously described (5). In brief, 500 μg of protein of crude extracts was purified by phenol extraction (51) and separated by isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were stained with colloidal Coomassie brilliant blue and dried in a stream of unheated air. Protein spots of interest were excised from two-dimensional gels, digested with trypsin (43), and analyzed with an AP (atmospheric pressure)-MALDI/TRAP XCT mass spectrometer (Agilent Technologies) in automatic tandem mass spectrometry (MS/MS) mode. MS/MS ion searches (MASCOT) (36) were carried out against the NCBI nr database.
PCR and sequencing.
The 16S rRNA genes of strains L10 and L108 were PCR amplified by using universal eubacterial primers and were bidirectionally sequenced as previously described (8). Sequences of 1,451 nucleotides (28 to 1491 bp, E. coli numbering) were used for similarity searches against the EMBL and GenBank databases. The icmA and icmB genes of strain L108 were PCR amplified by using primers derived from the corresponding sequences of strain PM1 (NZ_AAEM01000010, gene 85642.87330 and gene 82790.83200). DNA of L108 was prepared as previously described (24). For icmA, the following primers were used: ICMA_f (5′-ATGACCTGGCTTGAGCCGCA-3′), ICMA_r (5′-GCGAGACGCCGGTCTTCTGA-3′), ICMA_M_f (5′-GAGAAGCGCGGCTACGACCT-3′), and ICM_M_r (5′-TCCTCGGTCGGGATCGCGAA-3′). PCR with ICMA_f and ICMA_r gave the complete fragment, whereas in combination with ICM_M_f and ICM_M_r overlapping intermediary fragments were produced. For icmB, the primers ICMB_f (5′-ATGGACCAAATCCCGATCCGC-3′) and ICMB_r (5′-TCAGCGGGCGCCGCGCGCGG-3′) were used, producing the complete fragment. Fragments were gel purified and sequenced according to the method of Sanger et al. (42) using an ABI Prism 310 genetic analyzer (PE Applied Biosystems). Only high-quality reads were considered, and each sequence was determined twice. The comparison of sequences with DNA and protein sequences in databases was performed with BLAST (2) (http://www.ncbi.nlm.nih.gov/blast). Multiple sequence alignments were performed with CLUSTAL W (23; http://www.ebi.ac.uk/clustalw/).
Chemical and other analyses.
The concentrations of MTBE, TBA, and carbonic acids were determined by gas chromatography (GC). In all cases, standards of the pure chemicals were used for calibration. MTBE and TBA were analyzed by using an HP 6890 GC system from Agilent Technologies (Waldbronn, Germany) with an HP-5 column (30 m by 0.25 mm by 0.25 μm) and a flame ionization detector. Vials (10-ml headspace) were filled with 2 ml of liquid sample and incubated at 70°C for 20 min before 1 ml of gas phase was removed and injected into the GC system by using an HP 7694 autosampler. The carrier gas was helium, and the oven and detector temperatures were 35 and 200°C, respectively. Carbonic acids were analyzed by the same GC system using liquid injection mode (1 μl) and an Optima-FFAP column (25 m by 0.25 mm by 0.25 μm; Macherey-Nagel, Düren, Germany). 2-HIBA, 3-hydroxybutyrate, and methylmalonate were quantified with a detection limit of 0.5 mg per liter as methyl esters by modifying a published method (30). Samples (800 μl) were incubated with acidic methanol (400 μl containing 3% [vol/vol] sulfuric acid) for 2 h at 95°C. The esters formed were extracted with chloroform (400 μl). γ-Butyrolactone was used as internal standard. The GC oven program was 90°C for 2 min, 90 to 220°C over 3.71 min, and finally 2 min at 220°C. Methacrylate, isobutyrate, butyrate, and crotonate were quantified as free acids (6) with a detection limit of 1 mg per liter. Samples (1,000 μl) were saturated with NaCl, acidified with 3 M sulfuric acid (50 μl), and extracted with ethyl acetate (600 μl). Hexanoate was used as internal standard. The GC oven profile was 90°C for 2 min, 90 to 175°C over 1.31 min, 175 to 220°C over 1 min, and finally 2 min at 220°C. The reproducibility of the GC concentration analyses was within a standard deviation (SD) of 5%. Generally for carbonic acid analysis, an NaOH solution was added to samples at a final concentration of 50 mM in order to stop reactions. In the case of growth experiments and short-term activity tests, cells were removed by centrifugation prior to further sample processing. Samples were then stored at −25°C until preparation for GC measurements. If butyl rubber-sealed glass bottles were used for experiments, sampling was performed by plastic syringes removing an appropriate volume from the liquid phase. Cell growth was monitored by measuring the optical density at 700 nm (OD700) of the culture broth in a 1-cm cuvette and by measuring the dry weight of cells (by incubation at 100°C until reaching constant weight). Biomass values were also calculated from OD700 data after determining the relation between the OD and the dry weight for the investigated strains (an OD700 value of 2.15 was equal to 1 g of dry weight per liter). Cell protein concentrations were determined by the Lowry protein assay (31) for two-dimensional electrophoresis and by the Bradford method for the enzyme assay (7).
Nucleotide sequence accession numbers.
The nearly complete 16S rRNA, icmA, and icmB gene sequences of strain L108 have been deposited in the GenBank/EMBL/DDBJ database under the accession numbers DQ436455, DQ436456, and DQ436457, respectively.
RESULTS
Properties of investigated strains.
Strain L108 was isolated from an MTBE-contaminated site in Germany and was able to grow on MTBE and ETBE as the sole source of carbon and energy. From this isolate a mutant, strain L10, was obtained by subcultivation on nonselective medium. Strain L10 was incapable of MTBE degradation but could still grow on TBA. Both strains showed identical 16S rRNA gene sequences, and there was a 99.9% identity to the sequence of the TBA degrader CIP I-2052 (AF244133), which was previously isolated from the activated sludge of a wastewater treatment plant near Paris (37) and, consequently, was included in the present study. All three strains showed similar physiological properties and phylogenetically belong to the Rubrivivax subgroup of the β-Proteobacteria with highest 16S rRNA gene sequence identity (97%) to type strains of Leptothrix mobilis (X97071), L. cholodnii (X97070), Rubrivivax gelatinosus (D16213), and Ideonella dechloratans (X72724). The nearest MTBE-degrading relative was M. petroleiphilum PM1 (AF176594), showing 95.6% identity to the 16S rRNA gene sequence of strain L108.
Cobalt/cobalamin dependence of tert-butyl moiety degradation.
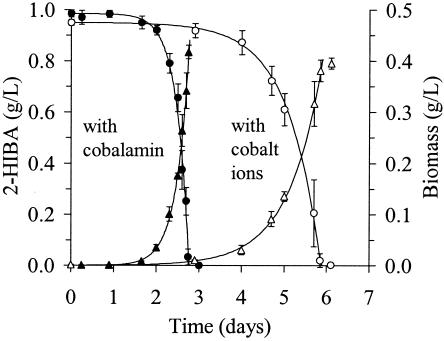
As already described for strain CIP I-2052 growing on TBA (37), all three strains—L108, L10, and CIP I-2052—generally showed an exceptional demand for the trace element cobalt when grown on substrates containing the tert-butyl moiety. Substrates tested in the present study were TBA and 2-HIBA for all strains and MTBE in the case of L108. In cobalt-deficient medium, degradation rates and growth yields were significantly reduced. However, such a cobalt dependence was not observed with simple growth substrates such as glucose, acetate, and 3-hydroxybutyrate (data not shown). Cobalt can be replaced by cobalamin, as was studied in detail for the growth of strain L10 on 2-HIBA (Fig. 2). Furthermore, compared to the cobalt-containing cultures, cobalamin addition decreased the doubling times on 2-HIBA from about 13 to 5 h and on TBA from about 13 to 9 h, whereas growth yields were not affected (Table 1). Only insignificant growth occurred in the absence of cobalt and cobalamin under these conditions (data not shown).
FIG. 2.
Growth on 2-HIBA in mineral salt medium supplemented with either 100 μg of cyanocobalamin or 50 μg of cobalt ions per liter (filled and open symbols, respectively). 2-HIBA concentrations (circles) and dry weight biomass (triangles) were measured for batch cultures of strain L10 using a cobalt- and cobalamin-deficient inoculum. The data represent the mean values and SD of four replicates.
TABLE 1.
Growth parameters of strains L10 and CIP I-2052 on 2-HIBA and TBA, incubated with either cobalt ions or cobalamin
| Growth parameter | Strain | Mean ± SDe
|
|||
|---|---|---|---|---|---|
| Growth on 2-HIBA witha:
|
Growth on TBA withb:
|
||||
| Cobalt | Cobalamin | Cobalt | Cobalamin | ||
| Doubling time (h)c | L10 | 13.0 ± 0.2 | 5.0 ± 0.1 | 12.9 ± 0.2 | 8.8 ± 0.3 |
| CIP I-2052 | 13.2 ± 0.2 | 5.8 ± 0.2 | ND | ND | |
| Growth yield (g biomass/g substrate)d | L10 | 0.41 ± 0.03 | 0.44 ± 0.02 | 0.44 ± 0.03 | 0.53 ± 0.09 |
| CIP I-2052 | 0.44 ± 0.05 | 0.44 ± 0.03 | ND | ND | |
Batch cultures with 1 g of 2-HIBA per liter containing either 50 μg of cobalt ions or 100 μg of cyanocobalamin per liter were inoculated with 3% (vol/vol) of a culture pregrown on TBA under cobalt- and cobalamin-deficient conditions.
Batch cultures with 0.5 g of TBA per liter containing either 50 μg of cobalt ions or 50 μg of cyanocobalamin per liter were inoculated with 3% (vol/vol) of a culture pregrown on TBA under cobalt- and cobalamin-deficient conditions.
Doubling times were determined for the exponential growth phase by monitoring the increase in the OD700.
Growth yields were determined by using biomass increase data at more than 50% substrate degradation.
Mean values of at least five replicates are shown. ND, not determined.
Accumulation of 2-HIBA during cobalamin-deficient TBA degradation.
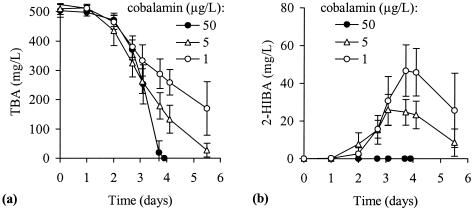
During the growth of all three strains on TBA at limiting cobalt or cobalamin concentrations, an accumulation of 2-HIBA was observed. While cultures, e.g., of strain L10 with 50 μg of cobalamin per liter showed the fastest TBA degradation and no 2-HIBA accumulation, decreasing oxidation rates and increasing amounts of temporary 2-HIBA were measured by gradually reducing the cobalamin concentrations (Fig. 3). Under these conditions, an accumulation of other possible carbonic acid intermediates such as methacrylate, isobutyrate, butyrate, crotonate, and 3-hydroxybutyrate (Fig. 6), as well as methylmalonate, was not observed.
FIG. 3.
Degradation of TBA (a) and concomitant accumulation of 2-HIBA (b) in mineral salt medium supplemented with 1, 5, or 50 μg of cyanocobalamin per liter and using a cobalt- and cobalamin-deficient culture of strain L10. The data represent the mean values and SD of four replicates.
Inducibility of 2-HIBA oxidation activity.
For testing the inducibility of the 2-HIBA oxidation activity, short-term degradation experiments were performed with resting cells of strains L10 and L108 grown on various substrates. Activity values of three replicates for each substrate showed quite high variation, with SD values of up to 50% (not shown). However, the following substrate classes could be discriminated. Cells grown on TBA, 2-HIBA, or isobutyrate showed the highest 2-HIBA degradation activity, with values between 10 and 40 nmol of 2-HIBA min−1 mg biomass−1. In contrast, with cells grown on methylmalonate, methacrylate, butyrate, or 3-hydroxybutyrate the activity was significantly reduced (values were between 1 and 2 nmol min−1 mg−1). Little if any 2-HIBA degradation (<1 nmol min−1 mg−1) was observed with cells grown on acetate or glucose, suggesting the 2-HIBA-degrading pathway was inducible.
Comparative proteome analysis.
Based on the findings of the inducibility tests, proteome analyses were done comparing total proteins of strain L108 grown on MTBE, TBA, 2-HIBA, and acetate. In two-dimensional sodium dodecyl sulfate-polyacrylamide gels, one small protein was identified occurring exclusively in crude extracts derived from MTBE-, TBA-, and 2-HIBA-grown cells (data not shown). An MS/MS ion search gave as the closest match, with a MASCOT score of 50, a gene sequence found in the finished genome of M. petroleiphilum PM1 (http://genome.jgi-psf.org/finished_microbes/metpe/metpe.home.html; AAEM00000000) coding for a 136-amino-acid protein. Automated GeneMark analysis had predicted this protein (ZP_00242467) to be a C-terminal domain/subunit of methylmalonyl-CoA mutase (MCM; EC 5.4.99.2) binding the coenzyme cobalamin. However, thus far identified prokaryotic MCMs are organized as homo- or heterodimers with a subunit size of about 700 amino acids and without having the substrate- and cobalamin-binding domain on different polypeptides (6, 48). Furthermore, sequence alignment analysis revealed a high similarity score (54% identity of amino acid residues) to the small subunit of isobutyryl-CoA mutase (ICM; EC 5.4.99.13) of Streptomyces cinnamonensis A3823.5 (AJ246005), a cobalamin-dependent mutase that is organized as a large substrate-binding 566- and a small cobalamin-binding 136-amino-acid subunit (IcmA and IcmB, respectively) (39, 52). This finding lets us assume that the identified protein was IcmB and not a subunit of MCM. Consequently, a sequence coding for an IcmA-like 562-amino-acid polypeptide (ZP_00242470) showed the closest match with BLAST analysis using IcmA of S. cinnamonensis A3823.5 (AAC08713) as a query sequence against the complete genome of M. petroleiphilum PM1.
Enzyme assay for 2-HIBA isomerization.
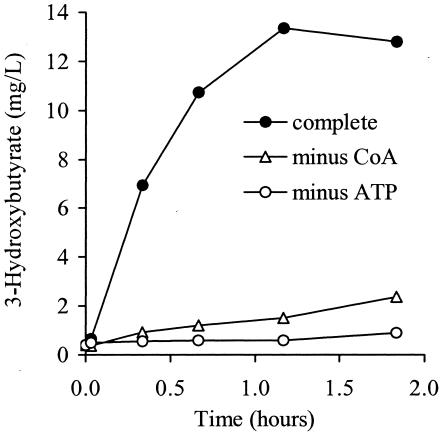
The identification of a cobalamin-binding protein being induced by 2-HIBA and with high similarity to the small subunit of ICM raised the question of whether a carbon skeleton rearrangement was involved in 2-HIBA degradation. To answer this question, an adequate enzyme assay was performed comparing the activity of cell extracts from induced and noninduced cells of strains L10 and L108. In this experiment, it was found that extracts from 2-HIBA-grown cells showed production of 3-hydroxybutyrate from 2-HIBA (Fig. 4), whereas a similar activity was not observed in material derived from acetate-grown cells (data not shown). Furthermore, transformation of 2-HIBA into 3-hydroxybutyrate was only achieved in the presence of ATP and free CoA, indicating the necessity of an activating thioester formation, i.e., 2-hydroxyisobutyryl-CoA, prior to the isomerization reaction. Formation of 3-hydroxybutyrate was not accompanied by the detection of possible intermediates (as indicated in Fig. 6) such as methacrylate, isobutyrate, butyrate, or crotonate.
FIG. 4.
Transformation of 2-HIBA into 3-hydroxybutyrate in cell extracts of 2-HIBA-grown cells of strain L108. The complete assay contained 2-HIBA, CoA, and ATP, whereas in the other cases either CoA or ATP was omitted. The SD of the replicates was within 5% (not shown).
Gene sequence of isobutyryl-CoA mutase of strain L108.
The two identified ICM-like sequences from M. petroleiphilum PM1 were used to derive PCR primers for the detection of orthologous sequences in strain L108. PCR analysis with total DNA and the primer pairs complementary to the 5′ and 3′ ends of the corresponding genes gave products with expected sizes of about 1,670 and 410 bp, respectively (data not shown). The corresponding almost complete peptide sequences showed high similarity to IcmA and IcmB of S. cinnamonensis A3823.5 (43 and 54% identity, respectively) and nearly 100% identity to the sequences found in M. petroleiphilum PM1 (for alignments of IcmA and IcmB amino acid sequences from S. cinnamonensis, strain PM1, and strain L108 and nucleotide sequences of icmA and icmB from strains PM1 and L108, see the supplemental material). The most significant difference was a 16-amino-acid segment at positions 424 to 439 (S. cinnamonensis numbering) of IcmA that is absent in PM1 and L108 but present in all three sequences of IcmA from Streptomyces spp. identified thus far (AAC08713, NP_629554, and NP_824008). A second interesting deviation was the replacement of Phe at IcmA position 80 in S. cinnamonensis by Ile in M. petroleiphilum PM1 and strain L108 sequences. This position corresponds to an absolutely conserved Tyr in MCMs and is thought to play an important role in substrate binding (39). A BLAST search with the complete IcmA amino acid sequence of strain PM1 as a query against the NCBI nr database (March 2006) resulted in only three ICM-like matches with an Ile and one with a Val at this position. In contrast, all other matches with more than 40% sequence identity (about 200) showed either a corresponding Phe residue or a corresponding Tyr residue (see the supplemental material). The highest similarities to IcmA of strain PM1 were found with predicted proteins of Rhodobacter sphaeroides ATCC 17029 (EAP67072) and Xanthobacter autotrophicus Py2 (EAS17594), with 83 and 80% sequence identities, respectively. For comparison, Fig. 5 shows a 30-amino-acid segment of IcmA of strain PM1, including the Ile position aligned with the corresponding sequences of the four closest BLAST matches, as well as with the S. cinnamonensis ICM and Propionibacterium shermanii MCM sequences.
FIG. 5.
A CLUSTAL W alignment of a 30-amino-acid segment of the ICM large subunit (IcmA) of Methylibium petroleiphilum PM1 (ZP_00242470), with the corresponding sequences of the four closest BLAST matches using the complete IcmA sequence of strain PM1 as a query against the NCBI database (in descending order): Rhodobacter sphaeroides ATCC 17029 (EAP67072), Xanthobacter autotrophicus Py2 (EAS17594), Nocardioides sp. strain JS614 (EAO08692), and Roseovarius sp. strain 217 (EAQ26421) (for complete search results, see the supplemental material). For comparison, alignment with the corresponding ICM and MCM sequences of S. cinnamonensis (AJ246005) and P. shermanii (X14965), respectively, is also shown. Amino acids that are conserved in all sequences are indicated under the sequence by asterisk. Residues in boldface represent the reactive site position proposed to play an important role in substrate binding and reaction mechanism of both ICM and MCM (32, 39, 50).
DISCUSSION
The three closely related β-proteobacterial strains L10, L108, and CIP I-2052 show an exceptional demand for cobalt while growing on tert-butyl moiety-containing compounds such as TBA and 2-HIBA, which are intermediates of the aerobic degradation of the fuel oxygenates MTBE and ETBE. Cobalamin can replace free cobalt ions under these conditions, indicating the involvement of a cobalamin-dependent enzymatic step. Since 2-HIBA is the most downstream tert-butyl compound tested in the present study, it can be concluded that the cobalamin-dependent reaction occurs at this position or later in the fuel oxygenate pathway. The most likely candidate for catalyzing such a reaction is the cobalamin-containing ICM, since the expression of an ICM-like subunit and 2-HIBA degradation activity is correlated and the genes of both ICM subunits are present in the genome of strain L108.
The involvement of ICM in the degradation of 2-HIBA may be covered by two possible pathways (Fig. 6). According to the original proposal of Steffan et al. (46), 2-HIBA can be dehydrated to methacrylate, presumably only after thioester formation as 2-hydroxy carbonic acids are quite resistant against dehydration (29). The resulting methacrylyl-CoA is then reduced to isobutyryl-CoA, which is isomerized to butyryl-CoA by ICM. Butyryl-CoA can be dehydrogenized to crotonyl-CoA and, after the addition of water, 3-hydroxybutyryl-CoA is formed. The alternative route uses only two steps. After thioester activation of 2-HIBA, 3-hydroxybutyryl-CoA is formed by ICM, moving the carbonic acid group from the tert-carbon atom to a primary one. Our results clearly favor the latter proposal, using a direct ICM-catalyzed isomerization of 2-HIBA, for the following reasons. (i) During cobalt- and cobalamin-deficient TBA degradation, only 2-HIBA temporarily accumulates but other possible carbonic acid intermediates are not detected. This indicates that directly the 2-HIBA transforming step is cobalamin dependent. (ii) The inducibility tests show only low 2-HIBA degradation activity in methacrylate- and methylmalonate-grown cells, ruling out the methacrylate pathway of Fig. 6 and the involvement of MCM, respectively. However, activity is significantly induced in cells grown on 2-HIBA or isobutyrate, which are possible substrates for ICM. (iii) We demonstrated the enzymatic transformation of 2-HIBA to 3-hydroxybutyrate in cell extracts of induced cells without the formation of any intermediates.
A notable consequence of the use of a cobalamin-containing enzyme for a dissimilatory pathway is the demand for cobalt. From the amount of cobalamin necessary for an unlimited growth on 2-HIBA, it can be calculated that about 5 μg of cobalt is sufficient for an efficient degradation of 1 g of a compound containing the tert-butyl group. Therefore, it can be recommended that at least this amount of cobalt has to be bioavailable when MTBE or ETBE contaminations should be rehabilitated by using strain L108 or bacteria with a similar pathway. In addition, at least on TBA and 2-HIBA, the biosynthesis of cobalamin determines the growth rate, as demonstrated by the increasing doubling times when the strains grow in the presence of free cobalt instead of cobalamin. This finding can be explained by the enormous metabolic burden of cobalamin de novo synthesis, requiring the activity of more than 30 genes (33), when it has to be produced for a dissimilatory enzymatic step.
The discovery of a carbon skeleton rearrangement in the degradation pathway of 2-HIBA identified a 2-HIBA-transforming step at the biochemical and genetic level. As well, it is only the second enzyme besides the monooxygenase found in R. ruber IFP 2001 (9) proved to be involved in the aerobic degradation of MTBE and ETBE. Thus far, the cobalt demand of other fuel oxygenate-degrading isolates has not been sufficiently studied. In most cases, which include M. petroleiphilum PM1, the growth medium contains excess cobalt (20, 21). Only the MTBE degrader M. austroafricanum IFP 2012 demonstrates similar cobalt dependence. However, the authors of that study (18) stated that cobalt cannot be replaced by cobalamin, thus probably excluding the involvement of the mutase pathway in this strain. On the other hand, the fact that M. petroleiphilum PM1 possesses ICM genes nearly identical to the ones of strain L108 let us postulate that in both strains 2-HIBA is processed via the described mutase reaction. In the future, this assumption has to be proved by physiological and biochemical analyses, including the testing of ICM defective mutants. In addition, the high similarity of the ICM sequences implies that a horizontal gene transfer has occurred between the two strains. This may also suggest that the ICM pathway plays a widespread role in fuel oxygenate degradation since PM1-like bacteria have been detected at several MTBE-contaminated sites (26, 28). Consequently, further investigations now under way will elucidate the distribution of the ICM genes and activities among other fuel oxygenate-degrading strains.
Although 2-HIBA is rarely found in nature, it is not strictly related to MTBE and ETBE metabolism. The plant cyanoglycoside linamarin (17) is synthesized and decomposed via 2-hydroxyisobutyronitrile, which normally hydrolyzes to cyanide and acetone (3, 4). Theoretically, by the action of nitrilase or a combination of nitrile hydratase and amidase (4), 2-HIBA could be produced from the nitrile. However, there is no evidence for a bacterium capable of growing on hydroxyisobutyronitrile and using the mentioned nitrile-degrading enzymes. A second oxygenate-independent source of 2-HIBA is the industrial synthesis of methacrylate. In the classical acetone cyanohydrin process, 2-HIBA is a by-product and, consequently, is found in wastewaters from methacrylate-producing plants. Interestingly, there exists one study on the bacterial conversion of 2-HIBA and its methyl ester from such a wastewater to polyhydroxybutyrate (25), which is generally synthesized by polymerization from 3-hydroxybutyrate. The authors of that study proposed a degradation pathway via pyruvate and acetyl-CoA, though without any physiological or biochemical evidence. However, exactly for the transformation of 2-HIBA to 3-hydroxybutyrate, we describe here the responsible mutase reaction which is probably also involved in the bacterial process previously described by Holowach et al. (25). Industrial methacrylate production started in the mid-1930s in Great Britain, Germany, and the United States (10), giving bacterial evolution more time to adapt for 2-HIBA degradation than to degrade fuel oxygenates. In this scenario, a carbon skeleton mutase like the ICM of Streptomyces spp. broadens its substrate spectrum for 2-HIBA. The preliminary results presented in the present study suggest that the replacement of the Phe residue (39) with Ile at position 80 (Fig. 5) could be one of the mutations necessary for the capability to react with 2-HIBA. Interestingly, this replacement is also found in a few other ICM-like sequences (Fig. 5). However, it is not known whether other ICMs or related enzymes can transform 2-HIBA. Thus far, substrate binding has been studied only for MCM in detail. Here the corresponding Tyr residue interacts via its hydroxyl group with the free carbonic acid of the substrate methylmalonyl-CoA (32, 50). In the ICM of Streptomyces spp., a similar role is proposed for the hydrophobic phenyl group of the Phe residue in binding isobutyryl-CoA (39). In contrast to this, the additional hydroxyl group in 2-hydroxyisobutyryl-CoA possibly resulted in a requirement of smaller residues such as those of Ile or Val. Consequently, at the moment we are investigating the reactivity of Streptomyces ICMs, as well as of other ICM-like enzymes against 2-HIBA. In addition, the contribution of the active-site residue Ile in the ICM of strain L108 to the transformation of 2-HIBA will be evaluated by site-directed mutagenesis converting this Ile residue to Phe.
Supplementary Material
Acknowledgments
This study was supported by the German Federal Ministry of Education and Research (02 WN 0348) and by the UFZ (SAFIRA2) within the METLEN project.
We thank F. Fayolle and F. Monot (IFP) for providing strain CIP I-2052. In addition, we are indebted to D. Brodkorb and A. Sturm (University Halle-Wittenberg) for 16S rRNA gene analysis; to K. Czamperla, R. Pfeiffer, and A. Steude (UFZ) for technical assistance in the enzyme assay experiments; and to B. Würz (UFZ) for excellent analytical advice. Finally, we thank U. Karlson (NERI) for help with sampling the Leuna groundwater.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achten, C., A. Kolb, and W. Puttmann. 2002. Occurrence of methyl tert-butyl ether (MTBE) in riverbank filtered water and drinking water produced by riverbank filtration. 2. Environ. Sci. Technol. 36:3662-3670. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, M. D., P. K. Busk, I. Svendsen, and B. L. Møller. 2000. Cytochromes P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. J. Biol. Chem. 275:1966-1975. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, A., R. Sharma, and U. C. Banerjee. 2002. The nitrile-degrading enzymes: current status and future prospects. Appl. Microbiol. Biotechnol. 60:33-44. [DOI] [PubMed] [Google Scholar]
- 5.Benndorf, D., I. Davidson, and W. Babel. 2004. Regulation of catabolic enzymes during long-term exposure of Delftia acidovorans MC1 to chlorophenoxy herbicides. Microbiology 150:1005-1014. [DOI] [PubMed] [Google Scholar]
- 6.Birch, A., A. Leiser, and J. A. Robinson. 1993. Cloning, sequencing, and expression of the gene encoding methylmalonyl-coenzyme A mutase from Streptomyces cinnamonensis. J. Bacteriol. 175:3511-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein, utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Breitenstein, A., J. Wiegel, C. Härtig, N. Weiss, J. R. Andreesen, and U. Lechner. 2002. Reclassification of Clostridium hydroxybenzoicum as Sedimentibacter hydroxybenzoicus gen. nov., comb. nov., and decription of Sedimentibacter saalensis sp. nov. Int. J. Syst. Evol. Microbiol. 52:801-807. [DOI] [PubMed] [Google Scholar]
- 9.Chauvaux, S., F. Chevalier, C. Le Dandec, F. Fayolle, I. Miras, F. Kunst, and P. Béguin. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chisholm, M. S. 2000. Artificial glass—the versatility of poly(methyl methacrylate) from its early exploitation to the new millennium. J. Chem. Edu. 77:841-845. [Google Scholar]
- 11.Church, C. D., P. G. Tratnyek, and K. M. Scow. 2000. Pathways for the degradation of MTBE and other fuel oxygenates by isolate PM1. ACS Preprints Extended Abstr. 40:261-263. [Google Scholar]
- 12.Deeb, R. A., K. M. Scow, and L. Alvarez-Cohen. 2000. Aerobic MTBE biodegradation: an examination of past studies, current challenges and future research directions. Biodegradation 11:171-186. [DOI] [PubMed] [Google Scholar]
- 13.European Union. 2003. On the promotion of the use of biofuels or other renewable fuels for transport. DIRECTIVE 2003/30/EC. Official J. Eur. Union 123:42-46. [Google Scholar]
- 14.Fayolle, F., A. François, L. Garnier, D. Godefroy, H. Mathis, P. Piveteau, and F. Monot. 2003. Limitations in MTBE biodegradation. Oil Gas Sci. Technol. 58:497-504. [Google Scholar]
- 15.Fayolle, F., G. Hernandez, F. Le Roux, and J.-P. Vandecasteele. 1998. Isolation of two aerobic bacterial strains that degrade efficiently ethyl t-butyl ether (ETBE). Biotechnol. Lett. 20:283-286. [Google Scholar]
- 16.Fayolle, F., J.-P. Vandecasteele, and F. Monot. 2001. Microbial degradation and fate in the environment of methyl tert-butyl ether and related fuel oxygenates. Appl. Microbiol. Biotechnol. 56:339-349. [DOI] [PubMed] [Google Scholar]
- 17.Forslund, K., M. Morant, B. Jørgensen, C. E. Olsen, E. Asamizu, S. Sato, S. Tabata, and S. Bak. 2004. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.François, A., H. Mathis, D. Godefroy, P. Piveteau, F. Fayolle, and F. Monot. 2002. Biodegradation of methyl tert-butyl ether and other fuel oxygenates by a strain, Mycobacterium austroafricanum IFP 2012. Appl. Environ. Microbiol. 68:2754-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.François, A., L. Garnier, H. Mathis, F. Fayolle, and F. Monot. 2003. Roles of tert-butyl formate, tert-butyl alcohol, and acetone in the regulation of methyl tert-butyl ether degradation by Mycobacterium austroafricanum IFP 2012. Appl. Microbiol. Biotechnol. 62:256-262. [DOI] [PubMed] [Google Scholar]
- 20.Hanson, J. R., C. E. Ackerman, and K. M. Scow. 1999. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl. Environ. Microbiol. 65:4788-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzinger, P. B., K. McClay, S. Vainberg, M. Tugusheva, C. W. Condee, and R. J. Steffan. 2001. Biodegradation of methyl tert-butyl ether by a pure bacterial culture. Appl. Environ. Microbiol. 63:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Perez, G., F. Fayolle, and J.-P. Vandecasteele. 2001. Biodegradation of ethyl tert-butyl ether (ETBE), methyl tert-butyl ether (MTBE) and tert-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 23.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 25.Holowach, L. P., G. W. Swift, S. W. Wolk, and L. Klawiter. 1994. Bacterial conversion of a waste stream containing methyl-2-hydroxyisobutyric acid to biodegradable polyhydroxyalkanoate polymers, p. 202-211. In M. L. Fishman, R. B. Friedman, and S. J. Huang (ed.), Polymers from agricultural coproducts. ASC Symposium Series 575. ASC, Washington, D.C.
- 26.Hristova, K., B. Gebreyesus, D. Mackay, and K. M. Scow. 2003. Naturally occurring bacteria similar to the methyl tert-butyl ether (MTBE)-degrading strain PM1 are present in MTBE-contaminated groundwater. Appl. Environ. Microbiol. 69:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman, M., K. Glover, A. House, E. Johnson, and C. Smith. 2004. Physiological and enzymatic diversity of aerobic MTBE biodegradation processes, p. 39-43. In Proceedings of the Second European Conference on MTBE. CSIC, Barcelona, Spain.
- 28.Kane, S. R., H. R. Beller, T. C. Legler, C. J. Koester, H. C. Pinkart, R. U. Halden, and A. M. Happel. 2001. Aerobic biodegradation of methyl tert-butyl ether by aquifer bacteria from leaking underground storage tank sites. Appl. Environ. Microbiol. 67:5824-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J., M. Hetzel, C. D. Boiangiu, and W. Buckel. 2004. Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of α-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 28:455-468. [DOI] [PubMed] [Google Scholar]
- 30.Lee, I.-Y., S. L. Nissen, and J. P. N. Rosazza. 1997. Conversion of β-methylbutyric acid to β-hydroxy-β-methylbutyric acid by Galactomyces reessii. Appl. Environ. Microbiol. 63:4191-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurements with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 32.Mancia, F., G. A. Smith, and P. R. Evans. 1999. Crystal structure of substrate complexes of methylmalonyl-CoA mutase. Biochemistry 38:7999-8005. [DOI] [PubMed] [Google Scholar]
- 33.Martens, J.-H., H. Barg, M. J. Warren, and D. Jahn. 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58:275-285. [DOI] [PubMed] [Google Scholar]
- 34.Moran, M. J., J. S. Zogorski, and P. J. Squillace. 2005. MTBE and gasoline hydrocarbons in ground water of the United States. Ground Water 43:615-627. [DOI] [PubMed] [Google Scholar]
- 35.Nakatsu, C. H., K. Hristova, S. Hanada, X.-Y. Meng, J. Hanson, K. M. Scow, and Y. Kamagata. 4. March 2005. Methylibium petrophilum PM1T gen. nov., sp. nov., a new methyl tert-butyl ether (MTBE) degrading methylotroph of the beta-Proteobacteria. Int. J. Syst. Evol. Microbiol. [Online.] doi: 10.1099/ijs. 0.63524-0. [DOI] [PubMed]
- 36.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 37.Piveteau, P., F. Fayolle, J. P. Vandecasteele, and F. Monot. 2001. Biodegradation of tert-butyl alcohol and related xenobiotics by a methylotrophic bacterial isolate. Appl. Biotechnol. Microbiol. 55:369-373. [DOI] [PubMed] [Google Scholar]
- 38.Poitrat, E. 1999. The potential of liquid biofuels in France. Renew. Energy 16:1084-1089. [Google Scholar]
- 39.Ratnatilleke, A., J. W. Vrijbloed, and J. A. Robinson. 1999. Cloning and sequencing of the coenzyme B(12)-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J. Biol. Chem. 274:31679-31685. [DOI] [PubMed] [Google Scholar]
- 40.Rohwerder, T., and W. Sand. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699-1709. [DOI] [PubMed] [Google Scholar]
- 41.Salanitro, J. P. 1995. Understanding the limitations of microbial metabolism of ethers used as fuel octane enhancers. Curr. Opin. Biotechnol. 6:337-340. [Google Scholar]
- 42.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos, P. M., D. Benndorf, and I. Sá-Correia. 2004. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4:2640-2652. [DOI] [PubMed] [Google Scholar]
- 44.Smith, C. A., K. T. O′Reilly, and M. R. Hyman. 2003. Characterization of the initial reactions during the cometabolic oxidation of methyl tert-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl. Environ. Microbiol. 69:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Squillace, P. J., J. F. Pankow, N. E. Korte, and J. S. Zogorski. 1997. Review of the environmental behavior and fate of methyl tert-butyl ether. Environ. Toxicol. Chem. 16:1836-1844. [Google Scholar]
- 46.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stocking, A. J., R. A. Deeb, A. E. Flores, W. Stringfellow, J. Talley, R. Brownell, and M. C. Kavanaugh. 2000. Bioremediation of MTBE: a review from a practical perspective. Biodegradation 11:187-201. [DOI] [PubMed] [Google Scholar]
- 48.Trevor, C. C., and A. Punita. 1999. Methylmalonyl-CoA mutase encoding gene of Sinorhizobium meliloti. Gene 226:121-127. [DOI] [PubMed] [Google Scholar]
- 49.U.S. Environmental Protection Agency. 1997. Drinking water advisory: consumer acceptability advice and health effects analysis on methyl tert-butyl ether (MTBE). EPA-822-F-97-008. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 50.Vlasie, M. D., and R. Banerjee. 2003. Tyrosine 89 accelerates Co-carbon bond homolysis in methylmalonyl-CoA mutase. J. Am. Chem. Soc. 125:5431-5435. [DOI] [PubMed] [Google Scholar]
- 51.Wang, W., M. Scali, R. Vignali, A. Spadafora, E. Sensi, S. Mazzuca, and M. Cresti. 2003. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24:2369-2375. [DOI] [PubMed] [Google Scholar]
- 52.Zerbe-Burkhardt, K., A. Ratnatilleke, N. Philippon, A. Birch, A. Leiser, J. W. Vrijbloed, D. Hess, P. Hunziker, and J. A. Robinson. 1998. Cloning, sequencing, expression, and insertional inactivation of the gene for the large subunit of coenzyme B12-dependent isobutyryl-CoA mutase from Streptomyces cinnamonensis. J. Biol. Chem. 273:6508-6517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.