Abstract
We employed culture-dependent and -independent techniques to study microbial diversity in Lake Chaka, a unique hypersaline lake (32.5% salinity) in northwest China. It is situated at 3,214 m above sea level in a dry climate. The average water depth is 2 to 3 cm. Halophilic isolates were obtained from the lake water, and halotolerant isolates were obtained from the shallow sediment. The isolates exhibited resistance to UV and gamma radiation. Microbial abundance in the sediments ranged from 108 cells/g at the water-sediment interface to 107 cells/g at a sediment depth of 42 cm. A major change in the bacterial community composition was observed across the interface. In the lake water, clone sequences affiliated with the Bacteroidetes were the most abundant, whereas in the sediments, sequences related to low G+C gram-positive bacteria were predominant. A similar change was also present in the archaeal community. While all archaeal clone sequences in the lake water belonged to the Halobacteriales, the majority of the sequences in the sediments were related to those previously obtained from methanogenic soils and sediments. The observed changes in the microbial community structure across the water-sediment interface were correlated with a decrease in salinity from the lake water (32.5%) to the sediments (approximately 4%). Across the interface, the redox state also changed from oxic to anoxic and may also have contributed to the observed shift in the microbial community.
Hypersaline lakes are considered extreme environments for microbial life (39) because of the effects of salt on water activity and balance. Saline environments are globally distributed on Earth. Halophiles thrive in hypersaline niches and include prokaryotes and eukaryotes (11). Among halophilic microorganisms are found a variety of heterotrophic and methanogenic archaea; photosynthetic, lithotrophic, and heterotrophic bacteria; and photosynthetic and heterotrophic eukaryotes. Previous studies have shown that the taxonomic diversity of microbial populations in terrestrial saline and hypersaline environments is low (11, 35) and that, in general, microbial diversity decreases with increased salinity (36).
The study on microbial diversity in saline environments is important for two reasons. First, some of the earliest microbial life on Earth might have been halophilic because of high salt and organic compound concentrations in evaporitic environments, and thus research on microbial survivability and adaptation in saline environments bears relevance to our understanding of the early evolution of life and the biosphere on Earth (25). Understanding diversity within an environmental context is a necessary first step in studying the survivability and adaptation of halophiles at different levels of tolerance. Second, because of the presence of hypersaline conditions on Mars (7, 32), studies of microbial diversity in terrestrial saline environments may have implications for the possibility of extinct and/or extant life on Mars.
Microbial diversity in most hypersaline environments is often studied using culture-dependent and -independent (small-subunit [SSU] rRNA gene analysis) methods (40). A variety of hypersaline environments have been surveyed for microbial diversity such as the Great Salt Lake in Utah, the Great Salt Plains of Oklahoma, the Dead Sea, the Mediterranean Sea, the Solar Lake in Sinai, Egypt, Antarctic hypersaline lakes, deep-sea brine sediments, and various salterns (evaporation ponds for salt recovery) (31, 38, 48). These previous studies have established that halophiles are distributed in both the Archaea and Bacteria domains. Within the domain Archaea, halophiles are classified in the Halobacteriaceae, the Methanospirillaceae, and the Methanosarcinaceae. All members of the family Halobacteriaceae are extreme halophiles (3 to 4 M salt) and are chemoheterotrophic, and most members are aerobic. Halophiles are also widely spread within the domain Bacteria. Unlike archaea, most halophilic bacteria can live only at moderate salinity (up to 2.5 M salt). Bacterial halophiles vary widely in physiological properties, including aerobic and anaerobic chemoheterotrophs, photoautotrophs, photoheterotrophs, and chemolithotrophs.
Despite these previous studies, our understanding of microbial diversity in hypersaline environments is still limited, especially in athalassohaline lakes (a saline lake not of marine origin but evolved from the evaporation of freshwater) at high elevation. Lake Chaka in northwestern China represents an ideal site for studying halophile diversity in such an environment. The lake is located on the Tibetan Plateau at an elevation of 3,214 m above sea level and possesses a salinity of 32.5% (or greater, depending on the season). The high salinity is developed via progressive evaporation of freshwater in the lake. The combination of high elevation (and thus high UV intensity) and salinity makes it an extreme environment. In addition, there exists a salinity gradient in the lake sediments, 32.5% or higher at the water-sediment interface to 0% at the 8-m depth (H. Jiang et al., unpublished data). This natural gradient is a result of progressive evaporation in the region over the past 50,000 years.
The goal of this research was to assess microbial diversity and abundance in the lake water and the shallow sediments (top 42 cm) in Lake Chaka and to correlate it with the dynamic environmental conditions. We integrated geochemical and microbiological approaches, including lake and pore water chemistry, sediment mineralogy and geochemistry, and culture-independent (SSU rRNA gene analysis) and -dependent microbiology. Bacterial halophiles were isolated under salinities that were similar to those measured in the lake water and sediment. We observed major differences in the microbial community structure between the lake water and the sediments, and these differences could be correlated with a gradient of geochemical characteristics.
MATERIALS AND METHODS
Description of the study site.
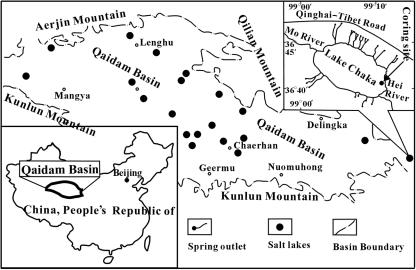
Lake Chaka (36°18′ to 36°45′N, 99°02′ to 99°12′E) (Fig. 1) is a shallow salt lake in northwestern China at an elevation of 3,214 m above sea level. It possesses high salinity (32.5%). It is an elliptic and closed drainage basin, trending northwest-southeast in parallel to nearby mountains, and is located at the southern corner of the Qaidam Basin, where a semiarid continental climate dominates. The Lake Chaka basin is approximately 80 km long and 30 km wide. Strong evaporation and little precipitation in this area (2,264 mm of evaporation versus 224 mm of rainfall/year) have resulted in a nearly dry lake and high salinity (28). The water depth and coverage of water in the lake vary seasonally. The area of liquid water in the high-water period (summer) can reach 104 km2, with an average water depth of 2 to 3 cm. The area of liquid water and water depth significantly decrease in the low-water period (winter). Average water temperature is 4.2°C with −6 to −8°C in winter and 6 to 20°C in summer. Upper Pleistocene and Holocene rocks occur widely in the basin. The lake became progressively saline in the last 50,000 years. Liu et al. (29) reported that the majority of the water supply (80%) to the lake is from rivers and spring water in the surrounding area. The authors reported that the source waters contain high concentrations of cations and anions as follows (mg/liter): Na+ (145 to 343), K+ (3 to 9), Mg2+ (26 to 72), Ca2+ (52 to 81), Cl− (188 to 386), SO42− (112 to 330), CO32− (10 to 12), and HCO3− (218 to 270).
FIG. 1.
Location of Lake Chaka, northwestern China. Lake Chaka is a hypersaline lake on the southeastern corner of the Qaidam Basin. The coring site is shown on the map.
Field measurements and sampling.
Field measurements and sampling were conducted in August 2003. pH, temperature, and salinity were measured with pH and conductivity probes (water depth of 2 to 3 cm). Field colorimetric Hach kits were used to measure soluble Fe (Fe2+), sulfide, sulfate, phosphate, nitrite, and nitrate concentrations. Water samples were subsequently collected with 50-ml sterile centrifuge tubes. A sediment core of 42 cm by 8 cm (length by diameter) was collected using a gravity coring device. After collection, the samples were immediately stored in a refrigerator at 4°C. Within 2 days, the samples were shipped cold (4°C) to China University of Geosciences in Beijing and then shipped in a cooler (regular ice) to Miami University in Ohio. For the lake water sample, enrichments were set up and clone libraries were constructed. The sediment core was dissected into 2-cm-long sediment subsamples inside a glove box filled with 95% N2 and 5% H2 (Coy Laboratory Products, MI). The external layers of the sediment subsamples were removed using sterile tools. Five subsamples, designated LCKS0 (the water-sediment interface), LCKS10 (depth of 10 to 12 cm), LCKS20 (20 to 22 cm), LCKS30 (30 to 32 cm), and LCKS40 (40 to 42 cm), were geochemically and microbiologically analyzed. The analyses included measurements of total organic carbon (TOC), mineralogy by X-ray diffraction (XRD), total microbial counts by acridine orange direct counting (AODC), and microbial diversity by SSU rRNA gene analysis. Phospholipid fatty acid (PLFA) analysis was performed for LCKS0, LCKS20, and LCKS40. Enrichments and isolations were performed for LCKS0. Samples were coded as follows, with LCKS20 as an example: LCKS, Lake Chaka sediment; 20, depth in centimeters. Lake water samples were coded as LCKW.
Laboratory chemical analyses of water samples.
Anion and cation compositions of the lake water were analyzed by high-performance liquid chromatography (HPLC) and direct current plasma emission spectrometry (DCP). For determination of TOC content, the lake water sample was acidified to pH 3 to remove inorganic carbon, followed by analysis with an organic carbon analyzer (TOC-5000A; Shimadzu). Because of the paucity of pore water in the sediments, “artificial pore water” was created and analyzed for acetate, lactate, formate, and sulfate concentrations. The “artificial pore water” was created by leaching 1 g of each sediment subsample with 50 ml of deionized water for 4 h, followed by centrifugation. The leaching step was repeated until all chemical species were leached. Subsequent analyses showed that one step was sufficient to completely leach acetate, lactate, and formate, but multiple steps (three to four steps) were required to leach sulfate. For analyses, the supernatants from multiple steps were combined. Acetate, lactate, formate, and sulfate concentrations were measured using HPLC. An IonPac AS11-HC column (4 by 250 mm) was used for acetate, lactate, and formate, and an IonPac AS14 column (4 by 250 mm) was used for sulfate. The concentrations were reported as micromolars per gram of wet sediment and were assumed to be proportional to those in natural pore water.
Sediment geochemistry.
XRD was employed to analyze the five sediment subsamples (LCKS0, LCKS10, LCK20, LCKS30, and LCK40) for mineralogy by following a previously used procedure. Concentrations of TOC, total nitrogen, bioavailable phosphorus, and soluble salt were determined in the Service Testing and Research laboratory of the Ohio State University. TOC content was analyzed with the dry combustion method, and total inorganic carbon was determined by U.S. Environmental Protection Agency method 9060A. These methods are available online at http://www.oardc.ohio-state.edu/starlab/references.htm. Bioavailable phosphorus was analyzed by following a previously published method (26). The amount of soluble salts in the sediments was measured as the conductivity of the solution (mS/cm) when a certain amount of sediment (typically, 5 g) was mixed with an equal amount of water (5 g), following the methods described in Rhoades (41).
Total microbial counts.
AODC was performed for both the lake water and the sediments to determine the total microbial counts. For the lake water sample, 10 ml of water was stained and counted (12). For the sediment samples, microbial cells were first detached from the sediments according to a previous protocol (5) and then stained and counted as described above.
PLFA analyses of the sediment samples.
Three sediment subsamples (LCKS0, LCKS20, and LCKS40) were chosen for PLFA analysis and shipped frozen (−80°C) to Microbial Insights, Inc. (Rockford, TN). PLFAs were analyzed after extraction of the total lipid (53) and separation of the polar lipids by column chromatography (16). The polar lipid fatty acids were derivatized to fatty acid methyl esters, which were quantified using gas chromatography (42). Fatty acid structures were verified by chromatography-mass spectrometry and equivalent-chain-length analysis.
Enrichment and isolation of microbes present in the lake water and the sediment.
Enrichment experiments were performed for the lake water and one sediment sample (LCKS0, the water-sediment interface sample) with a modified Kauri medium (22) for halophiles. Diluted modified R2A (DMR2A) medium was used for nitrate reducers, and modified MB (MMB) medium (Difco) was used for general heterotrophs in the lake water. The enrichment experiments were performed in 28-ml capacity Balch culture tubes incubated at 30°C in a water bath with 60 rpm shaking. The pH value for all three media was 7.0. Gradients of NaCl salt (5%, 10%, 15%, 20%, and 25%) were used in multiple tubes to target halophiles with the modified Kauri medium. DMR2A was made as previously described except that all final concentrations were diluted fivefold (13). MMB medium contained the following, per liter: 0.5 g of sodium acetate, 0.5 g of yeast extract, 4.7 g of Middlebrook 7H9 (Difco), 0.5 g of Casamino Acids, 0.5 g of sodium thiosulfate, 10 ml of mineral solution, and 2 g of NaCl. The mineral solution was the same as that used for DMR2A. Positive enrichments were transferred three times. The enrichment cultures were streaked onto agar plates consisting of the original enrichment medium supplemented with 2% agar. The plating step was repeated three times for isolates to ensure purity. The colonies from the final set of agar plates were grown in liquid medium, preserved in 35% glycerol, and frozen at −80°C for later analyses.
Physiological testing of halophilic isolates.
The influence of temperature on growth was studied by incubation of inoculated medium (the modified Kauri medium) at temperatures between 10°C and 50°C with shaking (210 rpm) for 96 h. The influence of pH on growth was studied in the same medium, with the pH value varying from 4.0 to 9.0 (adjusted by HCl or NaOH). The culture tubes were incubated at 37°C (the optimum growth temperature for the isolates) with shaking (210 rpm). The cell growth was monitored with a spectrophotometer (600 nm). Experiments were performed to test for the NaCl and MgCl2 requirement of the isolates by changing the NaCl and MgCl2 concentrations in the Kauri medium (0 to 35% with 5% increments for the NaCl test and 0 to 25% with 2.5% increments for the MgCl2 test). The culture tubes were incubated at 37°C and pH 7.0 with shaking (210 rpm). The cell growth was monitored as above.
To test UV resistances of the halophilic isolates, two UV light sources with different wavelengths were used: 312 nm (UVB; EB28OC, 620 μm/cm2 at a 6-in. distance; Spectronics Corp.) and 254 nm (UVC; UVG-11, 120 μm/cm2 at a 3-in. distance; VCP Inc.). Cells (108 cells/ml) were irradiated at different doses of UV radiation using a previously described method (2). The viable cell numbers were determined by plate counts on agar plates (the Kauri growth medium supplied with 2% agar). The survival rate was calculated by dividing the number of the remaining cells by the initial number of cells. To test the resistance of the isolates to gamma radiation, cells were exposed to 0.0 to 7.0 kGy of gamma rays at room temperature using a 3,600-Ci 60Co source located at the Ohio State University Nuclear Reactor Laboratory at a dosage rate of 1.29 kGy/h. The number of viable cells was counted immediately postirradiation with plate counts (the Kauri growth medium supplied with 2% agar). The survival rate was calculated in a similar manner.
PCR amplification and sequence determination of the isolates.
For the isolates obtained from the DMR2A and MMB media, the following PCR amplification procedure was used. Cell lysates were made of isolated microorganisms for the PCR amplification of the SSU rRNA genes by boiling cells suspended in Tris-EDTA buffer for 5 min at 100°C. PCRs were treated with SeqMix (Q-Biogene, Irvine, CA) prior to sequencing reactions according to the manufacturer's instructions. The SSU rRNA gene was amplified with the universal primers FD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1540R (5′-GGAGGTGWTCCARCCGC-3′) as previously described (54).
For the isolates obtained using the modified Kauri medium, the following procedure was used. Freshly grown isolates were suspended in boiling water for 10 min. The SSU rRNA gene was amplified with the bacterial forward primer Bac27F (5′-AGAGTTTGGATCMTGGCTCAG-3′) and universal reverse primer Univ1492R (5′-CGGTTACCTTGTTACGACTT-3′) or with archaeon-specific primers Arch21F (5′-TTCYGGTTGATCCYGCCRGA-3′), 925R (5′-CCGTCAATTCMTTTRAGTTT-3′), and Univ1492. A typical PCR mixture (25 μl in volume) contained the following components: 10 mM Tris, pH 8.3, 50 mM KCl, 1.5 nM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.2 μM concentration of each primer, and 1.25 U of Taq DNA polymerase. The following standard conditions were used for bacterial 16S rRNA gene amplification: initial denaturation at 95°C for 5 min; 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 55°C), and extension (2 min at 72°C); and a final extension at 72°C for 7 min. The standard conditions for amplification of archaeal 16S rRNA genes were the following: initial denaturation at 95°C for 5 min; 45 cycles of denaturing (30 s at 94°C), annealing (30 s at 54°C), and extension (2 min at 72°C); and a final extension at 72°C for 10 min. The PCR products were purified with a GeneClean Turbo kit (Qbiogene Inc., Irvine, CA) according to the manufacturer's suggested protocol.
For the sequence determination with ET Dye chemistry (Amersham Pharmacia Biotech Inc., Piscataway, NJ), primers FD1, 529R (5′-CGCGGCTGCTGGCAC-3′) and 1540R were used for the isolates obtained with DMR2A and MMB media. Primers Bac27F and Arch21F were used for the isolates obtained with the modified Kauri medium. All sequences were determined with an automatic 3100 DNA sequencer. The sequences were tested for chimeras by using the Ribosomal Database Project Chimera-Check program and were aligned with ClustalW. Phylogenetic analyses of partial 16S rRNA gene sequences were conducted using the MEGA (molecular evolutionary genetics analysis) program, version 2.1. Neighbor-joining phylogenies were constructed from dissimilar distances and pairwise comparisons with the Jukes-Cantor distance model.
Clone library construction for lake water and sediment samples.
Genomic DNA in the lake water and the sediment samples (0.5 to 0.7 g) (LCKS0, LCKS10, LCKS20, LCKS30, and LCKS40) was extracted and purified with an Ultra Clean Soil DNA Isolation Kit (MoBio Laboratories, Inc., Solana Beach, CA). Purified DNA was PCR amplified according to the procedure of the Failsafe Kit (Epicenter Biotechnologies, Madison, WI). The PCR conditions were the same as those for the isolates (from the modified Kauri medium). Primer sequences for bacteria were Bac27F and Univ1492 and those for archaea were Arch21F and Arch958R (5′-YCCGGCGTTGAMTCCATTT-3′).
The PCR product was ligated into the pGEM-T vector (Promega Inc., Madison, WI) and transformed into Escherichia coli DH5α competent cells. The transformed cells were plated on Luria-Bertani plates containing 100 μg/ml of ampicillin, 80 μg/ml of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated overnight at 37°C. Gene clone libraries of 16S rRNA were constructed, and 40 to 50 randomly chosen colonies per sample were analyzed for insert 16S rRNA gene sequences. Plasmid DNA containing inserts of the 16S rRNA gene was prepared using a QIAprep Spin miniprep kit (QIAGEN, Valencia, CA). Sequencing reactions were carried out with primer Bac27F for bacteria and Arch21F for archaea with a DYEnamic ET terminator cycle sequencing ready reaction kit (Amersham Biosciences, Piscataway, NJ). The 16S rRNA gene sequence was determined with an ABI 3100 automated sequencer. Sequences were typically ∼600 to 700 bp long. Phylogenetic analyses were carried out in the same manner as above.
Statistical analysis and sequence population diversity.
We followed the approach of Humayoun et al. (17) for these analyses. One major assumption was that sequences with similarities of greater than 97% were considered to represent the same phylotypes for the reasons stated in that study. Coverage (C) was calculated as follows: C = 1 − (n1/N), where n1 is the number of phylotypes that occurred only once in the clone library and N is the total number of clones analyzed. Rarefaction curves were constructed using software available online at http://www.uga.edu/∼strata/software.html.
LIBSHUFF (version 1.2) analysis was performed to compute the homologous and heterologous coverage within and between clonal libraries (45). The analysis estimates the similarity between clonal libraries from two different samples based upon evolutionary distances of all sequences. Thus, the sampled diversity of a community can be directly compared to another community. The predicted coverage of a sampled library is denoted by the homologous coverage, and the heterologous coverage is the observance of a similar sequence in a separate library. The values are reported over a sequence similarity range or evolutionary distance based upon a distance matrix. Analyses were performed according to specified directions given at the LIBSHUFF website (http://www.arches.uga.edu/∼whitman/libshuff.html).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers DQ129871 to DQ129877 and DQ247815 to DQ247820 for the bacterial isolate sequences from the lake water, DQ395131 for the bacterial isolate sequence from the sediment (LCKS0), DQ129878 to DQ129952 for the bacterial clone sequences, and DQ129953 to DQ129989 for the archaeal clone sequences.
RESULTS
Lake water chemistry.
Field measurements in August 2003 showed that salinity was 32.5%, with a pH of 7.4 and temperature of 16 to 17°C. The colorimetric measurements indicated the following concentrations in the lake water (μg/g): Fe2+, 0.3; sulfide, 1; phosphate, 3; and nitrite, 2. The DCP analyses of the lake water determined the concentrations of major cations (mg/liter) as follows: Li+ (11), Na+ (73 and 311), NH4+ (25), Mg2+ (33 and 179), K+ (4 and 957), Ca2+ (326), and Fe (2). HPLC analyses determined the concentrations of major anions (mg/liter) as Cl− (181 and 586), SO42− (31 and 350), Br− (72), F− (292), HCO3− (170), NO3− (7), and PO43− (2).
Sediment properties and pore water geochemistry.
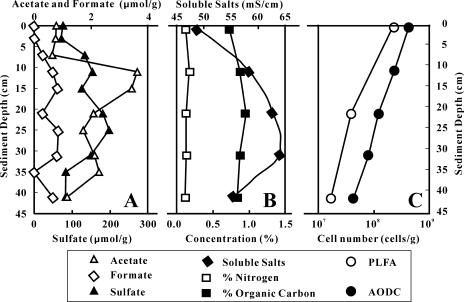
Although a direct measurement of the redox state was not possible, visual observation indicated that the lake sediments were anaerobic. Gas bubbles were seen to emerge from the sediment, and an odor of hydrogen sulfide was detected. The sediments were dark. Thus, we inferred that the redox boundary was at the water-sediment interface (further confirmed below). XRD analysis of the sediment samples identified the major minerals as halite, quartz, kaolinite, and muscovite. Acetate, formate, and sulfate concentrations in the artificial pore water were 0.5 to 3.8, 0 to 1, and 70 to 200 μmol/g (wet sediment), respectively (Fig. 2A). Lactate was not detectable. The TOC and total nitrogen content in the sediments were 0.8 to 1% and 0.11 to 0.17%, respectively (Fig. 2B). Bioavailable phosphorus was <1 μg/g except for LCK0 (∼1 μg/g). Soluble salt concentration in the sediments was in the range of 49 to 64 mS/cm (or ∼3 to 4%, given the conversion of 1 mS/cm as 640 ppm).
FIG. 2.
(A) Depth distributions of acetate, formate, and sulfate concentration in the Lake Chaka sediments as determined in the “artificial pore water” created by leaching the lake sediments with distilled water. (B) Distribution of total nitrogen and organic carbon in the lake sediments. (C) Microbial abundance distribution in the lake sediments as determined by AODC and PLFA analysis. Single samples were used, and so sample-to-sample variability assessment was not possible. Single samples were measured more than once, and analytical errors were smaller than the symbol sizes.
AODC and PLFA data.
Total cell counts in the lake water were 4.8 × 106 cells/ml. The total cell counts were higher in the sediments, ranging from 4.0 × 108 cells/g (dry weight) at the water-sediment interface to 4.2 × 107 cells/g at the 42-cm depth (Fig. 2C). Viable bacterial abundance in the sediments as determined by the total PLFA concentration (assuming a laboratory-determined conversion factor of 20,000 cells/pmol [9]) was consistently lower than the AODC-determined cell abundance, ranging from 2.1 × 108 cells/g of dry weight for LCKS0 to 1.7 × 107 cells/g for LCKS40 (Fig. 2C). The difference between the AODC counts and PLFA bacterial abundance could be taken to indicate the archaeal abundance.
PLFA profiles for the sediment samples indicated that the proportion of terminally branched saturated fatty acids, indicative of Firmicutes or anaerobic gram-positive bacteria, increased from approximately 22% in LCKS0 to 33% in LCKS40 (Table 1). The proportion of monoenoic fatty acids, indicative of gram-negative bacteria, decreased from approximately 30% in LCKS0 to 20% in LCKS40. The proportion of the characteristic biomarker for anaerobic metal reducers (branched monoenoic PLFA) increased from 0.8% in LCKS0 to 1.3% in LCKS40. Sulfate-reducing biomarkers (mid-chain branched PLFA) remained the same at approximately 3 to 4% among the three samples. Eukaryotic biomarkers in LCKS0 and LCKS40 decreased from 6% in LCKS0 to 3% in LCKS40. Physiological status biomarkers indicated that the samples were undergoing a similar and moderate level of starvation. Sample LCKS40 showed a moderate level of microbial response to environmentally induced stress (i.e., trans/cis ratio of 0.57).
TABLE 1.
PLFA composition of sediment samples from Lake Chaka
| Sample | Abundance (no. of cells/g) | % of total PLFAsa
|
Physiological status
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TerBrSat | Mono | BrMono | MidBrSat | Nsat | Polyenoic | Starved (cy/cis ratio)b | Stress (trans/cis ratio) | ||
| LCK0 | 2.05 × 108 | 22.1 | 29.7 | 0.8 | 3.6 | 37.4 | 5.9 | 0.47 | 0.17 |
| LCK20 | 3.80 × 107 | 22.5 | 28.1 | 0.7 | 3.1 | 43.1 | 2.5 | 0.47 | 0.29 |
| LCK40 | 1.65 × 107 | 32.5 | 19.2 | 1.3 | 3.7 | 41.0 | 2.9 | 0.43 | 0.57 |
PLFA type: TerBrSat, terminally branched saturated; Mono, monoenoic; BrMono, branched monoenoic; MidBrDat, mid-chain branched saturated; Nsat, normal saturated.
cy, cyclopropyl.
Isolate characteristics.
Bacteria were isolated from both the lake water (n = 10) and the sediment (n = 8, from LCKS0) with three different media. Archaea were isolated from the lake water (n = 3) with one medium (the modified Kauri medium). Bacterial isolates with significant sequence similarity to predominant clones were not obtained from the lake water sample. Archaeal isolates with moderate sequence similarity (∼90% similarity) to clones were obtained. All eight bacterial isolates from the sediment sample LCKS0 were similar to each other, and they were closely related (∼97% similarity) to several clones from the same sample. Six bacterial isolates from the lake water and eight from the sediment were related to the genus Holomonas of the Gammaproteobacteria group (Table 2). Two bacterial isolates from the water sample could be classified as Firmicutes, and another was classified as Actinobacteria. Three archaeal isolates from the water sample were closely affiliated with several species of the genus Haloarcula (Table 2).
TABLE 2.
Microbial isolates obtained from the lake water and sediment samples
| Isolate name and type | % Similarity to closest match | Closest match (GenBank accession no.) |
|---|---|---|
| Bacteria | ||
| LCKW-IsolateE, I, V, and U | 99 | Halomonas variabilis (AY505527) |
| LCKW-Isolate10A and 10N | 99 | Halomonas salina (AY505525) |
| LCKW-Isolate15N | 98 | Bacillus vedderi (Z48306) |
| LCKW-IsolateG | 100 | Gracilibacillus sp. strain BH235 (AY762980) |
| LCKW-IsolateW | 98 | Gracilibacillus sp. strain BH235 (AY762980) |
| LCKW-Isolate25N | 100 | Arthrobacter sp. strain AS18 (AY371223) |
| LCKS0-Isolate1a | 99 | Halomonas hydrothermalis (AF212218) |
| Archaea | ||
| LCKW-Isolate15A | 99.5 | Haloarcula marismortui (X61689) |
| LCKW-Isolate20A | 99.5 | Haloarcula marismortui (X61689) |
| LCKW-Isolate20N | 100 | Haloarcula argentinensis (D50849) |
Eight bacterial isolates were obtained from LCKS0, and only one representative was listed.
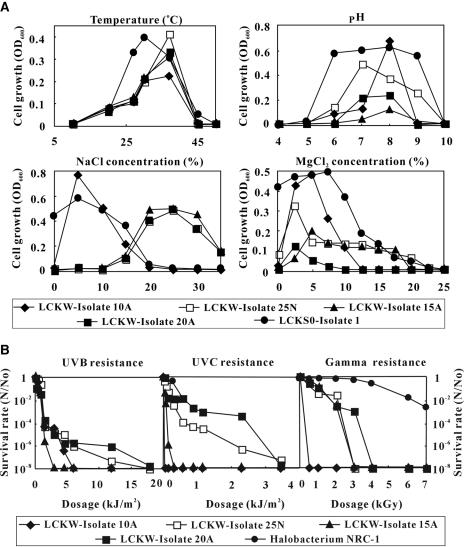
Physiological tests were performed for some representative isolates. All water isolates tested were aerobic and exhibited an optimum growth temperature of 37°C, pH of 7 to 8, and MgCl2 tolerance of 2.5 to 5% (Fig. 3A). The isolates exhibited a range of optimal salinity requirements. Whereas bacterium 10A exhibited the maximum growth rate at ∼5% salinity, bacterium 25N and archaea 15A and 20A exhibited an optimal salinity of ∼25%. Likewise, isolates E and W (representative of I, U, V, and G) could grow in the presence of up to 20% sodium chloride, but the maximal growth rate was observed in 5%. Growth was not observed when the sodium chloride concentration was above 25% (data not shown).
FIG. 3.
(A) Cell growth as a function of temperature, pH, and concentrations of NaCl and MgCl2. LCKW-Isolate10A and LCKW-Isolate25N are bacterial isolates from the lake water. LCKW-Isolate15A and LCKW-Isolate20A are archaeal isolates from the lake water. LCKS0-Isolate1 is a bacterial isolate from the sediment (LCKS0). (B) Survival rate of the bacterial and archaeal isolates in response to increasing UV and gamma radiation. The UV and gamma radiation resistance data for Halobacterium strain NRC-1 (2, 24) are plotted for comparison. N/N0, the number of cells remaining after irradiation/initial number of cells.
The isolates exhibited significant resistance to UV and gamma radiation (Fig. 3B), but the resistance levels were lower than that of Halobacterium strain NRC-1 (2, 24), an extremely halophilic archaeon. The bacterial isolate 10A, which exhibited a low optimum salinity requirement (5%), showed a low resistance to UV and gamma radiation. One representative bacterial isolate from the sediment showed a slightly lower optimum growth temperature of 30°C, optimum pH of 8, NaCl concentration of 5%, and MgCl2 concentration of 7.5%. In contrast to the isolates from the lake water, the sediment isolate did not require any salt for growth.
Bacterial diversity. (i) Phototrophic bacteria.
Three sequences from the LCKW library were related (99%) to phototrophic bacterium BN 9624 isolated from Abu Gabara Lake (Wadi Natrun, Egypt), which exhibits 36% salinity (19).
(ii) Alphaproteobacteria.
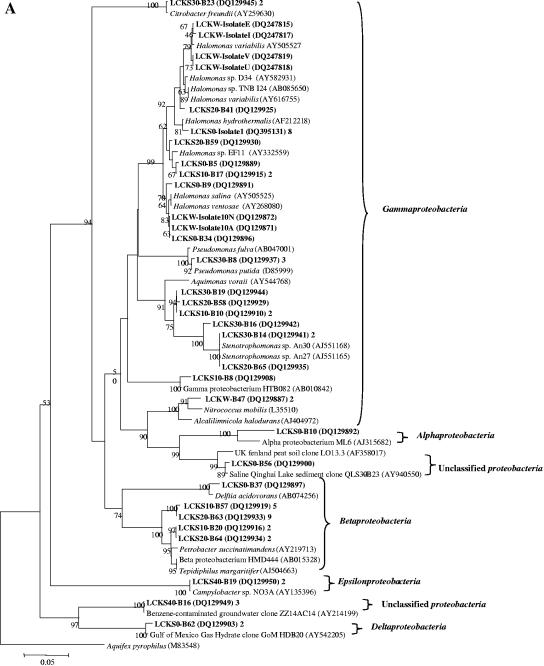
One sequence showed 94% similarity to Alphaproteobacteria strain ML6 (AJ315682) (Fig. 4A) from Mahoney Lake, south central British Columbia (56). Mahoney Lake is a meromictic saline lake (0.4 to 4%) with a surface pH of 9.0 and a pH of 8.0 near the chemocline. ML6 is an aerobic phototrophic bacterium that can use various organic carbon sources.
FIG.4.
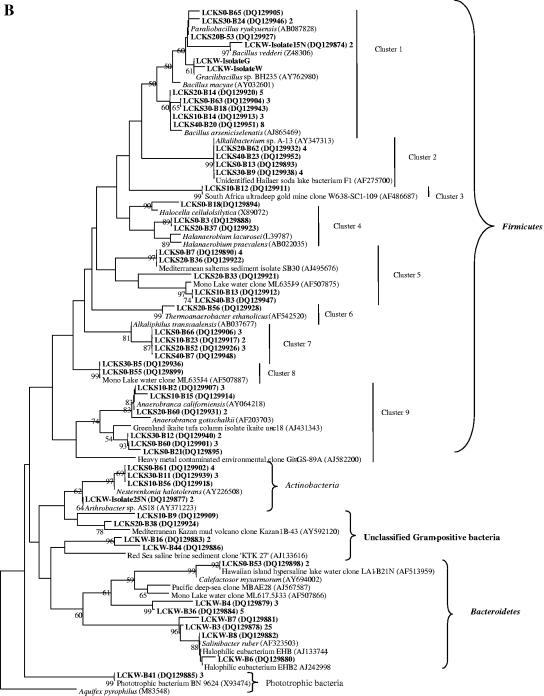
(A) Neighbor-joining tree (partial sequences, ∼600 bp) showing the phylogenetic relationships of bacterial 16S rRNA gene sequences cloned from the Lake Chaka samples to closely related sequences from the GenBank database. One representative clone type within each phylotype is shown, and the number of clones within each phylotype is shown at the end (after the GenBank accession number). If there is only one clone with a given phylotype, the number 1 is omitted. Clone sequences from this study are coded as follows for the example of LCKS30-B9: LCKS, Lake Chaka sediment; 30, sample depth in centimeters; B, bacterium; 9, clone number. The isolates were obtained from the lake water only. They are coded as described in the text: LCKW-Isolate25N, Lake Chaka water, isolate 25N. Scale bars indicate Jukes-Cantor distances. Bootstrap values of >50% (for 500 iterations) are shown. Aquifex pyrophilus is used as an outer group, and a single tree showing all bacterial sequences is created. Because of the large size of the tree, it is divided into two subtrees. Panel A is the first bacterial subtree showing the Alpha-, Beta-, Gamma-, Epsilon-, and Deltaproteobacteria. (B) This figure is the second subtree showing the Firmicutes (low G+C gram-positive bacteria), Bacteroidetes, and Actinobacteria.
(iii) Betaproteobacteria.
Nineteen sequences were affiliated with the Betaproteobacteria (Fig. 4A). One sequence (from LCKS0) was related (98%) to Delftia acidovorans strain B (AB074256), 7 (from LCKS10) were related (97%) to Petrobacter succinatimandens (AY219713) and Tepidiphilus margaritifer (AJ504663), and 11 (from LCKS20) were related (97%) to Petrobacter succinatimandens BON4 and strain HMD444 (AB015328). BON4 is a moderately thermophilic and nitrate-reducing bacterium that can grow optimally at pH 7.0 and 0.5% NaCl (tolerance up to 3% NaCl).
(iv) Gammaproteobacteria.
Twenty-eight sequences were related to the Gammaproteobacteria (Fig. 4A). Many sequences in this group were related to several species of the genus Halomonas. Halomonas, a family of gram-negative Proteobacteria, can tolerate or require a high salt concentration for growth (49).
Another group of clone sequences was related (98 to 99%) to Stenotrophomonas sp. strains An30 and 27 (AJ551168/5). An30 and 27 were obtained from deep-sea sediments in the west Pacific (GenBank description). One clone sequence (LCKS10-B8) was related (97% similarity) to bacterium HTB082 (AB010842) from deep-sea sediments from the Nankai Islands’ Iheya Ridge (1,050-m depth) (47). HTB082 can grow optimally at pH 7.6 and 3 M (17.6%) NaCl. Two clone sequences were related (96%) to Alcalilimnicola halodurans (AJ404972) isolated from a water-covered site of Lake Natron, Tanzania. Alcalilimnicola halodurans can grow in the presence of 0 to 28% NaCl (wt/vol) with an optimum growth at 3 to 8% NaCl (wt/vol) and a pH above 8.5 (54).
(v) Epsilonproteobacteria.
Two sequences were affiliated with the Epsilonproteobacteria (Fig. 4A), with 99% similarity to Campylobacter sp. strain NO3A (AY135396), which is capable of oxidizing lactate with nitrate or nitrite as the electron acceptor.
(vi) Unclassified Proteobacteria.
Three sequences (Fig. 4A) were related to an environmental clone (AY940550) recovered from Qinghai Lake, which is a saline (1.3% salinity) and alkaline (pH 9.4) lake in the same area as Lake Chaka (12).
(vii) Firmicutes (low G+C gram-positive bacteria).
The low G+C gram-positive clone sequences predominated the bacterial clone libraries (71 of 123 sequences) and were grouped into nine clusters (Fig. 4B). Sequences of 24 clones and three isolates formed cluster 1. The majority of the sequences in that cluster were closely related (98 to 99%) to Bacillus arseniciselenatis strain E1H (AJ865469), a moderate halophile and alkaliphile from Mono Lake, Calif. E1H is an obligate anaerobe that is able to use Se(VI), As(V), Fe(III), nitrate, and fumarate as electron acceptors (3). Mono Lake is an alkaline (pH 9.8) and hypersaline (84 to 94 g/liter) soda lake. Four sequences in cluster 1 were closely related (98 to 99%) to Paraliobacillus ryukyuensis (AB087828), a slightly halophilic, extremely halotolerant alkaliphilic anaerobe isolated from a marine alga. It can tolerate an NaCl concentration of 0 to 22% (optimum, 0.75 to 3.0%) and grow at pH 5.5 to 9.5 (optimum, 8.5) (20).
Ten clone sequences formed cluster 2 (Fig. 4B), showing 99% similarity to an unidentified Hailaer soda lake bacterium F1 (AF275700) and Alkalibacterium sp. strain A-13 (AY347313) isolated from hypersaline Tanzania soda lakes (GenBank description). Three clone sequences formed cluster 4 (Fig. 4B). These sequences were related (94 to 95% similarity) to Halocella cellulolsilytica (X89072) and Haloanaerobium lacurosei (L39787), two isolates from hypersaline lake sediments or lagoons (8, 44). Halocella cellulolsilytica and Haloanaerobium lacurosei are obligate anaerobes with optimal growth at pH 7.0 and NaCl concentrations of 15% and 20%, (wt/vol), respectively.
Eight clone sequences formed cluster 5 (Fig. 4B). Five sequences were related (99%) to an uncultivated low G+C gram-positive bacterium (AJ495676) obtained from anoxic sediments underlying cyanobacterial mats in two hypersaline ponds in Mediterranean salterns (33). These two hypersaline ponds have salinity similar to Lake Chaka (15 to 20% and 25 to 32% salinity, respectively). The rest of the sequences were related (90 to 99% similarity) to an uncultivated low G+C gram-positive bacterium (AF507875) from Mono Lake, CA.
In cluster 6, one sequence was closely related (98%) to Thermoanaerobacter ethanolicus strain X513 (AF542520), isolated from the deep subsurface environments of the Piceance Basin, Colorado (43). Strain X513 can use lactate, acetate, succinate, xylose, and glucose to reduce Fe(III) oxyhydroxide to form magnetite. Nine clone sequences formed cluster 7, and they were related (97%) to Alkaliphilus transvaalensis (AB037677), which is extremely alkaliphilic (optimum pH of 10) and halotolerant (∼4% sea salt) from mine water at 3.2 km below the land surface in an ultra-deep gold mine near Carletonville, South Africa (46). Ten clone sequences and their relatives formed cluster 9 (Fig. 4B). One of these relatives, Anaerobranca californiensis strain Paoha-1 (AY064218), was isolated from a hot spring in Mono Lake (15). It is an anaerobic, alkalithermophilic, fermentative bacterium with an ability to reduce elemental sulfur, Fe(III), and Se(IV) in the presence of organic matter.
(viii) Actinobacteria (high G+C gram-positive bacteria).
Sequences of eight clones and one isolate were affiliated with the Actinobacteria group (Fig. 4B); the two closest relatives were Nesterenkonia halotolerans (AY226508) from hypersaline soil in Xinjiang Province, China, and Arthrobacter sp. strain AS18 (AY371223) from lead-zinc mine tailings in Huize County, Yunnan Province, China (57).
(ix) Bacteroidetes.
Thirty-six clone sequences (34 from LCKW and 2 from LCKS0) were related to the Bacteroidetes group (Fig. 4B). They were 83 to 99% similar to cultivated and uncultivated Bacteroidetes, including Salinibacter ruber strain POLA 13 (AF323503), halophilic eubacterium EHB (AJ133744), and an uncultivated Flavobacteriaceae bacterium (AF513959). POLA 13 was isolated from saltern ponds in Mallorca, Balearic Islands, Spain, and can grow optimally in the presence of 150 to 300 g/liter total salt and at pH 6.5 to 8.0 (1). The Flavobacteriaceae bacterium clone was obtained from hypersaline Lake Laysan and a brackish pond on Pearl and Hermes Atoll, Hawaiian islands (GenBank description).
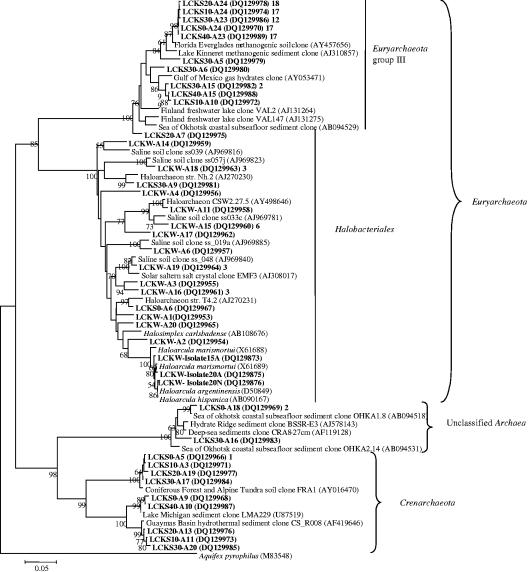
Archaeal diversity in water and sediment.
The majority of the archaeal sequences could be classified as the Euryarchaeaota and Crenarchaeota (Fig. 5). The Euryarchaeaota group consisted of two clusters. Eighty-nine clone sequences were grouped into a novel Euryarchaeota cluster within group III as defined by Jurgens et al. (21), and the majority were closely related (similarity of 97 to 99%) to an uncultivated archaeon (AY457656) from the Florida Everglades (6) and an uncultivated archaeon (AJ310857) associated with methanogenic sediment in subtropic Lake Kinneret (Israel) (34). Four clone sequences were related to an uncultivated archaeon (AY053471) associated with Gulf of Mexico gas hydrates (27).
FIG. 5.
Neighbor-joining tree (partial sequences, ∼600 bp) showing the phylogenetic relationships of archaeal 16S rRNA gene sequences cloned from the Lake Chaka samples to closely related sequences from the GenBank database. The same algorithms as those for the bacterial tree (Fig. 4) were used. Aquifex pyrophilus is used as an outer group. One representative clone type within each phylotype is shown, and the number of clones within each phylotype is shown at the end (after the GenBank accession number).
Sequences of three archaeal isolates (from the lake water) and 26 clones (24, 1, and 1 from LCKW, LCKS0, and LCKS30, respectively) formed the Halobacteriales cluster in the Euryarchaeaota group (Fig. 5). The isolates were phylogenetically related (97 to 99%) to several species of Haloarcula. Haloarcula argentinensis (D50849) is a halophilic archaeon isolated from the soils of the Argentine salt flats in Argentina (18) using a culture medium containing 16% NaCl. Haloarcula marismortui is another halophilic archaeon isolated from the Dead Sea. Its whole genome has been sequenced (2). Most of the clone sequences from the lake water were related to sequences obtained along a transient soil salinity gradient at Salt Spring in British Columbia, Canada (52). Several sequences were related (93%) to Halosimplex carlsbadense (AB108676). This organism was isolated from salt crystals from the 250-million-year-old Salado formation in New Mexico (51).
In the Crenarchaeota group, there were three small clusters. Four sequences formed the first and were closely related (∼98%) to an uncultured archaeon (AY016470) recovered from coniferous forest and alpine tundra soils in the Front Range of Colorado (GenBank description). The second cluster contained two sequences related to uncultured archaea (U87519) retrieved from Lake Michigan sediment (30). Three sequences formed the third. They were related to an uncultured archaeon (AF419646) from hydrothermal sediments in the Guaymas Basin, the Gulf of California, Mexico. Three sequences were related to unclassified sequences (AJ578143 and AF119128) retrieved from methane seep areas (GenBank description) and deep-sea sediments from several stations in the Atlantic Ocean.
Distribution of bacterial and archaeal groups.
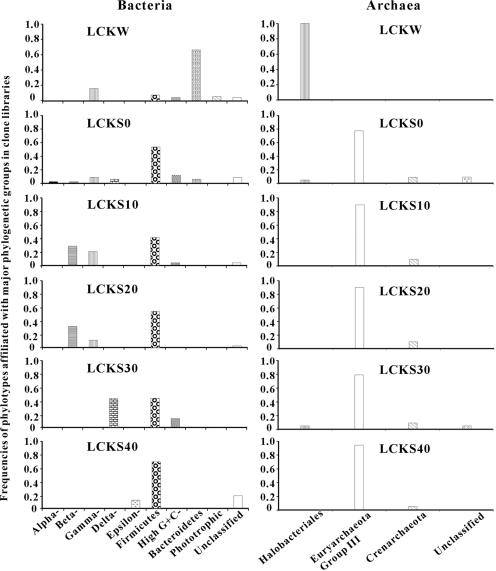
The relative abundances of different phylogenetic groups in the bacterial clone libraries were calculated for all samples (Fig. 6). The phylogenetic compositions were significantly different between the lake water and the sediments. Whereas the clone library for the lake water was dominated by sequences affiliated with Bacteroidetes (63%), those of the sediment samples were dominated by sequences affiliated with Firmicutes (low G+C gram-positive bacteria). Whereas the Betaproteobacteria were the second-most-abundant group in the LCKS10 and LCKS20 libraries, Gammaproteobacteria were the most abundant clones in the LCK30 library, followed by the low G+C gram-positive group.
FIG. 6.
Frequencies of phylotypes affiliated with the major phylogenetic groups in the bacterial and archaeal clone libraries for LCKW, LCKS0, LCKS10, LCKS10, LCKS20, LCKS30, and LCKS40. Alpha, Beta, Gamma, Delta, and Epsilon are Alpha-, Beta-, Gamma-, Delta-, and Epsilonproteobacteria, respectively. High G+C, high G+C gram-positive bacteria.
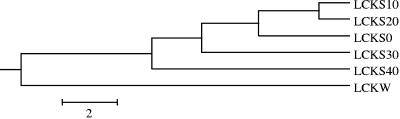
LIBSHUFF analysis was used to characterize the relationships between and among the different bacterial communities observed in the samples. The bacterial community in the water sample was markedly distinct from the communities in the sediment samples, and the communities in the sediment samples at 0-, 10-, and 20-cm depth were the most similar (Fig. 7). The data suggested that the water sample was distinct from the sediments at any depth; that the interface, 10-, and 20-cm depths were more similar; and that the 30- and 40-cm depths were distinct from the other samples and each other. The results were statistically significant (P = 0.001) with the number of clones analyzed; however, more clonal sequences need to be determined in order to elucidate possible relationships (e.g., succession) between the different depths and abiotic parameters.
FIG. 7.
Clustering of the different bacterial clone libraries based upon ΔCxy values determined from LIBSHUFF analysis. The tree was constructed with the unweighted-pair group method using average linkages in MEGA2. The parameter ΔCxy in the LIBSHUFF analysis represents the difference in coverage of the two sequence libraries (an increased ΔCxy represents greater dissimilarity between the given communities). The software for the analysis was used according to specified directions (http://www.arches.uga.edu/∼whitman/libshuff.html).
The relative abundances of different phylogenetic groups in the archaeal clone libraries were also calculated for all samples (Fig. 6). Again, there existed a distinct difference in the phylogenetic compositions of the clone libraries between the lake water and the lake sediments. Whereas all sequences in the library for the lake water were affiliated with the Halobacteriales group, most sequences in the sediment libraries were related to those sequences previously found in diverse environments (i.e., Euryarchaeota group III). Only a small percentage of sequences was related to the Crenarchaeota group.
Coverage of bacterial libraries.
Coverage values and the diversity index for the six bacterial clone libraries indicated that the sequence population from the interface sample was the most diverse and that that from the lake water sample was the least diverse. There was a large range in the sequence coverage and diversity index. In contrast, the archaeal clone libraries showed a different trend. Although there was only one major phylogenetic group in the LCKW archaeal clone library, the diversity within that group was the highest and much greater than the intergroup and intragroup diversity of the other archaeal libraries (Table 3).
TABLE 3.
Coverage and diversity indexes of the clone libraries of the lake water and sediments from Lake Chaka
| Sample (depth [cm]) | Bacteria
|
Archaea
|
||
|---|---|---|---|---|
| % Coverage | Nt/Nmaxa | % Coverage | Nt/Nmaxa | |
| LCKW (water) | 98 | 2 | 66.7 | 4.5 |
| LCKS0 (0-2) | 76.4 | 8.5 | 86.4 | 1.3 |
| LCKS10 (10-12) | 79.2 | 4.8 | 85 | 1.2 |
| LCKS20 (20-22) | 80 | 3.9 | 85.7 | 1.2 |
| LCKS30 (30-32) | 82.6 | 5.8 | 70 | 1.7 |
| LCKS40 (40-42) | 81.3 | 2 | 89.5 | 1.1 |
Nt/Nmax is the diversity index, where Nt is the total number of cells in each sample and Nmax is the number of cells of the most abundant phylotype in each sample.
DISCUSSION
Microbial abundance.
Comparisons between Lake Chaka and other hypersaline lakes indicated that the total abundance in the lake water was typical for such environments: total abundance was similar to that in the Dead Sea (2 × 106 to 2 × 107 cells/ml) and lower than that in the Great Salt Lake, Utah (∼7 × 107 cells/ml) (37). The small range in abundance in such diverse environments may suggest that salinity is the main factor in controlling microbial abundance. In general, abundance in unconsolidated sediments tends to be high, but in dry rock salt abundance can be low. Only the number of CFU has been reported in dry salt, ranging from 10 to 104 cells per g of dry salt (25).
Isolate characteristics.
The isolates from the water sample were obligately halophilic. Although the water isolates exhibited a range of salinity requirements, in general, either the optimum or the upper salinity limit (25 to 30%) was consistent with that in the lake water (32%). Phenotypic traits are difficult to predict from phylogenetic relationships, but the physiological characteristics of the isolates were in general accordance with presumptive phylogenetic positions. One exception was the bacterial isolate LCKW-Isolate25N, which exhibited an optimum salinity of 25%, whereas its closest relative, Arthrobacter sp. strain AS18, was a freshwater bacterium (57). Nonetheless, the isolates and the predominant clones from the water sample were consistent in showing a halophilic nature. In contrast, the isolates from the sediment showed a halotolerant nature with an optimum salinity of 5% or lower, consistent with the salinity in the sediment.
Bacterial diversity.
Although the bacterial community was largely dominated by halophilic and halotolerant microorganisms in all the samples studied, there was a distinct difference in the compositions of the community structure between the lake water and the sediments. Whereas the bacterial community for the lake water was dominated by sequences affiliated with the Bacteroidetes group (mostly related to halophilic bacteria), those for the sediments were dominated by sequences related to low G+C gram-positive bacteria. The water-sediment interface sample was most diverse and composed of a mixture of phylotypes that were present in both the lake water and the sediments. The distinct change in the bacterial assemblage across the water-sediment interface was likely caused by a difference in salinity and the redox state. Salinity in the lake water was 32.5%, and the precipitate on the lake surface was nearly pure salt. In the sediments, however, there were other minerals (quartz, calcite, and feldspars) in addition to halite salt. The quantitative measurement of soluble salts indicated that there was only about 3 to 4% soluble salts.
Although a direct measurement of the redox state was not possible, we infer from our qualitative observations that it also contributed to the observed difference in the microbial communities between the lake water and the sediments for the following reasons. First, visual observation indicated that the redox boundary was at the water-sediment interface (i.e., dark sediments with sulfide odor). Second, the LCKW clone libraries (both bacterial and archaeal) were dominated by sequences related to aerobic halophiles (such as Salinibacter rubber, Bacillus vedderi, halophilic eubacterium EHB, and Halosimplex carlsbadensis). Multiple aerobic halophiles were isolated in this sample. The dominant sequences in the LCKS0 clone library, which was immediately below the interface, were related to anaerobic bacteria (i.e., Bacillus arseniciselenatis, Halanaerobium lacurosei, Alkaliphilus transvaalensis, and Anaerobranca californiensis) and to environmental clones retrieved from anaerobic sediments. The isolate from LCKS0 (LCKS0-Isolate1) was closely related to a facultative bacterium Halomonas hydrothermalis isolated from deep-sea hydrothermal vent environments (23) Third, the highest diversity was observed for LCKS0, which showed a mixture of phylotypes observed for LCKW and LCKS0, consistent with the notion that the water-sediment interface was the redox boundary.
In addition to halophiles in Lake Chaka, clone sequences related to iron-reducing bacteria (i.e., Thermoanaerobacter ethanolicus) were also present. A group of clone sequences related to an anaerobic Fe(III) and Se(IV) reducer Anaerobranca californiensis strain Paoha-1 (15) suggested that Fe(III)- and Se(IV)-reducing activity may be present in the lake. The presence of selenate and selenite reducers has been reported in saline lakes (4) and alkaline environments, and these types of microorganisms may be important in such environments. The relatedness of a group of clone sequences to Alkaliphilus transvaalensis further suggested iron-reducing activity in Lake Chaka. The genus Alkaliphilus was recently established (46), and the type species was isolated from an alkaline, deep subsurface habitat. It has an optimal pH of 10 and optimal salt concentration of 0.5%. Other species of the genus Alkaliphilus have been shown to tolerate higher salt content (up to ∼10% NaCl) and to reduce metals such as Fe(III), Co(III), and Cr(VI) (55).
Archaeal diversity.
All archaeal isolate and clone sequences in the lake water were affiliated with the Halobacteriales, a group of extremely halophilic, aerobic archaea that have a salinity tolerance of 3 to 4 M salt. Representatives include Haloarcula species, Halosimplex carlsbadense, and haloarchaeon strain Nh.2. In contrast, all the archaeal clone libraries for the sediment samples were dominated by sequences that were grouped to form a distinct cluster (the Euryarchaeota group III) (Fig. 5). This shift was most likely caused by a change in salinity and the redox state across the water-sediment interface. A salinity-caused shift in archaeal community composition has been previously observed in saline soils (52). Clones among this group have been reported in a diverse range of environments, including marine sediments (50) and the deep sea (14). Most of the reference clone sequences in this group were obtained from methanogenic sediments, such as soils in the Florida Everglades (6) and sediments in Lake Kinneret (Israel) (34).
The fact that all the sequences in Euryarchaeota group III were closely related to environmental clones from a freshwater lake in Finland (Fig. 5, VAL2 and VAL147) and that those clones belonged to Thermoplasmales (21) suggests that this cluster may belong to Thermoplasmales. Interestingly, sequences that belong to Thermoplasmales have been reported to be present in another hypersaline lake, Solar Lake, Sinai, Egypt (10), and in saline soils (52). However, definitive identification must await acquisition of pure isolates and functional testing in future research.
In conclusion, we employed culture-dependent and -independent techniques to examine the difference in microbial diversity in the lake water, the water-sediment interface, and the sediments in a hypersaline lake in northwestern China. All these regions in the lake were inhabited by abundant microorganisms, which include representatives of Bacteria and Archaea. A significant difference in community structures was observed between the water, the water-sediment interface, and different depths of sediment. We attributed the observed differences to the high salinity in the water and the lower salinity in the sediments, and both predictions from phylogenetic relationships and phenotypic characteristics of field isolates corroborated this idea. Differences in the redox state, i.e., oxidizing in the lake water and reducing in the sediment, most likely contributed to the observed differences as well.
Acknowledgments
This research was partly supported by National Science Foundation grant EAR-0345307 to H.D. and National Science Foundation of China grants (40228004 and 40472064) to H.D. and B.Y.
We are grateful to Yu Shengsong and his students for their assistance in the field. We thank John Morton for his valuable assistance in DCP, XRD, and HPLC analyses. We are also grateful to W. Green for the use of his field sampling equipment and to Chris Wood for his help on the 16S rRNA gene analysis. Constructive comments from three anonymous reviewers significantly improved the quality of the manuscript.
REFERENCES
- 1.Anton, J., R. Rosselleo-Mora, F. Rodriguez-Valera, and A. Amann. 2000. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baliga, N. S., S. J. Bjork, R. Bonneau, M. Pan, C. Iloanusi, M. C. H. Kottemann, L. Hood, and J. DiRuggiero. 2004. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 14:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, J. S., A. B. Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp nov.: two haloalkaliphiles from Mono Lake, California, that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 4.Blum, J. S., J. F. Stolz, A. Oren, and R. S. Oremland. 2001. Selenihalanaerobacter shriftii gen. nov., sp nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch. Microbiol. 175:208-219. [DOI] [PubMed] [Google Scholar]
- 5.Bottomley, P. J. 1994. Light microscopic methods for studying soil microorganisms, p. 81-105. In R. W. Weaver (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. SSSA book series no. 5. Soil Science Society of America, Madison, Wis.
- 6.Castro, H., A. Ogram, and K. R. Reddy. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida everglades. Appl. Environ. Microbiol. 70:6559-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catling, D. C. 1999. A chemical model for evaporites on early Mars: possible sedimentary tracers of the early climate and implications for exploration. J. Geophys. Res. Planets 104:16453-16469. [Google Scholar]
- 8.Cayol, J. L., B. Ollivier, B. K. C. Patel, E. Ageron, P. A. D. Grimont, G. Prensier, and J. L. Garcia. 1995. Haloanaerobium lacusroseus sp. nov., an extremely halophilic fermentative bacterium from the sediments of a hypersaline lake. Int. J. Syst. Evol. Microbiol. 45:790-797. [DOI] [PubMed] [Google Scholar]
- 9.Chapelle, F. H. 2000. Ground-water microbiology and geochemistry. John Wiley & Sons, Inc., New York, N.Y.
- 10.Cytryn, E., D. Minz, R. S. Oremland, and Y. Cohen. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DasSarma, S., and P. Arora. 2001. Halophiles, p. 458-466. In J. R. Battista et al. (ed.), Encyclopedia of life sciences, vol. 8. Nature Publishing Group, London, United Kingdom. [Online.] http://els.wiley.com/els. [Google Scholar]
- 12.Dong, H., G. Zhang, H. Jiang, B. Yu, L. R. Chapman, C. R. Lucas, and M. W. Fields. 2006. Microbial diversity in sediments of saline Qinghai Lake: linking geochemical controls to microbial diversity. Microb. Ecol. 51:65-82. [DOI] [PubMed] [Google Scholar]
- 13.Fries, M. R., J. Zhou, J. Chee-Sanford, and J. M. Tiedje. 1994. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl. Environ. Microbiol. 60:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman, J. A., and A. A. Davis. 1997. Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar. Ecol. Prog. Series 150:275-285. [Google Scholar]
- 15.Gorlenko, V., A. Tsapin, Z. Namsaraev, T. Teal, T. Tourova, D. Engler, R. Mielke, and K. Nealson. 2004. Anaerobranca californiensis sp. nov., an anaerobic, alkalithermophilic, fermentative bacterium isolated from a hot spring on Mono Lake. Int. J. Syst. Evol. Microbiol. 54:739-743. [DOI] [PubMed] [Google Scholar]
- 16.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 17.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihara, K., S. Watanabe, and T. Tamura. 1997. Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int. J. Syst. Bacteriol. 47:73-77. [DOI] [PubMed] [Google Scholar]
- 19.Imhoff, J. F., F. Hashwa, and H. G. Truper. 1978. Isolation of extremely halophilic phototrophic bacteria from the alkaline Wadi Natrun, Egypt. Arch. Hydrobiol. 84:381-388. [Google Scholar]
- 20.Ishikawa, M., S. Ishizaki, Y. Yamamoto, and K. Yamasato. 2002. Paraliobacillus ryukyuensis gen. nov., sp nov., a new gram-positive, slightly halophilic, extremely halotolerant, facultative anaerobe isolated from a decomposing marine alga. J. Gen. Appl. Microbiol. 48:269-279. [DOI] [PubMed] [Google Scholar]
- 21.Jurgens, G., F. O. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 22.Kauri, T., R. Wallace, and D. J. Kushner. 1990. Nutrition of the halophilic archaebacteria Haloferax volcanni. Syst. Appl. Microbiol. 13:14-18. [Google Scholar]
- 23.Kaye, J. Z., M. C. Marquez, A. Ventosa, and J. A. Baross. 2004. Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54:499-511. [DOI] [PubMed] [Google Scholar]
- 24.Kottemann, M. C. H., A. Kish, C. Iloanusi, S. Bjork, and J. DiRuggiero. 2005. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219-227. [DOI] [PubMed] [Google Scholar]
- 25.Kunte, H. J., H. G. Truper, and H. Stan-Lotter. 2002. Halophilic microorganisms, p. 185-200. In G. Horneck and C. Baumstark-Khan (ed.), Astrobiology, the quest for the conditions of life. Springer, Koln, Germany.
- 26.Kuo, S. 1996. Phosphorus, p. 894-895. In D. L. Sparks et al. (ed.), Methods of soil analysis. Part 3: chemical methods. Soil Science Society of America, Madison, WI.
- 27.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, D., J. Chen, X. Xu, L. Zhao, and S. Gao. 1996. Study of physical chemistry in Lake Chaka. J. Salt Lake Sci. 4:20-41. [Google Scholar]
- 29.Liu, X., K. Cai, and S. Yu. 2004. Geochemical simulation of the formation of brine and salt minerals based on the Pitzer model in Chaka Lake. Sci. China Ser. D 47:720-726. [Google Scholar]
- 30.MacGregor, B. J., D. P. Moser, E. W. Alm, K. H. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancinelli, R. L. 2005. Microbial life in brines, evaporites and saline sediments: the search for life on Mars, p. 277-297. In T. Tokano (ed.), Water on Mars and life. Springer, Cologne, Germany.
- 32.Mancinelli, R. L., T. F. Fahlen, R. Landheim, and M. R. Klovstad. 2004. Brines and evaporites: analogs for Martian life. Adv. Space Res. 33:1244-1246. [Google Scholar]
- 33.Moune, S., P. Caumette, R. Matheron, and J. C. Willison. 2003. Molecular sequence analysis of prokaryotic diversity in the anoxic sediments underlying cyanobacterial mats of two hypersaline ponds in Mediterranean salterns. FEMS Microbiol. Ecol. 44:117-130. [DOI] [PubMed] [Google Scholar]
- 34.Nusslein, B., K. J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 35.Oren, A. 2001. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466:61-72. [Google Scholar]
- 36.Oren, A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28:56-63. [DOI] [PubMed] [Google Scholar]
- 37.Oren, A. 1993. Ecology of extremely halophilic microorganisms, p. 25-53. In R. H. Vreeland and L. I. Hochstein (ed.), The biology of halophilic bacteria. CRC Press, Boca Raton, Fla.
- 38.Oren, A. 2002. Halophilic microorganisms and their environments. Kluwer Academic, Dordrecht, The Netherlands.
- 39.Oren, A. 1999. Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, Fla.
- 40.Oren, A. 2003. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 39:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Rhoades, J. D. 1996. Salinity: electrical conductivity and total dissolved solids, p. 417-435. In D. L. Sparks et al. (ed.), Methods of soil analysis. Part 3: chemical methods. Soil Science Society of America, Madison, Wis.
- 42.Ringelberg, D. B., G. T. Townsend, K. A. DeWeerd, J. M. Sulita, and D. C. White. 1994. Detection of the anaerobic dechlorinating microorganism Desulfomonile tiedjei in environmental matrices by its signature lipopolysaccharide branch-long-chain hydroxy fatty acids. FEMS Microbiol. Ecol. 14:9-18. [Google Scholar]
- 43.Roh, Y., S. V. Liu, G. S. Li, H. S. Huang, T. J. Phelps, and J. Z. Zhou. 2002. Isolation and characterization of metal-reducing Thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado. Appl. Environ. Microbiol. 68:6013-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simankova, M. V., N. A. Chernych, G. A. Osipov, and G. A. Zavarzin. 1993. Halocella cellulolytica gen. nov., sp. nov., a new obligately anaerobic, halophilic, cellulolytic bacterium. Arch. Microbiol. 181:163-170. [Google Scholar]
- 45.Singleton, I., G. Merrington, S. Colvan, and J. S. Delahunty. 2003. The potential of soil protein-based methods to indicate metal contamination. Appl. Soil Ecol. 23:25-32. [Google Scholar]
- 46.Takai, K., D. P. Moser, T. C. Onstott, N. Spoelstra, S. M. Pfiffner, A. Dohnalkova, and J. K. Fredrickson. 2001. Alkaliphilus transvaalensis gen. nov., sp. nov., an extremely alkaliphilic bacterium isolated from a deep South African gold mine. Int. J. Syst. Evol. Microbiol. 51:1245-1256. [DOI] [PubMed] [Google Scholar]
- 47.Takami, H., K. Kobata, T. Nagahama, H. Kobayashi, A. Inoue, and K. Horikoshi. 1999. Biodiversity in deep-sea sites located near the south part of Japan. Extremophiles 3:97-102. [DOI] [PubMed] [Google Scholar]
- 48.Ventosa, A. 2004. Halophilic microorganisms. Springer, Berlin, Germany.
- 49.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vetriani, C., A. L. Reysenbach, and J. Dore. 1998. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol. Lett. 161:83-88. [DOI] [PubMed] [Google Scholar]
- 51.Vreeland, R. H., S. Straight, J. Krammes, K. Dougherty, W. D. Rosenzweig, and M. Kamekura. 2002. Halosimplex carlsbadense gen. nov., sp. nov., a unique halophilic archaeon, with three 16S rRNA genes, that grows only in defined medium with glycerol and acetate or pyruvate. Extremophiles 6:445-452. [DOI] [PubMed] [Google Scholar]
- 52.Walsh, D. A., R. T. Papke, and W. F. Doolittle. 2005. Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 7:1655-1666. [DOI] [PubMed] [Google Scholar]
- 53.White, D. C., R. J. Bobbie, J. D. King, J. S. Nickels, and P. Amoe. 1979. Lipid analysis of sediments for microbial biomass and community structure, p. 87-103. In C. D. Litchfield and P. L. Seyfried (ed.), Methodology for biomass determination and microbial activities in sediments: a symposium sponsored by ASTM Committee D19 on Water. ASTM, Philadelphia, Pa.
- 54.Yakimov, M. M., L. Giuliano, T. N. Chernikova, G. Gentile, W. R. Abraham, H. Lunsdorf, K. N. Timmis, and P. N. Golyshin. 2001. Alcalilimnicola halodurans gen. nov., sp nov., an alkaliphilic, moderately halophilic and extremely halotolerant bacterium, isolated from sediments of soda-depositing Lake Natron, East Africa Rift Valley. Int. J. Syst. Evol. Microbiol. 51:2133-2143. [DOI] [PubMed] [Google Scholar]
- 55.Ye, Q., Y. Roh, B. B. Blair, C. Zhang, J. Zhou, and M. W. Fields. 2004. Alkaline anaerobic respiration: isolation and characterization of a novel, alkaliphilic and metal-reducing bacterium. Appl. Environ. Microbiol. 70:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yurkova, N., C. Rathgeber, J. Swiderski, E. Stackebrandt, J. T. Beatty, K. J. Hall, and V. Yurkov. 2002. Diversity, distribution and physiology of the aerobic phototrophic bacteria in the mixolimnion of a meromictic lake. FEMS Microbiol. Ecol. 40:191-204. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, H., C. Duan, Q. Shao, W. Ren, T. Sha, L. Cheng, Z. Zhao, and H. Bin. 2004. Genetic and physiological diversity of phylogenetically and geographically distinct groups of Arthrobacter isolated from lead zinc mine tailings. FEMS Microbiol. Ecol. 49:333-341. [DOI] [PubMed] [Google Scholar]