Abstract
In this study we identified a putative virulence-associated DNA methyltransferase (MTase) gene carried on a novel 22.79-kb pathogenicity island-like element (VPAI) in V. parahaemolyticus. The V. parahaemolyticus MTase gene was shown by PCR to be prevalent (>98%) in pandemic thermostable direct hemolysin gene-positive isolates, which suggests that VPAI may confer unique virulence traits to pandemic strains of V. parahaemolyticus.
Vibrio parahaemolyticus, which is widely distributed in coastal and marine waters, has been the major cause of seafood-borne gastroenteritis in areas where raw or uncooked seafood is consumed (10). Although most V. parahaemolyticus strains are harmless to humans and animals, V. parahaemolyticus infections are often associated with strains that produce the thermostable direct hemolysin (TDH) and/or TDH-related hemolysin (TRH) (10, 22, 24). While sporadic cases of V. parahaemolyticus infections prior to 1996 were caused mainly by diverse serotypes, several major outbreaks of gastroenteritis that were reported since 1996 in many parts of the world, including the United States (2), Europe (19), Asia (5, 6, 22), and Africa (1), were associated with the emergence and pandemic spread of V. parahaemolyticus O3:K6 strains. Moreover, an increasing number of V. parahaemolyticus outbreaks have been linked to a number of clonal derivatives of O3:K6, including the O4:K68, O1:K25, and O1:KUT serovars (6, 8), which together constitute the so-called “pandemic group.” These serovars can be distinguished from nonpandemic strains by the presence of the TDH gene (but not the TRH gene) (22) and by molecular techniques such as pulsed-field gel electrophoresis (7), arbitrarily primed PCR and group-specific PCR of the toxRS gene (20), orf8-PCR of the f237 filamentous phage (21), and multilocus sequence typing (8). Nevertheless, the occurrence of group-specific PCR-positive but orf8-negative pandemic isolates has recently been reported (4, 8, 14), and the group-specific toxRS marker has been detected in TDH gene-negative O3:K6 strains of V. parahaemolyticus (23).
To date, only a limited number of virulence-associated factors have been described for pandemic strains of V. parahaemolyticus (17, 25, 26). Comparison of genomic differences between O3:K6 and nonpathogenic strains by subtractive hybridization could reveal novel mechanisms relevant to the study of the enteropathogenicity of V. parahaemolyticus. Here we describe identification of a putative virulence-associated DNA methyltransferase (MTase) gene on a 22.79-kb pathogenicity island-like element on chromosome 1 of V. parahaemolyticus.
Identification of a unique genomic locus.
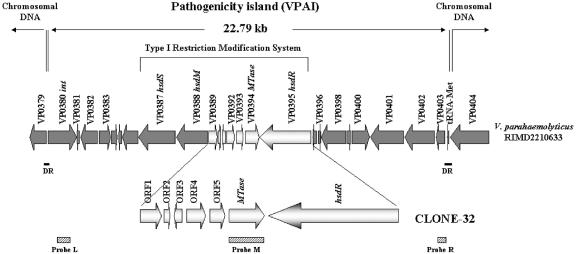
The differential subtraction chain method of Luo et al. (16) was used to identify unique genomic sequences of a clinical V. parahaemolyticus O3:K6 isolate (strain QM98284) by subtractive hybridization with DNA from an environmental isolate (strain CECT611). Genomic DNAs of clinical strain QM98284 (tester) and environmental strain CECT611 (driver) were digested with SphI, and tester DNA was ligated to adaptors A and B (Table 1); this was followed by 10 PCR cycles consisting of 94°C for 30 s, 60°C for 1.5 min, and 72°C for 1.5 min. Driver fragments (100-fold excess) were added to adaptor-ligated tester fragments, and two rounds of subtractive hybridization were performed. Tester fragments were then amplified by PCR with adaptor-specific primers RDSCA and RDSCB (Table 1) for 40 cycles consisting of 94°C for 30 s, 65°C for 30 s, and 72°C for 1.5 min, and the products were cloned into the pCR2.1-TOPO vector (Invitrogen). Genomic Southern hybridization showed that clone 32 (containing a 254-bp insert) was present only in the tester strain (QM98284) and not in the driver strain (CECT611) (data not shown), and the clone was extended using a universal GenomeWalker kit (Clontech, United States) to produce a 5,195-bp fragment (designated CLONE-32) (Fig. 1). Comparison of CLONE-32 to the genome sequence of V. parahaemolyticus RIMD2210633 (17) showed that it exhibited >99% identity with a homologous region on chromosome 1 of this strain at positions 390994 to 396188 (accession no. NC_004603).
TABLE 1.
Gene-specific PCR primers and adaptor sequences used in this study
| Target gene | Primer | Sequence (5′-3′) | Expected product size (bp) | Reference or source |
|---|---|---|---|---|
| TDH | Tdh-F | ATCTGTCCCTTTTCCTGCCC | 457 | 26 |
| Tdh-R | TTCTTTGTTGGATATACACATTACC | |||
| TRH | Trh-F | CTACTTTGCATTCAGTTTGC | 355 | 12 |
| Trh-R | CTCTGATTTTGTGAAGACCG | |||
| MTase | MTase-F | GTCTTGTCGAATAGAACTCTGA | 683 | This study |
| MTase-R | TAAGCTCCAAAATCCATACG | |||
| TTSS2 | TTSS-F | GGAAGCCATTGCGAAAGATA | 1,250 | 17 |
| TTSS-R | CACTCTCTTGTTGTTGCGTGA | |||
| 16S-23S rRNA gene intergenic spacer | Vpara-F | GCTGACAAAACAACAATTTATTGTT | 174 | 13 |
| Vpara-R | GGAGTTTCGAGTTGATGAAC | |||
| Putative ABC transporter (VP0379) and RpoD (VP0404) | VP0379-F | CTTGTTTTGCCGCTTCTTTC | 900 | 17 |
| VP0404-R | CAACAGCAACAAGCCTGAA | |||
| Differential subtraction chain fragment | RDSCA | GTCGCGGCCGCTAATACGACTC | ||
| RDSCB | GCCGGCAGATCGATACAGATG | |||
| Adaptor A | Longer strand | GTCGCGGCCGCTAATACGACTCACTATAGGGACATG | ||
| Shorter strand | AGGGATATCACTCAGCATAATC | |||
| Adaptor B | Longer strand | GCCGGCAGATCGATACAGATGTGGGACATG | ||
| Shorter strand | AGGGTGTAGACAGTAGCTAGA |
FIG. 1.
Physical map of the pathogenicity island (VPAI) on chromosome 1 of V. parahaemolyticus encompassing the CLONE-32 sequence. Predicted ORFs are indicated by arrows, and the orientations show the directions of transcription. Gene designations assigned by the sequencing project of Makino et al. (17) are indicated above the arrows. Open arrows indicate ORFs in CLONE-32. DR indicates the 47-bp direct repeat sequences flanking VPAI. The positions of gene probes L, M, and R for Southern hybridization are indicated. Abbreviations: MTase, DNA methylase gene; hsdM, gene encoding type I restriction-modification system methylation subunit M; hsdS, gene encoding type I restriction-modification system specificity subunit S; hsdR, gene encoding type I restriction-modification system restriction subunit R.
Characterization of the V. parahaemolyticus MTase gene.
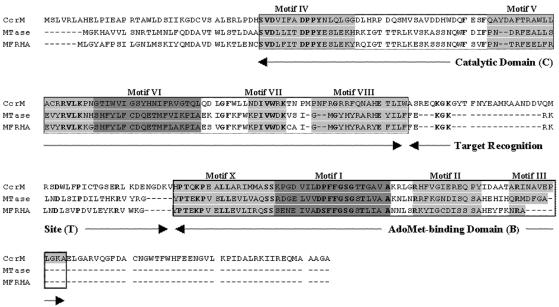
The V. parahaemolyticus MTase gene encodes a 233-amino-acid protein (Fig. 2), and a homology search showed that this protein exhibits a high level of amino acid identity (72%) with the mannose-fucose-resistant hemagglutinin (MFRHA) of Vibrio cholerae O1 (9). Analysis of the V. parahaemolyticus MTase and MFRHA deduced proteins using an HMM (hidden Markov models) search (at http://pfam.wustl.edu/hmmsearch.shtml) against the Pfam protein family database (3) showed that both proteins contain nine highly conserved sequence motifs (Fig. 2) typical of N4-N6 MTases (18). Specifically, motif I (DXFXGXG, where X is any amino acid residue) and motif IV [(D/N/S)PP(Y/F)] are the two most highly conserved motifs, and they are essential for S-adenosyl-l-methionine (AdoMet) binding and catalytic activity, respectively. Based on the arrangement of the nine conserved motifs, the linear order of the AdoMet-binding, catalytic, and target recognition domains, and alignment with the well-known cell cycle-regulated methyltransferase (CcrM; a group β MTase found so far only in α-proteobacteria [15]), it seems likely that V. parahaemolyticus MTase and MFRHA are members of the β class of m6-N-adenine MTases (11, 18). The fact that a mutant of V. cholerae defective in MFRHA exhibited significant attenuation in virulence potential (9) suggests that the MTase gene has an important role in virulence, and by extrapolation, it is likely that the V. parahaemolyticus MTase gene also has a similar pathogenic role in V. parahaemolyticus.
FIG.2.
Sequence alignment and domain organization of the V. parahaemolyticus MTase, MFRHA, and CcrM proteins. Conserved motifs are indicated by a gray background. The catalytic domain (C; consisting of motifs X, I, II, and II) and the AdoMet-binding domain (B; consisting of motifs IV, V, VI, VII, and VIII) are enclosed in boxes. The order of these domains is typical for β-class methyltransferases (18). Amino acids are designated by single-letter codes. Dashes indicate gaps inserted into the sequences for improved alignment. The proteins are highly conserved (72% identity, 90% similarity). The accession numbers for the CcrM (Brucella abortus), MTase (V. parahaemolyticus), and MFRHA (V. cholerae) protein sequences are AAB71351, VP0394, and X64097, respectively.
Association of the V. parahaemolyticus MTase gene with a pathogenicity island.
Inspection of the sequences flanking the CLONE-32 locus in the V. parahaemolyticus RIMD2210633 genome resulted in identification of a putative phage-like integrase (int) gene (VP0380) 9,936 bp upstream of hsdS (VP0387) and a methionine-specific tRNA (tRNAMet) gene sequence 7,656 bp downstream of hsdR (VP0395). Further analysis of this 22.79-kb genomic region revealed that it is flanked by a 47-bp direct repeat sequence (Fig. 1). These features suggest that CLONE-32 is carried on a pathogenicity island-like element (designated VPAI) that contains 25 open reading frames (ORFs) (Table 2). Ten of these ORFs were homologous to genes having known functions, including the genes encoding the type I restriction-modification complex (VP0387, VP0388, and VP0395), V. parahaemolyticus MTase (VP0394), phage P4-like integrase (VP0380), and proteins involved in DNA replication (VP0400), transcription regulation (VP0399), signal transduction (VP0382), and general metabolism (VP0386 and VP0392). Further analysis showed that the mean G+C content of the 25 coding genes of VPAI (43.79% ± 3.93%) is significantly lower than (but within 1 standard deviation of) the mean G+C content of the ca. 5,000 coding genes of the complete genome (accession no. NC_004603 and NC_004605; 4,992 genes; G+C content, 45.8% ± 4.19%) of V. parahaemolyticus (P < 0.05, as determined by a one-tailed t test), which suggests that VPAI was recently acquired by lateral gene transfer.
TABLE 2.
Characteristics of the 25 ORFs on VPAI of V. parahaemolyticus O3:K6
| ORF | Length of protein (amino acids) | Homology as determined by BLASTP search | Amino acid identity (%) | E value |
|---|---|---|---|---|
| VP0379 | 267 | ABC-type metal ion transport system, periplasmic component/surface antigen (Rhodospirillum rubrum) | 48 | 9e-51 |
| VP0380 | 455 | Phage integrase (Polaromonas sp. strain JS666) | 25 | 3e-29 |
| VP0381 | 33 | No significant match | ||
| VP0382 | 263 | Metallophosphoesterase (Shewanella amazonensis SB2B) | 25 | 2e-11 |
| VP0383 | 179 | Hypothetical protein plu3931 (Photorhabdus luminescens subsp. laumondii TTO1) | 58 | 2e-47 |
| VP0384 | 95 | Hypothetical protein (Dictyostelium discoideum) | 25 | 0.14 |
| VP0385 | 39 | No significant match | ||
| VP0386 | 245 | Putative inner membrane protein (Salmonella enterica serovar Typhimurium LT2) | 60 | 3e-81 |
| VP0387 | 583 | Type I restriction specificity (S) subunit (Vibrio vulnificus CMCP6) | 45 | 8e-98 |
| VP0388 | 496 | Type I restriction DNA methylase (M) subunit (Vibrio vulnificus CMCP6) | 99 | 0.0 |
| VP0389 | 132 | Hypothetical protein VV12032 (Vibrio vulnificus CMCP6) | 92 | 3e-49 |
| VP0390 | 36 | Hypothetical protein MG00497.4 (Magnaporthe grisea 70-15) | 42 | 3.3 |
| VP0391 | 49 | Hypothetical protein VFA0532 (Vibrio fischeri ES114) | 61 | 4e-08 |
| VP0392 | 121 | Translation elongation factor Ts (Vibrio vulnificus CMCP6) | 94 | 3e-52 |
| VP0393 | 94 | ORF23 (Vibrio cholerae) | 83 | 5e-40 |
| VP0394 | 221 | Putative DNA methyltransferase X64097 (Vibrio cholerae)a | 72 | 2e-101 |
| VP0395 | 814 | Type I restriction enzyme (Vibrio vulnificus CMCP6) | 97 | 0.0 |
| VP0396 | 61 | Hypothetical protein ENSANGP00000010383 (Anopheles gambiae strain PEST) | 31 | 9.7 |
| VP0397 | 33 | No significant match | ||
| VP0398 | 385 | Hypothetical protein VV12042 (Vibrio vulnificus CMCP6) | 24 | 3e-23 |
| VP0399 | 54 | Putative transcriptional regulator (Vibrio vulnificus CMCP6) | 61 | 4e-09 |
| VP0400 | 271 | DNA topoisomerase I:restriction endonuclease (Polaromonas sp. strain JS666) | 36 | 1e-37 |
| VP0401 | 529 | Conserved hypothetical protein (Nitrosomonas eutropha C71) | 23 | 7e-17 |
| VP0402 | 473 | Hypothetical protein lpp1915 (Legionella pneumophila strain Paris) | 38 | 2e-75 |
| VP0403 | 111 | Hypothetical protein lpg1935 (Legionella pneumophila subsp. pneumophila strain Philadelphia 1) | 31 | 1e-06 |
The identity of the putative X64097 protein is based on the analysis described in this study.
To define the chromosomal insertion site of VPAI, primers VP0379-F and VP0404-R flanking VPAI (Fig. 1 and Table 1) were used in PCRs with a number of V. parahaemolyticus MTase gene-negative and -positive isolates of V. parahaemolyticus. As expected, no PCR product was detected for the V. parahaemolyticus MTase gene-positive isolates examined (since VPAI is too large to be amplified), while a 0.9-kb PCR product was obtained for 10 of the 15 V. parahaemolyticus MTase gene-negative isolates tested (Table 3). DNA sequencing showed that the 22.79-kb VPAI is precisely inserted into the 47-bp direct repeat sequence at the 3′ end of the methionine tRNA gene (Fig. 1), and this was corroborated by Southern blot analysis of V. parahaemolyticus MTase gene-positive clinical (pandemic and nonpandemic) isolates and V. parahaemolyticus MTase gene-negative clinical and environmental isolates (Table 3).
TABLE 3.
Distribution of VPAI in V. parahaemolyticus MTase gene-positive and V. parahaemolyticus MTase gene-negative strains of V. parahaemolyticus
| Strain | Serotype | V. parahaemolyticus MTase gene PCR product (683 bp) | Presence of 0.9-kb PCR product obtained with VPAI-screening primersa | Southern hybridization result forb:
|
||
|---|---|---|---|---|---|---|
| VP0380 (probe L) | V. parahaemolyticus MTase (probe M) | VP0403 (probe R) | ||||
| QM98284 | O3:K6 | +c | − | + | + | + |
| QM115996 | O3:K6 | + | − | + | + | + |
| AN-2416 | O3:K6 | + | − | + | + | + |
| AO-97 | O3:K6 | + | − | + | + | + |
| AP-24659 | O1:K56 | − | + | − | − | − |
| AP-19569 | O5:KUT | − | + | − | − | − |
| TW-271 | K8 | − | + | − | − | − |
| TW-288 | K6 | − | + | − | − | − |
| TW-421 | K29 | − | + | − | − | − |
| TW-491 | K15 | − | + | − | − | − |
| QM-113892 | Unknown | − | + | − | − | − |
| QM-113890 | Unknown | − | + | − | − | − |
| T8-9 | Unknown | − | + | − | − | − |
| T1-41 | Unknown | − | + | − | − | − |
| QM-112998 | Unknown | − | − | − | − | − |
| AP-33593 | O4:K55 | − | − | − | − | − |
| TW-414 | K29 | − | − | − | − | − |
| TW-435 | K60 | − | − | − | − | − |
| T7-25 | Unknown | − | − | − | − | − |
VPAI-screening primers (VP0379-F and VP0404-R) located outside VPAI (Fig. 1) are expected to amplify a 0.9-kb fragment from V. parahaemolyticus strains that lack VPAI.
See Fig. 1 for the locations of the three VPAI hybridization probes.
+, gene amplimer or Southern hybridization signal present; −, gene amplimer or Southern hybridization signal absent.
Distribution of the genes encoding V. parahaemolyticus MTase, TDH, TRH, and type III secretion system 2 (TTSS2).
The clinical isolates of V. parahaemolyticus used in this study were kind gifts from W. C. Yam (University of Hong Kong, Hong Kong), H. C. Wong (Soochow University, Taiwan), and G. B. Nair (International Centre for Diarrheal Disease Research, Bangladesh). V. parahaemolyticus strains were confirmed by PCR using the species-specific primers Vpara-F and Vpara-R (Table 1) (13). Serotyping of V. parahaemolyticus was performed using commercial antisera purchased from Denka Seiken Co., Ltd., Tokyo, Japan. PCRs were carried out in 100-μl mixtures containing 50 ng DNA, each primer at a concentration of 0.2 to 0.25 μM, each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP) at a concentration of 0.2 mM, 1.5 mM MgCl2, and 5 U of Taq DNA polymerase. Each PCR consisted of an initial denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and elongation at 72°C for 1 min. The PCR products were analyzed by agarose gel electrophoresis.
A total of 108 clinical isolates of V. parahaemolyticus (55 pandemic strains consisting of 34 O3:K6, 13 O4:K68, 3 O1:K25, and 5 O1:KUT isolates and 53 nonpandemic strains having diverse serotypes) and 39 environmental isolates (Table 4) were tested by PCR for the presence of the V. parahaemolyticus MTase gene and the three known virulence genes, the genes encoding TDH, TRH, and TTSS2 of V. parahaemolyticus (17, 25). As reported previously (17), the TTSS2 gene was detected by PCR in all TDH gene-positive pandemic and nonpandemic strains. The V. parahaemolyticus MTase gene was detected in 98.2% (54/55) of TDH gene-positive pandemic strains and in 55.2% (16/29) of TDH gene-positive nonpandemic strains but was not detected in any of the TDH gene-negative clinical strains examined. Intriguingly, all TDH gene-positive “prepandemic” clinical isolates (collected before 1996) from Taiwan (H. C. Chung, personal communication) were PCR negative for the V. parahaemolyticus MTase gene (Table 4), and a few strains were confirmed to lack VPAI in the genome by PCR and Southern blot analysis (Table 3). The V. parahaemolyticus MTase gene was detected in only 2.56% (1/39) of the TDH gene-negative environmental isolates examined. Taken together, these results indicate that the V. parahaemolyticus MTase gene (and/or VPAI) may confer unique virulence or fitness traits to the TDH gene-positive pandemic strains of V. parahaemolyticus, enabling them to cause disease more frequently than strains belonging to other serovars. However, it is possible that some other ORFs (which encode hypothetical proteins whose functions are not known) in VPAI may code for virulence-associated factors. Clearly, further experiments are needed to understand the relevance of VPAI (on chromosome 1) and its relationship to the TDH gene (on chromosome 2) for the pathogenesis and virulence potential of V. parahaemolyticus.
TABLE 4.
Source and characteristics of the clinical and environmental V. parahaemolyticus isolates used in this study
| Source or strain (no. of strains) | Origin | Serovar | Genotype
|
|||
|---|---|---|---|---|---|---|
| TDH gene | TRH gene | TTSS2 gene | V. parahaemolyticus MTase gene | |||
| Pandemic strains (55) | ||||||
| Human (11) | Hong Kong | O3:K6 | + | − | + | + |
| Human (22) | Bangladesh | O3:K6 | + | − | + | + |
| Human | Bangladesh | O3:K6 | + | − | + | − |
| Human | Hong Kong | O4:K68 | + | − | + | + |
| Human (12) | Bangladesh | O4:K68 | + | − | + | + |
| Human (5) | Bangladesh | O1:KUT | + | − | + | + |
| Human (3) | Bangladesh | O1:K25 | + | − | + | + |
| Nonpandemic strains (53) | ||||||
| Human (2) | Bangladesh | O5:KUT | + | − | + | + |
| Human | Bangladesh | O5:KUT | + | − | + | − |
| Human | Bangladesh | O5:KUT | − | − | − | − |
| Human (3) | Bangladesh | O1:K56 | + | − | + | − |
| Human | Bangladesh | O1:K56 | − | + | − | − |
| Human | Bangladesh | O4:K55 | − | + | − | − |
| Human | Bangladesh | O4:K11 | − | + | − | − |
| Human (2) | Hong Kong | NDa | − | + | − | − |
| Human | Hong Kong | ND | + | + | − | − |
| Human (8) | Hong Kong | ND | + | − | + | − |
| Human (8) | Hong Kong | ND | + | − | + | + |
| Human | Taiwanb | K8 | + | − | + | − |
| Human (3) | Taiwan | K3 | + | − | + | − |
| Human (5) | Taiwan | K29 | + | − | + | − |
| Human (2) | Taiwan | K60 | + | − | + | − |
| Human (5) | Taiwan | K15 | + | − | + | − |
| Human | Taiwan | K10 | + | − | + | − |
| Human | Taiwan | K4 | + | − | + | − |
| Human | Bangladesh | O8:K22 | + | − | + | + |
| Human | Bangladesh | O4:K22 | + | − | + | + |
| Human | Bangladesh | O1:K38 | + | − | + | + |
| Human | Bangladesh | O4:K10 | + | − | + | + |
| Human | Bangladesh | O2:K3 | + | − | + | + |
| Human | Bangladesh | O3:K29 | + | − | + | + |
| Environmental strains (39) | ||||||
| CECT611 (marine) | Spain | ND | − | − | − | − |
| CECT612 (marine) | Spain | ND | − | − | − | − |
| CECT613 (plankton) | Spain | ND | − | − | − | − |
| Seawater (35) | Hong Kong | ND | − | − | − | − |
| T4-1 (marine) | Hong Kong | ND | − | − | − | + |
ND, serotype unknown.
All Taiwanese strains examined in this study were isolated before 1996.
Acknowledgments
This work was supported by an Area of Excellence grant from the University Grants Committee of the Hong Kong Special Administrative Region, China (project AoE/P-04/04).
REFERENCES
- 1.Ansaruzzaman, M., M. Lucas, J. L. Deen, N. A. Bhuiyan, X.-Y. Wang, A. Safa, M. Sultana, A. Chowdhury, G. B. Nair, D. A. Sack, L. von Seidlein, M. K. Puri, M. Ali, C.-L. Chaignat, J. D. Clemens, and A. Barreto. 2005. Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43:2559-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among the recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhuiyan, N. A., M. Ansaruzzaman, M. Kamruzzaman, K. Alam, N. R. Chowdhury, M. Nishibuchi, S. M. Faruque, D. A. Sack, Y. Takeda, and G. B. Nair. 2002. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40:284-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiou, C.-S., S.-Y. Hsu, S.-I. Chiu, T.-K. Wang, and C.-S. Chao. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38:4621-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, A., M. Ishibashi, V. D. Thiem, D. T. Tuyet, T. V. Tung, B. T. Chien, L. von Seidlein, D. G. Canh, J. Clemens, D. D. Trach, and M. Nishibuchi. 2004. Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48:319-327. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury, N. R., S. Chakraborty, B. Eampokalap, W. Chaicumpa, M. Chongsa-Nguan, P. Moolasart, R. Mitra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, Y. Takeda, and G. B. Nair. 2000. Clonal dissemination of Vibrio parahaemolyticus displaying similar DNA fingerprint but belonging to two different serovars (O3:K6 and O4:K68) in Thailand and India. Epidemiol. Infect. 125:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury, N. R., O. C. Stine, J. G. Morris, and G. B. Nair. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42:1280-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzon, V. L., A. Barker, and P. A. Manning. 1993. Nucleotide sequence encoding the mannose-fucose-resistant hemagglutinin of Vibrio cholerae O1 and construction of a mutant. Infect. Immun. 61:3032-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 11.Jeltsch, A. 2002. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 3:274-293. [DOI] [PubMed] [Google Scholar]
- 12.Kishishita, M., N. Matsuoka, K. Kumagai, S. Yamasaki, Y. Takeda, and M. Nishibuchi. 1992. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong, R. Y. C., S. K. Y. Lee, T. W. F. Law, S. H. W. Law, and R. S. S. Wu. 2002. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 36:2802-2812. [DOI] [PubMed] [Google Scholar]
- 14.Laohaprertthisan, V., A. Chowdhury, U. Kongmuang, S. Kalnauwakul, M. Ishibashi, C. Matsumoto, and M. Nishibuchi. 2003. Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in southern Thailand. Epidemiol. Infect. 130:395-406. [PMC free article] [PubMed] [Google Scholar]
- 15.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, J. H., J. A. Puc, E. D. Slosberg, Y. Yao, J. N. Bruce, T. C. Wright, Jr., M. J. Becich, and R. Parsons. 1999. Differential subtraction chain, a method for identifying differences in genomic DNA and rRNA. Nucleic Acids Res. 27:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 18.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Urtaza, J., A. Lozano-Leon, A. DePaola, M. Ishibashi, K. Shimada, M. Nishibuchi, and E. Liebana. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Tammamurthy, H. C. Wong, A. DePaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osawa, R., A. Iguchi, E. Arakawa, and H. Watanabe. 2002. Genotyping of pandemic Vibrio parahaemolyticus O3:K6 still open to question. J. Clin. Microbiol. 40:2708-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, T. Iida, and T. Honda. 2004. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48:313-318. [DOI] [PubMed] [Google Scholar]
- 25.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams, T. L., S. M. Musser, J. L. Nordstrom, A. DePaola, and S. R. Monday. 2004. Identification of a protein biomarker unique to the pandemic O3:K6 clone of Vibrio parahaemolyticus. J. Clin. Microbiol. 42:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]