Abstract
In this study, 231 strains of Yersinia enterocolitica, 25 strains of Y. intermedia, and 10 strains of Y. bercovieri from human and porcine sources (including reference strains) were analyzed using amplified fragment length polymorphism (AFLP), a whole-genome fingerprinting method for subtyping bacterial isolates. AFLP typing distinguished the different Yersinia species examined. Representatives of Y. enterocolitica biotypes 1A, 1B, 2, 3, and 4 belonged to biotype-related AFLP clusters and were clearly distinguished from each other. Y. enterocolitica biotypes 2, 3, and 4 appeared to be more closely related to each other (83% similarity) than to biotypes 1A (11%) and 1B (47%). Biotype 1A strains exhibited the greatest genetic heterogeneity of the biotypes studied. The biotype 1A genotypes were distributed among four major clusters, each containing strains from both human and porcine sources, confirming the zoonotic potential of this organism. The AFLP technique is a valuable genotypic method for identification and typing of Y. enterocolitica and other Yersinia spp.
Yersinia enterocolitica, a member of the family Enterobacteriacae, is a well-known food-borne pathogen. Y. enterocolitica is isolated more often from children with diarrhea than from adults (11, 25). In children, infection with Y. enterocolitica can cause gastroenteritis with mild diarrhea, abdominal pain, low fever, and pseudoappendicitis. In adults, Y. enterocolitica can cause pharyngitis and various systemic infections and is frequently associated with autoimmune complications. In Germany, the incidence of Y. enterocolitica was reported to be 8.0 per 100,000 inhabitants in 2003 (26). In the same study, it was found that Yersinia spp. were the third most frequently isolated pathogens from patients with enteric diseases after Salmonella spp. and Campylobacter spp. An initial infection with Yersinia spp. is more often followed by postinfection reactive arthritis than an infection with Salmonella spp. or Campylobacter spp. is (16).
Y. enterocolitica can be differentiated into six biotypes (biotypes 1A, 1B, 2, 3, 4, and 5) and several serotypes. The most common bioserotypes associated with human disease are 1B/O:8, 2/O:5,27, 2/O:9, 3/O:3, and 4/O:3 (8). In Europe biotype 1B is only sporadically detected in France and Italy. The virulence spectra of the biotypes differ; bioserotype 1B/O:8 is considered highly pathogenic, while the pathogenicity of biotypes 2 to 4, including all commonly isolated serotypes, is lower, as shown in animal models (5). Y. enterocolitica biotype 1A lacks the Yersinia virulence plasmid pYV and therefore is considered avirulent (24). However, some evidence suggests that biotype 1A strains have some pathogenicity, as strains belonging to this biotype are frequently isolated from humans suffering from diarrhea. Furthermore, Y. enterocolitica biotype 1A strains carry chromosomally encoded virulence factors and are able to invade cultured epithelial cells by a mechanism different from the mechanism used by pYV-bearing strains (31). Furthermore, a recent study showed that with regard to virulence genes, biotype 1A strains isolated from clinical samples did not differ significantly from strains isolated from other sources (32). Moreover, since Y. enterocolitica is difficult to differentiate from other Yersinia species by routine phenotypic tests (3, 4), strains identified as Y. enterocolitica biotype 1A and biotype 3 may belong to related species, such as Y. intermedia or Y. bercovieri (this study). As these Yersinia species lack classical virulence factors, they are considered nonpathogenic. These isolates originated from either clinical samples or environmental and food samples (2, 12, 29). In Y. bercovieri an enterotoxin different from that in Y. enterocolitica has subsequently been found (30).
Pork meat is one of the potential sources of infection of humans by Y. enterocolitica. In Switzerland the prevalence of Y. enterocolitica in pork meat at the retail level was reported to be 15.4% (20). However, only 0.7% of the strains isolated belonged to the potentially pathogenic biotypes 2, 3, and 4, while the majority of the strains belonged to apparently apathogenic biotype 1A (20). At the farm level, the prevalence of Y. enterocolitica was 63%, and 36.8% of the strains were biotype 1A strains in 2001 (20). Thus, the importance of pork meat as a source of Y. enterocolitica infection in Switzerland is unclear, given the assumptions concerning the virulence spectra of the different biotypes.
The aim of this study was to elucidate the genetic relatedness among Y. enterocolitica strains belonging to various biotypes originating from human stool samples, swine feces, and pork meat with the amplified fragment length polymorphism (AFLP) technique (33).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains were collected from swine feces (n = 41), pork meat (n = 99), and human stool samples (n = 113). All strains from humans were isolated from individuals suffering from diarrhea and were kindly provided by the Swiss Federal Office of Public Health. The pork meat and swine feces isolates were obtained during an investigation of the prevalence of latent zoonoses in pigs and pork from animal-friendly farms performed by the Federal Veterinary Office (20). All isolates originated from healthy pigs or from pork meat derived from healthy animals. They were collected in various geographical areas of Switzerland during 2000 to 2002. Reference strains and collection strains are listed in Tables 1 and 2. Four Y. enterocolitica biotype 1B strains were obtained from abroad as this biotype was not represented in our sample collection.
TABLE 1.
Reference Y. enterocolitica strains used in this study
| Straina | Biotype | Serotype |
|---|---|---|
| IP Ye 106 | 1A | O:7, O:8 |
| IP Ye 21991 | 1A | O:5 |
| IP Ye 102 | 1A | O:6, O:30 |
| IP Ye 1501 | 1A | O:34 |
| ATCC 23715 | 1B | O:8 |
| ATCC 9610 | 1B | O:8 |
| CIP 80.27 | 1B | O:8 |
| ATCC 55075 | 2 | O:9 |
| CIP 81.42 | 2 | O:9 |
| CCTM 3247b | 3 | O:5, O:27 |
| IP Ye 134 | 4 | O:3 |
| IP Ye 1105 | 4 | O:8 |
| CCUG 8233 | 4 | O:3 |
ATCC, American Type Culture Collection; CIP, Collection de l'Institut Pasteur; IP, Institut Pasteur; CCUG, Culture Collection of the University of Göteborg, Göteborg, Sweden; CCTM, Centre de Collection de Types Microbiens, Université de Lausanne, Lausanne, Switzerland.
This strain clusters with biotype 2 strains.
TABLE 2.
Sample strains used in this study
| Species | No. of strains
|
Biotype | Serotype | |||
|---|---|---|---|---|---|---|
| Human feces | Swine feces | Pork meat | Total | |||
| Y. enterocolitica | 46 | 16 | 65 | 127 | 1A | NT or diversea |
| Y. enterocolitica | 4 | 4 | 1Bb | O:8 | ||
| Y. enterocolitica | 26 | 4 | 6 | 36 | 2 | O:9 |
| Y. enterocolitica | 2 | 8 | 3 | 13 | 2 | O:5 |
| Y. enterocolitica | 2 | 2 | 3 | O:3 | ||
| Y. enterocolitica | 29 | 7 | 36 | 4 | O:3 | |
| Y. intermedia | 25 | 25 | ||||
| Y. bercovieri | 10 | 10 | ||||
| Total | 113 | 41 | 99 | 253 | ||
NT, not typeable.
All biotype 1B strains were obtained from countries other than Switzerland.
Strains were stored by using a bacterial bead storage system (Technical Services Consultants Limited, Lancashire, United Kingdom) at −70°C and were cultured on Columbia agar (Biomérieux, Marcy l”Etoile, France) containing 5% sheep blood at 30°C for 24 h. All strains were identified with the API 20E system (Biomérieux) and were biotyped and serotyped by using a previously published protocol (32). Further biochemical identification was performed by the Swiss National Center of Enteropathogens, using established procedures (7).
Isolation of chromosomal DNA.
For each strain analyzed a loopful from a fresh culture on solid medium was suspended in 200 μl of phosphate-buffered saline (0.14 M NaCl, 0.027 M KCl, 0.10 M phosphate) (pH 7.4). DNA was extracted with a High Pure PCR template preparation kit (Roche Diagnostics, Mannheim, Germany) used according to the manufacturer's protocol. The DNA concentration was determined by agarose gel electrophoresis using a XIV 100-bp DNA ladder (Roche Diagnostics) whose DNA concentration was known as the standard. The gels were scanned and the signals were analyzed with the GelDoc 2000 system (Bio-Rad Laboratories AG, Switzerland).
AFLP procedure.
For each strain, 60 to 150 ng genomic DNA was digested with 10 U of restriction enzymes BamHI and BspDI (New England BioLabs, United Kingdom) in NEB 4 buffer (New England BioLabs) containing 50 μg/μl bovine serum albumin for 2 h at 37°C. The adaptor oligonucleotides (5′CGGACTAGAGTACACTGTC3′) and (5′GATCGACAGTGTACTCTAGTC3′) (Microsynth Balgach, Switzerland) were preincubated for 10 min at 65°C and cooled for 15 min to allow annealing. Five microliters of the digested DNA was used for a 20-μl adaptor ligation mixture containing T4 buffer (Promega, Madison, WI) with 50 μg/μl bovine serum albumin (New England BioLabs), 1 U of T4 Ligase (Promega), and a double-stranded adaptor mixture (final concentration of each adaptor, 2 μM). The ligation reaction took place during overnight incubation at room temperature. Amplification was performed with 2 μl of 10-fold-diluted ligation mixture in 20 μl (final volume) of commercial Taq polymerase Master Mix (QIAGEN, Hilden, Germany) containing 0.25 μM BspDI primer (5′GTGTACTCTAGTCCGAT3′) and 0.25 μM 6-carboxylfluorescin-labeled BamHI primer (5′GAGTACACTGTCGATCC3′) (Microsynth). The amplification protocol, performed with a PE GeneAmp PCR System 9600, included 4 min of denaturation at 94°C, followed by 25 cycles of 60 s of denaturation at 94°C, 60 s of annealing at 56°C, and 90 s of elongation 72°C and then 10 min at 72°C. Products were verified by 1% agarose gel electrophoresis (5 μl at 100 V for 30 min). For the final analysis 2 μl of the product was denatured with 12 μl deionized formamide (AppliChem, Darmstadt, Germany), mixed with 1 μl of the internal GeneScan-500 ROX standard (Applied Biosystems, Foster City, CA), boiled for 3 min, and placed on ice immediately. Capillary gel electrophoresis was carried out with an ABI Prism 310 genetic analyzer (injection time, 12 s at 15 kV; run time, 37 min at 13 kV and 60°C). The reproducibility of the method was evaluated with 20 different runs using DNA from eight independent extracts from Y. enterocolitica biotype 1B serotype O:8 reference strain ATCC 23715. A reproducibility analysis in which 20 different runs were performed revealed a mean similarity (S) level of 92.07%.
Data analysis.
AFLP raw data were collected with GeneScan (PE Applied Biosystem, Boston, MA), and profiles were subsequently analyzed using the software BioNumerics 3.0 (Applied Maths, Kortrijk, Belgium). Only DNA fragments that were 80 to 450 bp long were considered in the comparisons. Briefly, after normalization, interprofile similarities were calculated with the Pearson product-moment correlation algorithm, and relationships were displayed in a dendrogram based on the unweighted pair group with mathematical average method. A threshold of 92%, as determined from the reproducibility analysis, was used to perform the cluster analysis.
RESULTS
AFLP-based discrimination of species and Y. enterocolitica biotypes and serotypes.
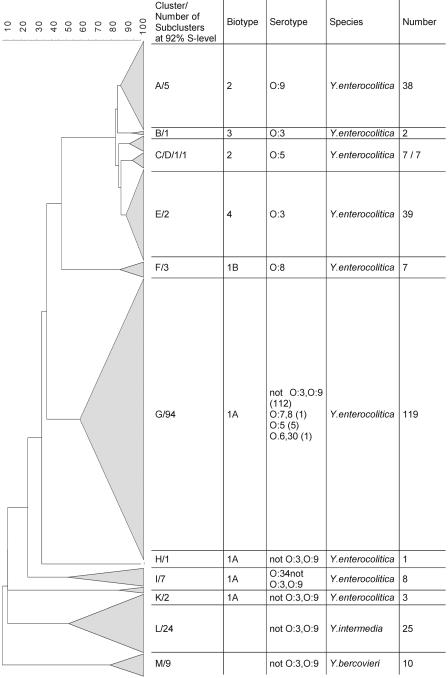
A total of 266 strains of Yersinia species were analyzed by the AFLP technique, including 231 Y. enterocolitica strains, 25 Y. intermedia strains, and 10 Y. bercovieri strains. The AFLP profiles of these species comprised between 50 and 120 bands, and the number and distribution of these bands varied and formed the basis of the cluster analysis (Fig. 1). At the 24% similarity level, four distinct clusters were formed, three of which represented the species Y. intermedia, Y. bercovieri, and Y. enterocolitica; the remaining cluster contained three strains from human and porcine sources that were identified initially as Y. enterocolitica biotype 1A but proved to be not serotypeable and yielded AFLP profiles clearly different from those of all other Y. enterocolitica strains examined (Fig. 1).
FIG. 1.
Dendrogram of 231 Y. enterocolitica strains belonging to different biotypes forming clusters A to K, 25 Y. intermedia strains forming cluster L, and 10 Y. bercovieri strains in cluster M.
At higher S levels, Y. enterocolitica strains formed smaller groups that were largely concordant with their bio- or serotype designations (Fig. 1). Strains assigned to biotype 1A were clearly distinguished from the other biotypes examined at an S level of 37%, and the biotype 1A strains were very heterogeneous and were distributed in four clusters. The biotype 1B strains were clearly distinguished at an S level of 47% from the other virulent biotypes, biotypes 2, 3, and 4. At S levels ranging from 83 to 87% strains representing bioserotypes 2/O:9, 3/O:3, and 4/O:3 clustered together; bioserotype 2/O:5 isolates were distributed in two clusters. Most of the 13 reference strains or collection strains were in the corresponding biotype clusters; the only exception was the bioserotype 3/O:5,O:27 strain, which clustered according to its serotype. A correlation with the source of the isolate and AFLP clustering was less apparent. Cluster D of biotype 2 contained only strains from porcine feces, but cluster C, also comprising biotype 2 isolates, contained strains from both porcine and human sources.
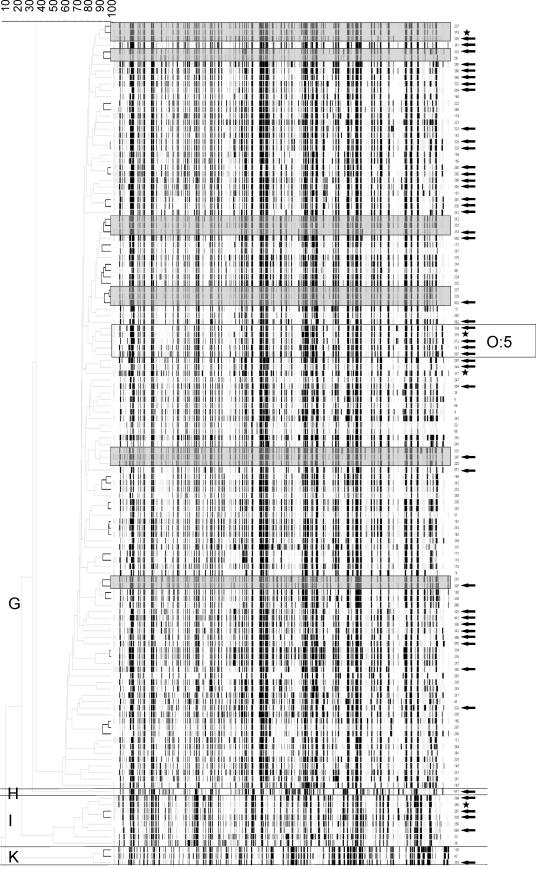
The 92% cutoff value was derived from the reproducibility analysis, and the data clearly indicated that matching biotypes and serotypes could be further differentiated by the AFLP results. This is shown in Fig. 2, which shows the AFLP profiles of all biotype 1A strains and the types formed at the 92% S level. All biotype 1A serotype O:5 strains were from human sources and represented the same AFLP type. Notably, four other AFLP types contained human isolates that were genotypically indistinguishable from pork meat isolates.
FIG. 2.
Dendrogram and AFLP banding patterns of 131 pork and human samples of Y. enterocolitica biotype 1A. The arrows indicate isolates from human sources. The stars indicate reference strains. The black lines indicate a similarity level of ≥92%. There are four major clusters, clusters G, H, I, and K. Six clusters of Y. enterocolitica biotype 1A strains from human and porcine sources exhibiting levels of similarity of ≥90% are highlighted by gray boxes in the banding pattern. The cluster containing biotype 1A, serotype O:5 strains is highlighted with a rectangle. The other serotypes are not indicated since the serotypes of several biotype 1A isolates could not be determined.
DISCUSSION
Our study showed that AFLP is a valuable method for species identification and epidemiological typing of Yersinia species at the molecular-genetic level. This was shown previously for other bacteria (1, 13, 18, 23), including Yersinia spp., as recently shown by Fearnley et al. (14). In our study we found that phenotypic identification of Yersinia spp. is problematic; all strains and isolates studied here were initially identified as Y. enterocolitica by an API 20E commercial identification kit. However, AFLP analysis clearly placed the species Y. enterocolitica, Y. bercovieri, and Y. intermedia in separate clusters. A subsequent thorough phenotypic analysis using specialized reference methods at the Swiss National Center for Enteropathogens confirmed the three species shown in Fig. 1. The taxonomic position of another group of Yersinia spp. that resembled Y. enterocolitica but had an AFLP type that was clearly distinct from that of Y. enterocolitica and the other known species examined requires further study. The Y. intermedia isolates used in this study were recovered only from pork meat, whereas the Y. bercovieri isolates were obtained only from human diarrhea samples. The method used for isolation of Yersinia spp. from pork meat involved the use of cefsulodin-irgasan-novobiocin agar, which has been reported to inhibit the growth of Y. bercovieri, and this may account for the absence of this species in porcine material. Alternatively, pigs may not be a host for Y. bercovieri (12).
As recently shown by Fearnley et al., the notable correlations between biotype, serotype, and AFLP type of Y. enterocolitica strains are consistent with the clonal nature of this species (14). In particular, the close relationship of Y. enterocolitica bioserotype 3/O:3 and bioserotype 2/O:9 demonstrated in this study has also been shown by using ribotype analysis (17). However, in the AFLP study of Fearnley et al. (14), a distinction was found only at the serotype level. This could have been due to the difficulty of identifying biotype 2, which differs from biotype 3 only in indole production. Moreover, the Y. enterocolitica bioserotype 2/O:5 strains form two distinct AFLP clusters. Both of these clusters are more closely related to Y. enterocolitica biotype 4 than to biotype 2, serotype O:9 strains. This is in agreement with the results of other genetic studies (17). For other enteric bacteria, AFLP results have been found to correlate with the results of multilocus sequence typing, an established tool for investigation of genetic population structures (21, 27). Our data suggest that AFLP analysis may be useful for inferring clonal relationships of Y. enterocolitica.
The data obtained in this study also show the high discriminatory potential of the AFLP technique for genotyping Yersinia spp., and 149 genotypes were defined by use of the cutoff for reproducibility, a common way by which strains are distinguished using this approach. In agreement with the results of Fearnley et al. (14), Y. enterocolitica biotypes 2, 3, and 4 are closely related; nonetheless, we found 10 AFLP types among the 83 strains examined. This compares favorably with the results of multilocus enzyme electrophoresis (12) and pulsed-field gel electrophoretic (PFGE) studies (9).
Strains of Y. enterocolitica bioserotype 4/O:3 are typically considered the most common enteropathogenic type in this species (6, 19, 28). The closely related strains of Y. enterocolitica bioserotype 4/O:3 from human and porcine sources confirmed the importance of swine as a reservoir for pathogenic Y. enterocolitica, as also shown by PFGE (15). However, bioserotype 4/O:3 was not isolated from pork meat obtained at the retail level in Switzerland. Therefore, pork meat was not considered an important source of infection (20), a conclusion which contradicts the findings of a recent study in Finland (15). In our study, we found porcine strains of Y. enterocolitica biotype 1A that were genotypically indistinguishable from isolates from human sources. This observation supports the hypothesis that pigs and pork products are likely sources of human infection in Switzerland and that Y. enterocolitica bioserotypes other than the classical pathogenic type 4/O:3 are potential causes of food-borne yersiniosis.
Y. enterocolitica biotype 1A strains appeared to be particularly diverse as determined by AFLP analysis, an observation in agreement with ribotyping, multilocus enzyme electrophoresis, and PFGE typing data (10, 12, 17, 22). The high percentage of biotype 1A strains recovered from porcine samples could have been due to the cold enrichment procedure used for isolation of the porcine samples, which increased the likelihood of detecting Y. enterocolitica biotype 1A strains (31). Furthermore, the low storage temperatures used for meat could facilitate the growth of biotype 1A strains. One AFLP-based cluster comprised only Y. enterocolitica bioserotype 2/O:5 strains from porcine samples. We hypothesized that these strains might be less virulent. However, the remaining cluster of this bioserotype comprised both human and porcine isolates, indicating that bioserotype alone is not reliable as a predictive marker of pathogenic potential.
In conclusion, AFLP was found to be a reliable method for identification and epidemiological subtyping of Yersinia spp. A correlation with phenotypic markers was observed, indicating a clonal relationship. The results demonstrated that several indistinguishable genotypes were obtained from human diarrhea and porcine material and suggested that pigs and their products are a source of human infections in Switzerland, as demonstrated previously for pigs and sheep in England and Wales (14). The indistinguishable genotypes are distributed among several bioserotypes of Y. enterocolitica.
.
Acknowledgments
This study was supported by the Swiss Federal Veterinary Office.
We thank Denise Howald, Elisabeth Luethi, and Christian Kohler of the Federal Veterinary Office and Herbert Haechler and Grethe Saegesser of the Swiss National Centre of Enteropathogenic Bacteria (NENT) for excellent technical support. We also thank Marianne Kueffer, Swiss Federal Office of Public Health, for collecting human isolates of Yersinia. In the context of a case-control study, these strains were kindly made available by D. Buhl of the Institute für klinische Mikrobiologie und Immunologie, St. Gallen, Switzerland, O. Dubuis, P. Friedrich, and F. Mueller of Viollier AG, Basel, Switzerland, B. Lowsky of Enzymlabor Dr. H. Weber AG, St. Gallen, Switzerland, and R. Zbinden of the Department of Medical Microbiology, University of Zurich, Switzerland.
REFERENCES
- 1.Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95-102. [DOI] [PubMed] [Google Scholar]
- 2.Agbonlahor, D. E. 1986. Characteristics of Yersinia intermedia-like bacteria isolated from patients with diarrhea in Nigeria. J. Clin. Microbiol. 23:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, J. R., R. F. Schell, D. R. Pennell, and P. D. Wick. 1987. Identification of Yersinia spp. with the API 20E system. J. Clin. Microbiol. 25:2398-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, T., H. Neubauer, K. Nikolaou, U. Roesler, and A. Hensel. 2004. Identification of Yersinia enterocolitica in minced meat: a comparative analysis of API 20E, Yersinia identification kit and a 16S rRNA-based PCR method. J. Vet. Med. Ser. B 51:23-27. [DOI] [PubMed] [Google Scholar]
- 5.Aulisio, C. C., W. E. Hill, J. T. Stanfield, and R. L. Sellers, Jr. 1983. Evaluation of virulence factor testing and characteristics of pathogenicity in Yersinia enterocolitica. Infect. Immun. 40:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissett, M. L., C. Powers, S. L. Abbott, and J. M. Janda. 1990. Epidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency, and serogroup distribution. J. Clin. Microbiol. 28:910-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bockemuhl, J., and S. Aleksic. 2003. Yersinia, p. 672-683. In Manual of clinical microbiology, 8th ed. American Society for Microbiology. Washington, DC.
- 8.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser, C., S. D. Weagant, and C. W. Kaspar. 1994. Molecular characterization of Yersinia enterocolitica by pulsed-field gel electrophoresis and hybridization of DNA fragments to ail and pYV probes. Appl. Environ. Microbiol. 60:4371-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnens, A. P., A. Frey, and J. Nicolet. 1996. Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol. Infect. 116:27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control. 2004. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—selected sites, United States, 2003. Morb. Mortal. Wkly. Rep. 53:338-343. [PubMed] [Google Scholar]
- 12.Dolina, M., and R. Peduzzi. 1993. Population genetics of human, animal, and environmental Yersinia strains. Appl. Environ. Microbiol. 59:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duim, B., T. M. Wassenaar, A. Rigter, and J. Wagenaar. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearnley, C., S. L. On, B. Kokotovic, G. Manning, T. Cheasty, and D. G. Newell. 2005. Application of fluorescent amplified fragment length polymorphism for comparison of human and animal isolates of Yersinia enterocolitica. Appl. Environ. Microbiol. 71:4960-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredriksson-Ahomaa, M., S. Hallanvuo, T. Korte, A. Siitonen, and H. Korkeala. 2001. Correspondence of genotypes of sporadic Yersinia enterocolitica bioserotype 4/O:3 strains from human and porcine sources. Epidemiol. Infect. 127:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill Gaston, J. S., and M. S. Lillicrap. 2003. Arthritis associated with enteric infection. Best Pract. Res. Clin. Rheumatol. 17:219-239. [DOI] [PubMed] [Google Scholar]
- 17.Iteman, I., A. Guiyoule, and E. Carniel. 1996. Comparison of three molecular methods for typing and subtyping pathogenic Yersinia enterocolitica strains. J. Med. Microbiol. 45:48-56. [DOI] [PubMed] [Google Scholar]
- 18.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kontiainen, S., A. Sivonen, and O. V. Renkonen. 1994. Increased yields of pathogenic Yersinia enterocolitica strains by cold enrichment. Scand. J. Infect. Dis. 26:685-691. [DOI] [PubMed] [Google Scholar]
- 20.Ledergerber, U., G. Regula, B. Danuser, B. Bissig-Choisat, T. Jemmi, and K. C. D. Stärk. 2003. Prävalenz latenter Zoonoseerreger in tierfreundlicher Schweineproduktion. Arch. Lebensmittelhyg. 54:90-94. [Google Scholar]
- 21.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najdenski, H., I. Iteman, and E. Carniel. 1994. Efficient subtyping of pathogenic Yersinia enterocolitica strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.On, S. L., and C. S. Harrington. 2000. Identification of taxonomic and epidemiological relationships among Campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol. Lett. 193:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Portnoy, D. A., and R. J. Martinez. 1985. Role of a plasmid in the pathogenicity of Yersinia species. Curr. Top. Microbiol. Immunol. 118:29-51. [DOI] [PubMed] [Google Scholar]
- 25.Robert Koch-Institut. 2003. Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2002. Robert Koch-Institut, Berlin, Germany.
- 26.Robert Koch-Institut. 2004. Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2003. Robert Koch-Institut, Berlin, Germany.
- 27.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolk-Engelaar, V. M., and J. A. Hoogkamp-Korstanje. 1996. Clinical presentation and diagnosis of gastrointestinal infections by Yersinia enterocolitica in 261 Dutch patients. Scand. J. Infect. Dis. 28:571-575. [DOI] [PubMed] [Google Scholar]
- 29.Sulakvelidze, A., K. Dalakishvili, E. Barry, G. Wauters, R. Robins-Browne, P. Imnadze, and J. G. Morris. 1996. Analysis of clinical and environment Yersinia isolates in the Republic of Georgia. J. Clin. Microbiol. 34:2325-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulakvelidze, A., A. Kreger, A. Joseph, R. M. Robins-Browne, A. Fasano, G. Wauters, N. Harnett, L. DeTolla, and J. G. Morris. 1999. Production of enterotoxin by Yersinia bercovieri, a recently identified Yersinia enterocolitica-like species. Infect. Immun. 67:968-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tennant, S. M., T. H. Grant, and R. M. Robins-Browne. 2003. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. Microbiol. 38:127-137. [DOI] [PubMed] [Google Scholar]
- 32.Thoerner, P., C. I. Bin Kingombe, K. Bogli-Stuber, B. Bissig-Choisat, T. M. Wassenaar, J. Frey, and T. Jemmi. 2003. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Appl. Environ. Microbiol. 69:1810-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]