Abstract
The siderophore and virulence factor yersiniabactin is produced by Pseudomonas syringae. Yersiniabactin was originally detected by high-pressure liquid chromatography (HPLC); commonly used PCR tests proved ineffective. Yersiniabactin production in P. syringae correlated with the possession of irp1 located in a predicted yersiniabactin locus. Three similarly divergent yersiniabactin locus groups were determined: the Yersinia pestis group, the P. syringae group, and the Photorhabdus luminescens group; yersiniabactin locus organization is similar in P. syringae and P. luminescens. In P. syringae pv. tomato DC3000, the locus has a high GC content (63.4% compared with 58.4% for the chromosome and 60.1% and 60.7% for adjacent regions) but it lacks high-pathogenicity-island features, such as the insertion in a tRNA locus, the integrase, and insertion sequence elements. In P. syringae pv. tomato DC3000 and pv. phaseolicola 1448A, the locus lies between homologues of Psyr_2284 and Psyr_2285 of P. syringae pv. syringae B728a, which lacks the locus. Among tested pseudomonads, a PCR test specific to two yersiniabactin locus groups detected a locus in genospecies 3, 7, and 8 of P. syringae, and DNA hybridization within P. syringae also detected a locus in the pathovars phaseolicola and glycinea. The PCR and HPLC methods enabled analysis of nonpathogenic Escherichia coli. HPLC-proven yersiniabactin-producing E. coli lacked modifications found in irp1 and irp2 in the human pathogen CFT073, and it is not clear whether CFT073 produces yersiniabactin. The study provides clues about the evolution and dispersion of yersiniabactin genes. It describes methods to detect and study yersiniabactin producers, even where genes have evolved.
Iron is essential for life in nearly all microorganisms. However, it is not readily available because the solubility of ferric ions at neutral pH is very low, and generally iron exists precipitated or chelated to iron-binding proteins in a host and to various compounds in the environment (7, 34, 48, 67). A frequent mechanism used by bacteria to meet their needs for iron is the secretion of low-molecular-mass iron chelating compounds called siderophores. Siderophores are able to solubilize iron and translocate it back to the bacterial cytosol via a specific outer membrane receptor and via transport proteins located in the periplasm and in the inner membrane (7, 67).
Yersiniabactin (YBT) is a bacterial siderophore with a very high stability constant for iron (4 × 1036) that was characterized in Yersinia pestis and Yersinia enterocolitica (15, 24, 33, 63). It has been extensively studied because it is a virulence factor widespread among human- and animal-pathogenic enterobacteria. In Yersinia spp. (14, 64), the YBT iron uptake system, called the YBT locus (∼30 kb), is located in the 36-kb (Yersinia pseudotuberculosis and Y. pestis) or 43-kb (Y. enterocolitica) genomic high-pathogenicity island (HPI). The YBT locus contains one regulatory gene, three genes involved in transport, and the YBT synthesis genes (reviewed in reference 20). The synthesis genes encode Irp4/YbtT of poorly characterized function (32, 54) and, in order of their intervention, the salycilate synthase Irp9/YbtS (62), the salycil-AMP ligase Irp5/YbtE, the peptide synthetase high-molecular-weight protein 2 (HMWP2) (encoded by irp2), the polyketide synthase/peptide synthetase HMWP1 (encoded by irp1), and the thiazoline reductase Irp3/YbtU. The HPI is found in high-pathogenicity strains of Y. pestis, the causal agent of bubonic plague, and of the enteropathogenic species Y. pseudotuberculosis and Y. enterocolitica. But it is never found in low-pathogenicity or nonpathogenic strains and species. The HPI is mobile due either to its excision mediated by an HPI-encoded P4-like integrase (frequency, ∼10−4) or to an insertion sequence (IS)-mediated deletion of the 102-kb pgm locus containing the HPI (frequency, 2 × 10−3). The HPI is transmissible by horizontal transfer, and it was detected in strains of the human-pathogenic enterobacteria Escherichia coli, Citrobacter spp., Klebsiella spp., Salmonella enterica, and Enterobacter spp. (4, 55, 58, 71). Modifications were found in these species in the HPI, but the YBT locus was highly conserved. In E. coli, the HPI is generally present in human-pathogenic strains of the phylogenetic groups B2 and D, but it is also found in nonpathogenic strains of the phylogenetic groups A and B1 (16, 38). The YBT locus was sequenced in several strains, including three Y. pestis strains (21, 60, 80) and E. coli CFT073 (84). Recently, genes that are similar to YBT genes have been found in Corynebacterium diphtheriae (45), in the insect pathogen Photorhabdus luminescens (25), and in two pathovars of the plant pathogen Pseudomonas syringae (8, 36).
P. syringae is divided into about 50 pathovars grouped into 9 genospecies; it belongs to the fluorescent pseudomonads, which produce peptide siderophores called pyoverdins (PVDs) (30). Efficient and specific iron-supplying systems appear to be important for fluorescent pseudomonad fitness and competitiveness. Indeed, a diversifying selection occurred in the genes encoding the Pseudomonas aeruginosa PVD peptide chain (79), and numerous siderophore outer membrane receptors are present in the genomes of P. aeruginosa, Pseudomonas putida, Pseudomonas fluorescens, and P. syringae (19, 50). P. syringae, Pseudomonas viridiflava, and Pseudomonas ficuserectae produce the same PVD with a stability constant for iron of 1025 which is not incorporated by other pseudomonads, except for Pseudomonas cichorii (10, 11, 12, 13, 17, 40, 53). This PVD is not essential for growth and virulence of P. syringae pv. syringae (18, 47), but it might be useful in competition with other microorganisms under nutritionally poor conditions encountered on the plant surface (10). Since P. syringae is an efficient epiphyte and an important plant pathogen (35), a good knowledge of its potential ecological benefits is necessary for developing new methods of population and disease control. Since siderophores play a role in the iron competition-mediated antagonisms between bacteria, the production of a siderophore with a high affinity for iron could be an ecological benefit for a strain.
In this study, methods of producing and detecting YBT (genes) in P. syringae and E. coli are described. The production of YBT by P. syringae is shown to be correlated with the possession of the irp1 gene, which is located in a recently detected putative YBT locus (8). The P. syringae YBT locus is described and compared with similar loci in Y. pestis and P. luminescens, and it is shown to differ from the usual HPI. The study provides clues about the evolution and dispersion of the YBT genes and describes techniques to detect YBT production (genes) that help in the study of known YBT producers and in finding new ones.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Pseudomonas strains are listed in Table 1. The E. coli reference (ECOR) strains (57) were provided by J. R. Johnson, E. Denamur, and B. Picard (Table 2). Precultures were grown at 28°C on medium 2 agar (9) or on nutrient agar. In general, siderophore production was carried out over 3 days in solid-liquid cultures grown unshaken at 20°C in petri dishes containing 10 ml of liquid GASN medium (10) or King's medium B (42) and one block of the corresponding agar medium (10). The culture media were not deferrated, because it has been shown in a previous study (11) that reducing the concentration of iron resulted in the highest activation of siderophore production but reduced total siderophore production because of reduced bacterial growth. The most important total siderophore production was observed when 5 μM Fe(III)-EDTA was added to a Fe-depleted culture medium, when bacteria were slightly repressed in siderophore production but grew abundantly. More importantly, siderophore repression was observed when 10 or 20 μM Fe(III)-EDTA was added to the Fe-depleted culture medium. In this study, procedures to obtain high siderophore production using standard medium constituents were selected. Important siderophore production was observed in solid/liquid GASN and King's B media using the following: osmosed water; Bacto agar and Bacto peptone (Becton Dickinson); l-asparagine, 99% thin-layer chromatography (Sigma); D(+)-glucose anhydrous for biochemistry and KH2PO4 and Na2HPO4 for analysis (Merck); glycerol bidistilled, 99.5% (wt/vol), K2HPO4, and MgSO4 · 7H2O for analysis (Prolabo).
TABLE 1.
Characteristics of Pseudomonas strains
| Straina | Origin | Country | Strain genospeciesb (pathotype) | Yersiniabactin analysisc
|
||||
|---|---|---|---|---|---|---|---|---|
| HPLC result and MS- measured iond | PCR test result for the following gene and primers:
|
Dot blot resulte | ||||||
| fyuA; FyuA f′, FyuA r | irp1; irp1up, irp1lp | irp1; PSYE2, PSYE2R | ||||||
| P. syringae pv. aceris LMG 2106T | Unknown | Unknown | 1 | − | NT | NT | − | − |
| P. syringae pv. aptata LMG 5059T | Sugar beet | United States | 1 | − | NT | NT | − | − |
| P. syringae pv. atrofaciens LMG 5095T | Wheat | New Zealand | 1 | − | NT | NT | − | − |
| P. syringae pv. dysoxyli LMG 5062T | Dysoxylum spectabile | New Zealand | 1 | − | NT | NT | − | − |
| P. syringae pv. japonica LMG 5068T | Barley | Japan | 1 | − | NT | NT | − | − |
| P. syringae pv. lapsa LMG 2206T | Zea sp. | Unknown | 1 | − | NT | NT | − | − |
| P. syringae pv. panici LMG 2367T | Unknown | Unknown | 1 | − | NT | NT | − | − |
| P. syringae pv. papulans LMG 5076T | Apple | Canada | 1 | − | NT | NT | − | − |
| P. syringae pv. pisi LMG 5079T | Pea | New Zealand | 1 | − | NT | NT | − | − |
| P. syringae pv. syringae LMG 1247T | Lilac | England | 1 | − | NT | NT | − | − |
| P. syringae pv. ciccaronei LMG 5541T | Ceratonia siliqua | Italy | 2 | − | NT | NT | − | − |
| P. syringae pv. eriobotryae LMG 2184T | Eriobotrya japonica | United States | 2 | − | NT | NT | − | − |
| P. syringae pv. glycinea LMG 5515 | Soybean | Canada | ND (2) | − | NT | NT | − | + |
| P. syringae pv. mellea LMG 5072T | Tobacco | Japan | 2 | − | NT | NT | − | − |
| P. syringae pv. mori LMG 5074T | White mulberry | Hungary | 2 | − | NT | NT | − | − |
| P. syringae pv. myricae LMG 5668T | Myrica rubra | Japan | 2 | − | NT | NT | − | − |
| P. syringae pv. phaseolicola LMG 2245T | Bean | Canada | 2 | − | − | − | − | + |
| P. syringae pv. savastanoi LMG 2209T | Common olive | Yugoslavia | 2 | − | NT | NT | − | − |
| P. syringae pv. sesami LMG 2289T | Sesame | Yugoslavia | 2 | − | NT | NT | −- | − |
| P. syringae pv. tabaci LMG 5393T | Tobacco | Hungary | 2 | − | NT | NT | − | − |
| P. syringae pv. ulmi LMG 2349T | Elm | Yugoslavia | 2 | − | NT | NT | − | − |
| P. syringae pv. antirrhini LMG 5057T | Snapdragon | England | 3 | − | − | − | + | NT |
| P. syringae pv. apii LMG 2132T | Celery | United States | 3 | +, 535 | − | − | + | NT |
| P. syringae pv. berberidis LMG 2147 | Barberry | New Zealand | ND (3) | +, 535 | − | − | + | NT |
| P. syringae pv. delphinii LMG 5381T | Larkspur | New Zealand | 3 | +, 535 | − | − | + | + |
| P. syringae pv. lachrymans LMG 5070T | Cucumber | United States | 3 | +, 535 | − | − | + | + |
| P. syringae pv. maculicola LMG 5295 | Radish | United States | ND (3) | − | NT | NT | − | − |
| P. syringae pv. passiflorae LMG 5185T | Passiflora edulis | New Zealand | 3 | − | − | − | + | NT |
| P. syringae pv. persicae LMG 5184T | Peach | France | 3 | +, 535 | − | − | + | NT |
| P. syringae pv. ribicola LMG 2276T | Currant | Unknown | 3 | − | NT | NT | − | − |
| P. syringae pv. tomato strains | ||||||||
| LMG 5093T | Tomato | England | 3 | +, 535 | − | − | + | + |
| LMG 5155 | Tomato | United States | ND | +, 535 | − | − | + | NT |
| LMG 5507 | Tomato | Canada | ND | +, 535 | − | − | + | NT |
| LMG 5508 | Tomato | Switzerland | ND | + | − | − | + | NT |
| LMG 5509 | Tomato | New Zealand | ND | + | − | − | + | NT |
| P. syringae pv. viburni LMG 2351T | Arrowwood | United States | 3 | − | − | − | + | NT |
| P. syringae pv. coronafaciens strains | ||||||||
| LMG 5060T | Oat | England | 4 | − | NT | NT | − | NT |
| LMG 2330 | Unknown | Unknown | ND | − | NT | NT | − | − |
| P. syringae pv. garcae LMG 5064T | Coffee | Brazil | 4 | − | NT | NT | − | − |
| P. syringae pv. oryzae LMG 10912T | Rice | Japan | 4 | − | NT | NT | − | − |
| P. syringae pv. primulae LMG 2252T | Primrose | United States | 6 | − | NT | NT | − | − |
| P. syringae pv. helianthi LMG 5067T | Mirasol | Mexico | 7 | +, 535 | − | − | + | + |
| P. syringae pv. tagetis LMG 5090T | Marigold | Zimbabwe | 7 | +, 535 | − | − | + | + |
| P. syringae pv. theae LMG 5092T | Tea | Japan | 8 | +, 535 | − | − | + | + |
| P. syringae pv. morsprunorum race 1 LMG 2222 | Sweet cherry | England | ND (2, 3) | − | NT | NT | − | − |
| P. syringae pv. morsprunorum race 2 strains | ND | |||||||
| CFBP 3798 | Prunus sp. | England | − | − | − | + | NT | |
| CFBP 3799 | Sour cherry | England | + | − | − | + | NT | |
| CFBP 3800 | Sour cherry | England | + | − | − | + | NT | |
| Pm2C69 | Sour cherry | Belgium | + | − | − | + | NT | |
| Pm2C76 | Sour cherry | Belgium | − | − | − | + | NT | |
| Pm2C86 | Sour cherry | Belgium | − | − | − | + | NT | |
| Pm2C92 | Sour cherry | Belgium | − | − | − | + | NT | |
| P. viridiflava LMG 2352T | Bean | Switzerland | 6 | − | NT | NT | − | − |
| P. meliae LMG 2220T | Melia azedarach | Japan | 2 | − | NT | NT | − | − |
| P. ficuserectae LMG 5694T | Ficuserecta | Japan | 2 | − | NT | NT | − | − |
| P. cichorii LMG 2162T | Endive | Germany | − | − | − | − | NT | |
| P. asplenii LMG 2137 | Fern | Unknown | − | NT | NT | − | NT | |
| P. fuscovaginae LMG 2158T | Rice | Japan | − | NT | NT | − | NT | |
| P. agarici LMG 2112T | Agaricus bisporus | New Zealand | − | NT | NT | − | NT | |
| P. marginalis LMG 14572 | Dahlia | Unknown | − | NT | NT | − | NT | |
| P. marginalis pv. marginalis LMG 5177 | Bean | Unknown | − | NT | NT | − | NT | |
| P. fluorescens LMG 1794T | Water | England | − | − | − | − | NT | |
| P. chlororaphis LMG 5004T | Contaminated plate | Unknown | − | NT | NT | − | NT | |
| P. putida LMG 2257T | Soil | United States | − | − | NT | − | NT | |
LMG, Laboratorium voor Microbiologie van Gent; CFBP, Collection Française de Bactéries Phytopathogènes; T, type or pathotype.
Data from reference 30 obtained by DNA-DNA hybridization; ND, not determined; the number in parentheses is the genomic species of the pathotype strain when it was not used in this study.
Data from this study; NT, not tested.
+ and −, results of the HPLC analyses; the number is the nominal ion (m/z) measured in ESI-MS positive-ion analyses after Fe-YBT purification, when analyzed.
Dot blot results using the most stringent washing conditions.
TABLE 2.
Characteristics of E. coli strains
| E. coli straina | Origin | Country | Phylogenetic groupb | Yersiniabactin analysisc
|
||||
|---|---|---|---|---|---|---|---|---|
| HPLC result | PCR test result for the following gene and primers:
|
Dot blot resulte | ||||||
| fyuA; FyuA f′, FyuA r | irp1; irp1up, irp1lp | irp1; PSYE2, PSYE2R | ||||||
| ECOR 1 | Human | United States | A | − | − | − | − | − (P) |
| ECOR 3 | Dog | United States | A | − | − | − | − | − (P) |
| ECOR 4 (P) | Human | United States | + | + | + | + | + | |
| ECOR 4 (D) (J) | Human | United States | A | − | − | − | − | NT |
| ECOR 10d | Human | United States | A | + | + | + | + | + (P) |
| ECOR 13d | Human | Sweden | A | − | − | − | − | − (P) |
| ECOR 18d | Celebes ape | United States | A | − | − | − | − | − (P) |
| ECOR 20d | Steer | India | A | − | − | − | − | − (P) |
| ECOR 22 | Steer | India | A | − | − | − | − | − (P) |
| ECOR 67 | Goat | Indonesia | B1 | − | − | − | − | − (J) |
| ECOR 69 (P) (D) | Celebes ape | United States | + | + | + | + | + (P) | |
| ECOR 69 (J)d | Celebes ape | United States | B1 | − | − | − | − | NT |
(P), obtained from B. Picard (University of Brest, France); (D), obtained from E. Denamur (Faculty of Medicine Xavier Bichat, Paris, France); (J), obtained from J. R. Johnson (University of Minnesota, Minneapolis); no notation, identical results for three origins.
Information from reference 38.
Data from this study.
Strain previously noted as possessing fyuA (38). However, recent reassessments using the same method as used previously confirmed the result presented here (J. R. Johnson, personal communication).
Dot blot results using the intermediate and lowest stringent washing conditions; NT, not tested; since only one origin was tested for each reference strain, the origin of the tested strain is specified with the result.
Production and HPLC detection of YBT.
The high-pressure liquid chromatography (HPLC) method of analyzing Fe(III)-chelated PVD (Fe-PVD) (12) was used to detect Fe(III)-chelated YBT (Fe-YBT) in the culture medium of Pseudomonas strains, but the chromatograms were also analyzed at 305 nm because Fe-YBT absorbs more at 305 nm than at 403 nm, which was the wavelength used for Fe-PVD. The HPLC program 2 (12) used was as follows: (A, 17 mM NaOH-acetic acid buffer, pH 5.3; B, acetonitrile, 0.9 ml/min): 100% A, 8 min; from 100% A to 98% A, 2 min; 98% A, 10 min; from 98% A to 95% A, 5 min; from 95% A to 30% A, 15 min; 30% A, 5 min. Before the injection at pH 5.0 to 5.3 in a Nucleosil C18 column, the liquid medium from one solid/liquid culture was supplemented with FeCl3, centrifuged, and filtrated; HPLC peaks were identified by their retention times and spectra, with reference to a control in the sample set.
E. coli ECOR 10 (D) and ECOR 20 (D) were grown at 37°C for 4 days in solid-liquid cultures in parafilm-sealed petri dishes. The media tested were as follows: King's medium B, T-medium (77), SN (74), GASN, and single-amino-acid-based media (not shown). E. coli strains were then grown in King's medium B for 2 or 4 days, as above, or in 25-ml-containing 100-ml shaken (220 rpm) Erlenmeyer flasks. The pH was set near 7.0 before the addition of FeCl3. YBT production was detected as described above or by using the HPLC program 3 (A, 17 mM NaOH-acetic acid buffer, pH 5.3; B, acetonitrile; 0.9 ml/min): from 95% A to 30% A, 15 min; and 30% A, 5 min.
Purification and characterization of YBT.
The recommended procedures for purifying Fe-PVDs and PVDs using a C18 column (10, 13) were followed, but the water-methanol fractions were excluded and YBT or Fe-YBT was collected in methanol. Cation-exchange chromatography was carried out for Fe-YBT with a type CM C25 Sephadex column eluted with a 30 mM NaOH-formic acid buffer (pH 4.2). Desalting was carried out in a C18 column. Purity was assessed using HPLC. A model Lambda 5 UV/VIS spectrophotometer (Perkin-Elmer) was used for spectrophotometry. YBT was analyzed in water-methanol (40:60, vol/vol); Fe-YBT was analyzed in water and in a 25 mM NaOH-phosphoric acid buffer (pH 7.0). Spectra were also obtained from the photodiode array detector in HPLC analyses. Electrospray-ionization-mass spectrometry (ESI-MS), tandem mass (MS/MS) and MS/MS/MS analyses were obtained using an Ion Trap Finnigan MAT LCQ mass spectrometer in the direct infusion mode.
PCR detection of YBT genes.
The cells were grown shaken (200 rpm) overnight in 4 ml of nutrient broth at 28°C. The lysed cells were prepared as previously described (9), but without the passages from −20°C to 70°C. The published primers used were (irp1) irp1up and irp1lp (41) and (fyuA) FyuA f′ and FyuA r (37). The PCR conditions were as follows: 5 × 105 lysed cells; 30 pmol of each primer; 2 U Taq DNA polymerase used with its buffer (Pharmacia); and a concentration of 200 μM of each deoxynucleoside triphosphate (Roche). The programs (iCycler; Bio-Rad) were as follows: an initial denaturation at 94°C for 5 min; 30 cycles at 94°C for 1 min, 58°C (fyuA) or 59°C (irp1) for 1 min, and 72°C for 1 min; and a final elongation at 72°C for 8 min.
In order to develop a general PCR detection test of irp1, irp1 of P. syringae pv. tomato DC3000 and Y. enterocolitica WA-C were aligned to find short conserved sequences. Checks were carried out to ensure that these sequences were conserved in known YBT producers and not found in genes encoding proteins from other species that were found with BLASTP to be similar to the irp1 product HMWP1. The proteins checked in this way were HMWP1 from P. syringae pv. tomato (NP_792409), Y. enterocolitica (CAA73127), and Y. pestis (NP_405471); the c_2427, c_2428, c_2429 and c_2460 proteins from E. coli (NP_754319, NP_754320, NP_754321, and NP_754352); NrpS from Proteus mirabilis (AAD10390); Plu2321 from P. luminescens (NP_929573); COG1020 from Desulfovibrio desulfuricans (ZP_00130212) and P. syringae pv. syringae (ZP_00124542); COG3321 from Nostoc punctiforme (ZP_00111186) and Trichodesmium erythraeum (ZP_00074380); MtaD from Stigmatella aurantiaca (AAF19812); EPOS B from Polyangium cellulosum (AAF26920); and PpsE from Mycobacterium tuberculosis (NP_217451). The selected sequences were evaluated as primers in PCR tests (not shown). The primers selected for screenings were PSYE2 (5′-GGCACCTGGAACAGG-3′) and PSYE2R (5′-GCCAGATCGTCCATCAT-3′) (product, 943 bp in P. syringae and 925 bp in E. coli); 25 pmol of each primer and 1 U Taq DNA polymerase were used, and the program was as follows: an initial denaturation at 94°C for 4 min; 37 cycles at 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min, and then a final elongation at 72°C for 6 min. The E. coli 925-bp and P. syringae 943-bp expected PCR products were 64.2% identical (Align).
To take account of a modification in P. syringae pv. phaseolicola 1448A at the last but one base in the 3′-end region of the primer PSYE2, the primer PSYE4 (5′-TGGCACCTGGAACA-3′) was tested with the reverse primers PSYE4R (5′-GCCAGATCGTCCATC-3′) (product, 896 bp; annealing temperature, 57°C) and PSYE5R (5′-GCCAGATCGTCCATC-3′) (product, 944 bp; annealing temperature, 57°C); nevertheless, there was still one base difference in the middle of the region corresponding to PSYE4 in P. syringae pv. phaseolicola 1448A.
DNA hybridization.
The 943-bp PCR product obtained with P. syringae pv. tomato LMG 5093 using the primers PSYE2 and PSYE2R was purified using a QIAquick PCR purification kit (QIAGEN) and used to construct an [α-32P]dCTP-marked radioactive probe using an RPN 1604 kit (Amersham). Dot blots were performed using about 5 μg of DNA per strain, purified as previously described (9), positively charged nylon membranes, and standard protocols (3). The strains analyzed were as follows: PCR-positive control strains of P. syringae from different genospecies, all the PCR-negative strains of P. syringae and of the genetically closely related species, and PCR-positive as well as PCR-negative E. coli strains. The most stringent final washing conditions were as follows: 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [3])-0.1% sodium dodecyl sulfate (SDS); 75°C. Intermediate final washing conditions were as follows: 0.1× SSC-0.1% SDS; 55.5°C. The lowest stringent final washing conditions were as follows: 0.2× SSC-0.1% SDS; 49°C. The membranes were read using a model Molecular ImagerR FX and the program Quantity oneR (Bio-Rad).
Comparisons of the YBT loci and map constructions.
The BLAST programs (1, 81), Conserved Domain Search, and annotated sequences were used or obtained from the National Center for Biotechnology Information. The sequences used to compare the YBT loci were as follows: P. syringae pv. tomato DC3000 AE016864, AE016865, and AE016866; P. syringae pv. syringae B728a NC_007005; P. syringae pv. phaseolicola 1448A NC_005773; Y. pestis 91001 AE017133; P. luminescens TTO1 BX571866; E. coli CFT073 AE016762; Y. enterocolitica L18881, Z35486 (8081), and Z29675 (WA-C); Y. pseudotuberculosis IA Z35107; C. diphtheriae NCTC13129 NC_002935; and P. aeruginosa PAO1 X82644. The GC content, identities, and similarities were obtained using Freqsq, Align, Ssearch (56, 78; http://www.infobiogen.fr), BLASTP, BLASTN, and BLAST 2 Sequences. The maps were constructed using Vector NTI (InforMax, Inc.) or PowerPoint.
Search for mutations in irp1 and irp2 in E. coli.
In order to determine whether the modifications in irp1, irp2, and irp5 found in E. coli CFT073 give functional proteins in E. coli, a search for the modifications in irp1 and irp2 was undertaken with YBT-producing ECOR strains. The irp1 segments modified in E. coli CFT073 were sequenced in six ECOR strains. Primer3 (http://www.be.embnet.org) was used to select the primers irp1122 (5′-GCGATTGTCGCGTTTGAAATC-3′) for c_2427 and irp1121R (5′-GCCAGTAATCCGCCTGGTTG-3′) for c_2428 (product, 540 bp in CFT073), and irp1232 (5′-AGTCATGGCTACGCGACGTG-3′) in c_2428 and irp1232R (5′-CATCACCGCCTGTTCCAGGT-3′) in c_2429 (product, 510 bp in CFT073). The PCR conditions were as for PSYE2/PSYE2R, but 1 ng of purified DNA, 30 pmol of each primer, and 2 U Taq DNA polymerase were used; the annealing temperature was 69°C. The products were purified using a QIAquick PCR purification kit (QIAGEN). Sequencing was carried out with 500 ng of DNA and IRD800-labeled primers (Biolegio) by using the DYEnamic Direct cycle sequencing kit (Amersham Biosciences). The products were loaded into a model 4200 IR2 DNA sequencer (LI-COR) and analyzed with e-Seq (LI-COR). The forward primers used to detect the CFT073 IS1541A-like due insertion in irp2 (i.e., caused by an IS element similar to an IS1541A element) were irp2121 (5′-CCTTACCGCTGACGGCTA-3′) and irp2122 (5′-ACCCCTGAAGCGGAAAAC-3′) for c_2424. The reverse primers were irp2121R (5′-CGCCTTGCTGGAAGAAGT-3′) and irp2122R (5′-CGCTTCATAACCTGCCTGA-3′) for c_2426. The expected product was 900 (2121/2121R), 959 (2121/2122R), 843 (2122/2121R), or 903 bp (2122/2122R) in the presence of the insertion and 189, 248, 132, or 192 bp in its absence. The PCR conditions were as for PSYE2/PSYE2R, but the annealing temperature was 61°C.
RESULTS
Production and HPLC detection of YBT.
Fe-YBT was detected during the Fe-PVD purification of P. syringae pv. tomato LMG 5093, because a reddish-orange compound remained adsorbed in the C18 column after Fe-PVD elution. In the absence of iron, the compound was colorless in the column and in methanol, but it readily turned reddish-orange following the addition of FeCl3. While YBT was characterized, the Fe-YBT peak was detected using the HPLC method for analyzing Fe-PVD production in fluorescent pseudomonads (Fig. 1A). It was then detected, using HPLC, in P. syringae genospecies 3, 7, and 8 and pathovar morsprunorum race 2 (Table 1). GASN medium was generally used, but strains of pathovars apii and persicae produced YBT only in King's medium B. PVD-nonproducers, such as P. syringae pv. persicae LMG 5184 and P. syringae pv. tomato LMG 5155, produced YBT, but others did not (not shown).
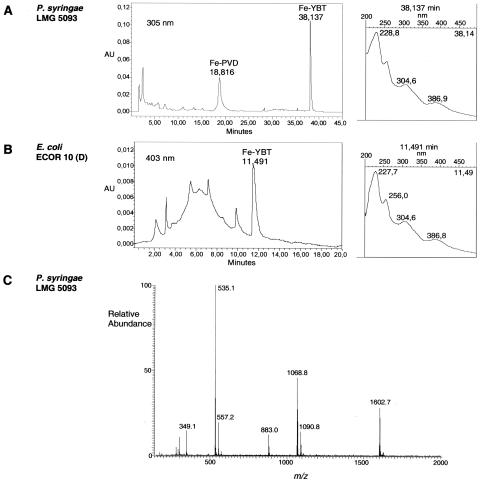
FIG. 1.
Detection (A) in GASN medium with the HPLC program 2 of Fe-PVD and Fe-YBT produced by P. syringae pv. tomato LMG 5093 and (B) in King's medium B with the HPLC program 3 of Fe-YBT produced by E. coli ECOR 10. For each strain, an HPLC analysis (on the left) and the spectral characteristics of Fe-YBT analyzed in line (on the right) are shown. Both HPLC programs can be used for each species. In King's medium B (B), Fe-YBT is more easily detected at 403 nm because medium components (visible between 2 and 10 min) absorb more at 305 nm. (C) ESI-MS positive-ion analysis of Fe-YBT of P. syringae pv. tomato LMG 5093.
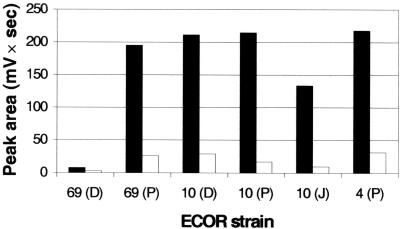
With regard to E. coli, the 10 ECOR strains were initially chosen because they belong to the nonpathogenic phylogenetic groups A and B1 and because 5 of them had been previously found to possess the YBT receptor gene fyuA (38; Table 2). Single-amino-acid-based media proved to be inefficient for inducing YBT production by E. coli. Growing E. coli at 37°C for 48 h in King's medium B was the best way to produce YBT (not shown). In one experiment, the Fe-YBT HPLC peak area at 403 nm was 2.4, 7.6, 7.2, 12.4, 13.4, or 7.0 times higher, depending on the strain, using the solid-liquid technique with petri dishes rather than shaken Erlenmeyer flasks (Fig. 1B and 2). However, the HPLC results did not always accord with published data, and the results with ECOR 4 and ECOR 69 obtained from three origins did not always agree with each other (Table 2).
FIG. 2.
YBT production in King's medium B by the E. coli strains ECOR 69, ECOR 10, and ECOR 4 received from different origins: (P), received from B. Picard; (D), received from E. Denamur; and (J), received from J. R. Johnson (Table 2). The strains were grown at 37°C for 2 days in either one still petri dish with one agar block (black bars) or one shaken Erlenmeyer flask (white bars). YBT production was assessed by the Fe-YBT HPLC peak area at 403 nm.
Purification of YBT.
Due to the YBT hydrophobicity, a one-step purification of YBT from the very simple GASN medium enabled us to characterize it. Ion-exchange chromatography further improved Fe-YBT purity and facilitated its purification from King's medium B. Purification was carried out for YBT of P. syringae pv. tomato LMG 5093 and for Fe-YBT of the strains for which the ion 535 is specified in Table 1.
Chemical characterization of YBT.
The spectrophotometric analyses accorded with data for YBT (15, 24, 33, 63): the free molecule (M) showed absorbance maxima near 207, 251, and 310 nm in water-methanol; and the Fe molecule showed absorbance maxima near 227, 255, 305, and 386 nm in water, in phosphate buffer (pH 7.0), and under the HPLC conditions. FeM from P. syringae and Fe-YBT from E. coli showed similar retention times and absorbance maxima (Fig. 1A and B) in HPLC analyses. MS analyses indicated that M and FeM had the same molecular masses and split at the same places to produce the same fragments as YBT. Indeed, ESI-MS of M produced an [M+H]+ ion of m/z 482.2, as related for YBT (24). Then, in their ESI-MS characterization of Fe-YBT, Drechsel et al. (24) observed the monomer (m/z 535), the dimer, and the trimer of the iron complex, as well as a dominant iron ion of an uncharged fragment of 295, which actually corresponds to an ion [295-2H+Fe]+ of m/z 349 resulting from the loss of a described uncharged 186 fragment from the monomer. As expected for YBT, in the present study (Fig. 1C), ESI-MS of FeM carried out using an ion trap mass spectrometer produced the ions [FeM+H]+ of m/z 535.1 and [FeM+H-186]+ of m/z 349.1, as well as the other characteristic ions (24, 63) of m/z 557.2 (monomer Na adduct), 1,068.8 (dimer), 1,090.8 (dimer Na adduct), and 1,602.7 (trimer). In addition, MS/MS confirmed that 883.0 (Fig. 1C) is the one-charge combination of 349.1 and 535.1, and MS/MS/MS analyses gave an ion of m/z 192.5 corresponding to the positive ion of an uncharged fragment of 191 also described by Drechsel et al. (24). Finally, MS/MS of 535.1 produced the described ion of m/z 489.0 (63) and other ions also found in the other analyzed pathovars (Table 1).
General PCR detection of irp1 and DNA hybridization.
Although efficient with E. coli (Table 2), the primers irp1up/irp1lp and FyuA f′/FyuA r were inefficient for all the YBT-producing P. syringae strains (Table 1).
Only four conserved 17-bp sequences were found between irp1 of Y. enterocolitica WA-C and irp1 of P. syringae pv. tomato DC3000 located in a putative YBT locus in the genome of this strain (8), and no sequence was also conserved in a similar locus in P. luminescens TTO1 (25). The PSYE2/PSYE2R primer pair was selected. PSYE2 is conserved in Y. pestis and E. coli, but there is one base substitution in PSYE2R in both species, whereas there are six and nine base substitutions in P. luminescens.
In P. syringae, the detection of YBT by HPLC correlated with the PCR detection of the irp1 gene located in the putative YBT locus of P. syringae DC3000 and 1448A, which accords with the putative function assigned to these loci (Table 1) (8, 36). Also, the concordant results obtained with YBT-positive strains using two irp1-based PCR tests were indicative of the similarity in the irp1 gene sequence in the different P. syringae pathovars. However, seven strains in pathovars antirrhini, passiflorae, viburni, and morsprunorum race 2 were negative using HPLC but positive using PCR. Among the 25 strains tested from genospecies 3, 7, and 8 and pathovar morsprunorum race 2, only P. syringae pv. maculicola LMG 5295 and P. syringae pv. ribicola LMG 2276 were negative using PCR. All the strains from other genospecies and from other Pseudomonas species were negative using PCR. A good conservation of irp1 was observed in the pathovars tomato and morsprunorum race 2.
For E. coli, the PCR results confirmed all the HPLC results (Table 2).
Dot blot using the most stringent washing conditions confirmed all the PCR results for P. syringae and the genetically closely related species, except that an irp1 homologue was detected in P. syringae pv. phaseolicola LMG 2245 and P. syringae pv. glycinea LMG 5515 (Table 1); the identity of P. syringae pv. phaseolicola 1448A irp1 with the probe is 87.5%. The PCR-positive E. coli ECOR 4 (P), ECOR 10 (P), and ECOR 69 (P) gave negative results in these conditions, as expected given the low sequence identity of E. coli irp1 with the probe (64.2%). The use of the intermediate or the lowest stringent washing conditions confirmed the results for the pathovars phaseolicola and glycinea, and it enabled the very probable detection of the irp1 homologues in E. coli ECOR 4 (P), ECOR 10 (P), and ECOR 69 (P) (Table 2). But only clearly weaker signals, or no signals, were observed for all the other PCR-negative Pseudomonas and E. coli strains analyzed, which indicated an identity clearly inferior to 64% and confirmed the very probable absence of a YBT locus in all these strains.
The new primers PSYE4, PSYE4R, and PSYE5R take account of the irp1 sequence in P. syringae pv. phaseolicola 1448A, and they were workable with both YBT-positive P. syringae and E. coli strains. But the attempts to find general PCR conditions that would also be workable with P. syringae pv. phaseolicola LMG 2245 and P. syringae pv. glycinea LMG 5515 remained surprisingly unsuccessful, which probably reflects additional sequence variations in both strains.
Sequence comparisons between YBT locus genes.
Comparisons were carried out between confirmed or putative YBT genes and proteins of different species, and three groups were identified. This is illustrated by irp2 (Align): irp2 from Y. pestis 91001 had 98% identity with its homologue in Y. enterocolitica 8081, 98.5% with that in E. coli CFT073 (without an IS1541A-like due 711-bp insertion), but only 59.9% with that in P. syringae DC3000 and 55.7% with that in P. luminescens TTO1, whereas irp2 homologues of the latter two species showed only 55.9% identity.
Functionally comparable predicable proteins were found in the YBT loci of Y. pestis, P. syringae, and P. luminescens. Similarities were found between these proteins, and names to use when referring to the Y. enterocolitica nomenclature are proposed for P. syringae because it produces YBT (Table 3). Both P. syringae and Y. pestis proteins differed about equally from those of the other species, which confirmed the existence of three YBT locus evolutionary groups. HMWP1 of P. syringae and that of Y. pestis were about 740 amino acids (aa) shorter than Plu2321 of P. luminescens (Table 3), and the similarity in the overlap was lower when Plu2321 was compared with HMWP1 of P. syringae (62.8%) or Y. pestis (65.5% similarity in a 2,923-aa overlap) than when HMWP1 of P. syringae and Y. pestis were compared to each other (76.0%). This was not observed for HMWP2 (Table 3).
TABLE 3.
Similarity search between P. syringae and Y. pestis or P. luminescens proteins
| Proteina (aab) of P. syringae DC3000 | Proposed new name | Homologous proteina (aab) of Y. pestis 91001 | % Identity-% similarity (amino acid overlap)c | Homologous proteina (aab) of P. luminescens TTO1 | % Identity-% similarity (amino acid overlap)c |
|---|---|---|---|---|---|
| PchA (469) | YbtS (434) | 27.6-52.9 (420) | Plu2324 (977) | 27.3-53.4 (457) | |
| Irp5 (522) | YbtE (525) | 57.5-81.0 (515) | Plu2324 (977) | 56.4-79.2 (514) | |
| Irp4 (271) | YbtT (267) | 50.8-71.0 (238) | Plu2323 (258) | 52.8-74.4 (250) | |
| Irp3 (360) | YbtU (386) | 49.9-72.9 (343) | Plu2322 (365) | 46.0-72.3 (361) | |
| HMWP1 (3,173) | HMWP1 (3,163) | 52.6-76.0 (3,187) | Plu2321 (3,908) | 41.3-62.8 (3,211) | |
| PSPTO2601 (408) | Irp8 | YbtX (467) | 29.2-60.1 (404) | Plu2317 (414) | 30.7-62.1 (378) |
| PSPTO2602 (2,057) | HMWP2 | HMWP2 (2,041) | 52.3-74.2 (2,045) | Plu2320 (2,049) | 51.2-74.6 (2,053) |
| PSPTO2603 (581) | Irp7 | YbtQ (600) | 33.1-65.8 (564) | Plu2319 (600) | 34.2-65.6 (582) |
| PSPTO2604 (593) | Irp6 | YbtP (600) | 37.9-66.6 (578) | Plu2318 (575) | 38.8-68.0 (557) |
| PSPTO2605 (685) | FyuA | FyuA (673) | 32.2-61.7 (652) | Plu2316 (668) | 35.3-66.4 (651) |
| PSPTO2606 (312) | YbtA | YbtA (319) | 30.8-60.9 (169) | Plu2315 (323) | 30.4-59.5 (289) |
Protein names accord with the annotated genomes.
No. of amino acids.
Data obtained using Ssearch.
HMWPI and HMWP2 of P. syringae DC3000 had a relatively weak similarity of 53.5% (2,413-aa overlap) and 51.0% (2,061-aa overlap) with DIP2160 and DIP2161, respectively, from C. diphtheriae NCTC13129, whose coding genes sidB and sidA are similar to Y. pestis irp1 and irp2 (45).
Gene organization.
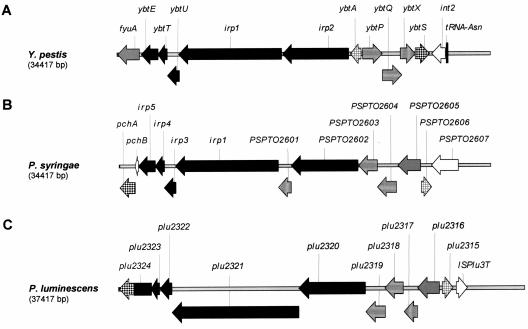
The YBT locus organization of P. syringae is closer to that of P. luminescens than that of Y. pestis (Fig. 3). Two differences are the position of irp8 of P. syringae (PSPTO2601) between irp1 and irp2 (PSPTO2602) and the fusion of the irp5 and pchA homologues in P. luminescens (plu2324). In contrast with Y. pestis, the latter two genes are also close together in P. syringae but separated by pchB. It is worth noting that in the P. syringae YBT locus there is no insertion in a tRNA locus, no integrase, and no IS element in its proximity (Fig. 4), whereas an integration in an asn tRNA locus, the integrase-coding int2, and IS elements on both sides of the represented genes are present in Y. pestis, and ISPlu3T is present in P. luminescens (Fig. 3). In P. syringae, PSPTO2607 encodes an additional putative siderophore receptor on the right border of the cluster (Fig. 3B). The GC content of the YBT locus of P. syringae DC3000 is 63.4%, which is higher than the mean value of 58.4% for the chromosome (8).
FIG. 3.
Map of the YBT locus in (A) Y. pestis 91001, (B) P. syringae DC3000, and (C) P. luminescens TTO1, according to annotated genomes. The genes are designed according to the encoded protein functions: black, biosynthetic enzymes, apart from ybtS homologues, represented by a dark squaring; gray, membrane receptors; horizontal lines, transport proteins; light squaring, regulatory proteins; and vertical lines, proteins of unknown function. The white-coded genes have no homologues in the other species.
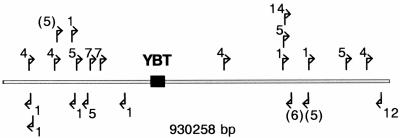
FIG. 4.
Map and orientation of IS elements around the YBT locus (black box) of P. syringae DC3000 according to the annotated genome: 1, ISPssy; 4, ISPsy4; 5, ISPsy5; 6, ISPsy6; 7, ISPsy7; 12, ISPsy12; 14, ISPsy14. The parentheses indicate disrupted IS elements.
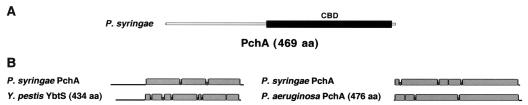
In Y. pestis, YbtS converts chorismate to salicylate in YBT synthesis (31, 62), but PchA and PchB are necessary for this conversion in P. aeruginosa in pyochelin synthesis (28, 29, 75). PchA of P. syringae DC3000 has 52.9% similarity in a 420-aa overlap with YbtS, but 64.3% similarity in a 462-aa overlap with PchA of P. aeruginosa PAO1, which converts chorismate to isochorismate. PchA of P. syringae is similar to YbtS essentially in the chorismate binding domain, whereas the entire protein shows a similarity with PchA of P. aeruginosa (Fig. 5). Also, PchB of P. syringae DC3000 has 81.2% similarity in a 96-aa overlap with PchB of P. aeruginosa PAO1, which converts isochorismate to salicylate. The GC content of pchA and pchB, however, is different in P. syringae DC3000 (63.18% and only 58.26%, respectively) and in P. aeruginosa PAO1 (70.44% and 67.31%, respectively).
FIG. 5.
(A) Location of the chorismate binding domain (CBD, black bar) detected in PchA of P. syringae DC3000 (gray bar) using the NCBI conserved domain search. (B) BLAST2 Sequences comparison of PchA of P. syringae DC3000 with either YbtS of Y. pestis 91001 or PchA of P. aeruginosa PAO1; the proteins are represented by thin lines, apart from zones with high similarity, which are represented by wide gray bars, and gaps, represented by intermediate-thickness dark gray bars.
Gene organization in CFT073, the only E. coli strain whose YBT locus is sequenced (84), is the same as that in Y. pestis, but irp1, irp2, and irp5/ybtE would be modified into three open reading frames (ORFs) (c_2427, c_2428, and c_2429), two ORFs (c_2424 and c_2426), and two ORFs (c_2433 and c_2434), respectively. Depending on the annotated genome, the protein c_2433 is an ortholog of the enterobactin synthetase component E of E. coli K12 (NP_415126, b0594; 69.4% similarity in a 408-aa overlap), which activates 2,3-dihydroxybenzoate in catecholate siderophore synthesis (83). However, since c_2433 (415 aa) and c_2434 (94 aa) have 100% similarity with residues 1 to 413 and 432 to 525 of YbtE, respectively, it is clear that they are parts of a previous irp5/ybtE gene, which activates salicylate in phenolate siderophore synthesis (54, 83).
Comparisons between P. syringae pathovars.
A YBT locus is present in P. syringae pv. tomato DC3000 (PSPTO2595, pchA, to PSPTO2606; Fig. 3B) and P. syringae pv. phaseolicola 1448A (PSPPH2904 to PSPPH2893) but absent in P. syringae pv. syringae B728a (8, 27, 36). The YBT locus gene organizations are totally identical in pathovars tomato and phaseolicola. The protein homologies range from 82.2% identity (93.5% similarity) for PchA to 95.1% identity (99.0% similarity) for PchB.
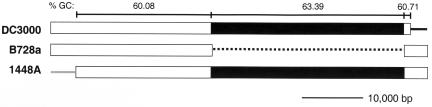
The YBT loci and the adjacent gene PSPTO2607 (Fig. 3B) or PSPPH2892 lie in a zone conserved in the three pathovars (Fig. 6): between homologues of the genes Psyr_2284 and Psyr_2285 of pathovar syringae. This intergenic region in pathovar syringae showed 88% identity in a 165-bp overlap with the side adjacent to PSPTO2608 in the 393-bp intergenic region between PSPTO2607 (Fig. 3B) and PSPTO2608 in pathovar tomato. In this common region, BLASTX indicated 49% similarity of a 59-aa sequence with a segment of the serine/threonine protein kinase MaK from Arabidopsis thaliana and 54% similarity of a 33-aa sequence with a segment of the NAD-dependent dehydrogenases COG0446 of P. syringae pv. syringae B728A. On the other hand, the 393-bp region between PSPTO2607 and PSPTO2608 showed 79% identity in, this time, a 371-bp overlap with the similar region in pathovar phaseolicola (between PSPPH2892 and PSPPH2891). The region common only to the pathovar phaseolicola contains a 70-bp AT-rich (84%) sequence that showed (BLASTN) 100% identity with 22-bp AT-rich sequences from Homo sapiens and A. thaliana and 90% identity with a 33-bp sequence from H. sapiens. The GC content is higher in the YBT locus than in the adjacent regions common in the three pathovars (Fig. 6).
FIG. 6.
Homologous regions around the YBT locus in P. syringae pv. tomato DC3000, P. syringae pv. syringae B728a, and P. syringae pv. phaseolicola 1448A and GC contents (P. syringae pv. tomato DC3000). Homologous regions are represented by rectangles; the YBT loci and the adjacent gene PSPTO2607 (DC3000) or PSPPH2892 (1448A) are black coded. Nonhomologous regions are shown either by a dark line when homologous proteins were found elsewhere in the genome of another strain or by a gray line when homologous proteins were not found anywhere in the genome of either of the other strains. The dotted line represents a gap. Homologous regions were determined by comparing protein or gene sequences using SSearch, BLASTP, or BLASTN and by comparing gene positions; protein homologies observed were higher than 80% identity. The last left and right genes represented that have a homologue in the same area in another pathovar are PSPTO2573 and PSPTO2609 (DC3000), Psyr_2264 and Psyr_2289 (B728a), and PSPPH2922 and PSPPH2887 (1448A); in 1448A, PSPPH2923 is an IS801 transposase.
Search for mutations in irp1 and irp2 in E. coli strains.
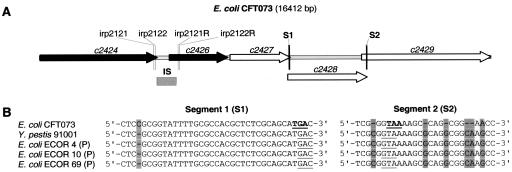
Tests were conducted to establish whether the E. coli CFT073 modifications in the irp2 and irp1 homologues (Fig. 7A) were present and, consequently, the resulting proteins functional in the six YBT-producing ECOR strains. In irp2, the four PCR tests gave PCR products of about 189, 248, 132, and 192 bp, which indicated the absence of the IS1541A-like due 711-bp insertion in the ECOR strains and the correct expression of the irp2 product HMWP2. In irp1, the sequenced segments were identical to the sequences of Y. pestis (Fig. 7B). We can conclude that there are differences in irp1 and irp2 between the strains investigated here and CFT073.
FIG. 7.
(A) Map of the irp2 and irp1 homologous zones in E. coli CFT073, as annotated in its genome. Shown in the irp2 zone are two ORFs (black arrows), a 711-bp IS1541A-like due insertion (IS), and the primers used in PCR analyses. Shown in the irp1 zone are three ORFs (white arrows) and the modified segments S1 and S2. (B) Comparisons of S1 and S2 in Y. pestis and E. coli. In E. coli CFT073, modifications (gray) induce the lecture of stop codons (bold); in the next ORFs, the correct translation would restart downstream of the last base modification. No indication of this was found in the six YBT-producing ECOR strains, which indicates that irp1 encodes HMWP1 in these strains.
DISCUSSION
The study showed that YBT is produced by P. syringae and confirmed the highly probable involvement of the predicted YBT locus of P. syringae pv. tomato DC3000 (8) in YBT production. However, in this study, irp1 was detected only in pathovars belonging to the genospecies 3, 7, and 8, as well as in the closely related pathovars phaseolicola and glycinea belonging to genospecies 2 (30, 49, 82). Although the presence of the YBT locus varies in a species (4, 14, 55, 58, 71), the separation observed within P. syringae was relatively clear. This may indicate the acquisition of the YBT locus by an ancestor of the producing pathovars and its stabilization in the chromosome or the acquisition of the YBT locus by an ancestor of P. syringae and a locus deletion in an ancestor of the nonproducing pathovars. This is confirmed by the identical locus organization and position in the genomes of the distant strains P. syringae pv. tomato DC3000 and P. syringae pv. phaseolicola 1448A, as well as by the protein heterogeneity in the YBT locus of these strains, which reflects a long evolution in P. syringae. A zone clearly involved in the insertion or the deletion of the YBT locus in P. syringae lies between PSPTO2607 and PSPTO2608 in pathovar tomato (between PSPPH2891 and PSPPH2892 in pathovar phaseolicola). The stability of the YBT locus in P. syringae was confirmed by the absence of integrase and IS elements in its proximity and by the observation that 92% of the strains from genospecies 3, 7, and 8 and pathovar morsprunorum race 2 possess the irp1 gene. The PCR test could then be used in the diagnostic of the YBT-positive pathovars.
The YBT locus was functional in 10 pathovars out of 15. The HPLC-negative strains could produce less YBT or have nonfunctional or differently regulated genes. The advantage of producing YBT is unclear for strains producing both PVD and YBT; the very high stability constant of Fe-YBT (4 × 1036 compared with 1025 for the PVD) could carry an adaptive advantage in the competition for iron on the plant surface, even perhaps against other pseudomonads. However, the existence of genospecies 1, 2, 4, and 6 within which almost all of the strains are YBT negative probably indicates that pathogenicity is possible for P. syringae without producing YBT. This seems all the more probable because the PVD was not involved in the virulence of P. syringae pv. syringae (a YBT-negative pathovar) (Table 1) on cherry fruits (18), but this should be further confirmed by the study of negative mutants. The YBT locus could be a genomic island involved in the fitness and competitiveness of P. syringae rather than a pathogenicity island sensu stricto (14, 23, 69).
Two evolutionary lineages were described in HPI-positive enterobacteria (66, 70), but the differences observed between Y. pestis, P. syringae, and P. luminescens are of a greater magnitude. It is clear that three YBT locus evolutionary groups exist, of which two have been shown to be functional; all the HPI-positive enterobacteria belong to the Y. pestis group. The P. syringae and P. luminescens YBT locus organizations are quite similar, and both lack integrase. Since it is unlikely that excision from the HPI occurred in the same way in two species, it is possible that this gene organization resembles that of the YBT locus before its insertion in the HPI, but the understanding of the HPI formation is still conjectural (73). However, it is not clear whether the final product of the P. luminescens YBT locus is YBT, because the final polyketide synthase/peptide synthetase Plu2321 shows little similarity with, and is 745 aa longer than, HMWP1 of Y. pestis. Consequently, a YBT-related compound could be the final product in P. luminescens, as observed with a pseudomonad producing micacocidin, which resembles YBT (43). The P. syringae YBT locus would then represent an evolutionary link between the two systems.
In P. syringae, the presence of the isochorismate pyruvate lyase gene pchB and the deduced sequence of PchA strongly suggest that chorismate is converted to salicylate in a two-step mechanism. On the other hand, the YbtS homologue of Y. enterocolitica (Irp9) converts chorismate directly to salicylate; a similar function is supposed for MbtI in mycobactin synthesis (62, 65). PchA of P. aeruginosa is unable to effect the direct conversion (29). Since PchA of P. syringae is more similar to PchA of P. aeruginosa than to YbtS, it is probably also unable to effect this conversion. The presence of pchB in the YBT locus of P. syringae could indicate that a pchB homologue was present in the ancestor of the YBT locus, but the low GC content of pchB suggests that it could have an external origin. Also, since PchA showed the lowest similarity with its homologue in the Y. pestis or P. luminescens YBT locus (Table 3), it is not clear whether these proteins, which probably have slightly different functions, have a common ancestor in the YBT locus.
Nonpathogenic E. coli strains were investigated to develop the general irp1-based PCR test and to confirm the adaptability of the HPLC method. The ECOR collection represents the diversity of E. coli (57), but variations were observed between the same strains from three origins. This should be noted by those studying the ECOR strains.
The YBT-producing nonpathogenic ECOR strains tested in this study lack the modification in irp1 and irp2 found in the sequences of CFT073, which belongs to the uropathogenic group, responsible for acute cystitis and pyelonephritis. The production of YBT by CFT073 is not clear, because irp5/ybtE, encoding the salicyl-AMP ligase, is also disrupted, and the resulting protein, c_2433, is 110 aa shorter than Irp5. The high occurrence of the HPI in pathogenic E. coli suggests that it could be a virulence factor (5, 6, 16, 22, 37, 38, 39, 41, 44, 59, 70, 71, 72), although this is still a matter of debate (46). But HMWP1, HMWP2, or YBT was not detected in parts of populations recorded as positive for irp1 and irp2 using DNA hybridization and/or PCR (70, 71, 72). The CFT073 modifications in irp1 are not detectable by the genetic methods used, and such modifications could be responsible for the observed results, since the inactivation of irp1 down-regulated HMWP2, FyuA, and YBT synthesis in Y. enterocolitica (61). All the HPI-positive ECOR strains were positive when using HPLC in this study, and HPLC combined with the genetic procedures described could be used to explore the ability of human-pathogenic strains to produce YBT.
YBT detection methods are based on the detection or expression of genes and proteins of the YBT locus or on cross-feeding tests (55, 70, 71), but no method allows the molecule to be visualized. And yet this does provide the most unambiguous information. It is the second time that the solid-liquid technique in petri dishes (10) has proved to be an efficient way of producing siderophores, and it probably has potential for other genera and metabolites. The HPLC method for detecting YBT in the culture medium is similar to that described for PVDs (12) and comparable to the HPLC methods used to detect salicylic acid-based siderophores in concentrated supernatant extracts (2, 26, 51, 52, 68, 76). The method seems to be useful because YBT production appears to be more widely dispersed in the environment than previously thought and because it can be adapted for every environmental microorganism without having to grow potentially human-pathogenic indicator strains. It could be used to find new YBT producers, such as P. luminescens, perhaps, and new YBT locus groups irrespective of gene sequences. In addition, the irp1 PCR test enables strains from two YBT locus groups to be simultaneously screened. The study of E. coli confirmed the usefulness of both methods.
Acknowledgments
We thank J. R. Johnson, E. Denamur, and B. Picard for providing strains, J. R. Johnson for exchanging data, R. Rozenberg for the MS analyses, and Y. Muhovski for advice on the DNA hybridization study.
The work was supported by the Ministry of the Walloon Region (A.B. and I.G.) and by the Belgian National Fund for Scientific Research (E.D.H.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankenbauer, R. G., T. Toyokuni, A. Staley, K. L. Rinehart, and C. D. Cox. 1988. Synthesis and biological activity of pyochelin, a siderophore of Pseudomonas aeruginosa. J. Bacteriol. 170:5344-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidlan, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bach, S., A. de Almeida, and E. Carniel. 2000. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183:289-294. [DOI] [PubMed] [Google Scholar]
- 5.Bingen-Bidois, M., O. Clermont, S. Bonacorsi, M. Terki, N. Brahimi, C. Loukil, D. Barraud, and E. Bingen. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonacorsi, S., O. Clermont, V. Houdouin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange, and E. Bingen. 2003. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J. Infect. Dis. 187:1895-1906. [DOI] [PubMed] [Google Scholar]
- 7.Budzikiewicz, H. 2004. Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non fluorescent Pseudomonas spp.), p. 81-237. In W. Herz, H. Falk, and G. W. Kirby (ed.), Progress in the chemistry of organic natural products, vol. 87. Springer, Vienna, Austria. [DOI] [PubMed] [Google Scholar]
- 8.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. Scott Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W.-L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultreys, A., and I. Gheysen. 1999. Biological and molecular detection of toxic lipodepsipeptide-producing Pseudomonas syringae strains and PCR identification in plants. Appl. Environ. Microbiol. 65:1904-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bultreys, A., and I. Gheysen. 2000. Production and comparison of peptide siderophores from strains of distantly related pathovars of Pseudomonas syringae and Pseudomonas viridiflava LMG 2352. Appl. Environ. Microbiol. 66:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bultreys, A., I. Gheysen, H. Maraite, and E. de Hoffmann. 2001. Characterization of fluorescent and nonfluorescent peptide siderophores produced by Pseudomonas syringae strains and their potential use in strain identification. Appl. Environ. Microbiol. 67:1718-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bultreys, A., I. Gheysen., B. Wathelet, H. Maraite, and E. de Hoffmann. 2003. High-performance liquid chromatography analyses of pyoverdin siderophores differentiate among phytopathogenic fluorescent Pseudomonas species. Appl. Environ. Microbiol. 69:1143-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bultreys, A., I. Gheysen, B. Wathelet, M. Schäfer, and H. Budzikiewicz. 2004. The pyoverdins of Pseudomonas syringae and Pseudomonas cichorii. Z. Naturforsch. Sect. C. 59:613-618. [DOI] [PubMed] [Google Scholar]
- 14.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 15.Chambers, C. E., D. D. McIntyre, M. Mouck, and P. A. Sokol. 1996. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. BioMetals 9:157-167. [DOI] [PubMed] [Google Scholar]
- 16.Clermont, O., S. Bonacorsi, and E. Bingen. 2001. The Yersinia high-pathogenicity island is highly predominant in virulence-associated phylogenetic groups of Escherichia coli. FEMS Microbiol. Lett. 196:153-157. [DOI] [PubMed] [Google Scholar]
- 17.Cody, Y. S., and D. C. Gross. 1987. Characterization of pyoverdinpss, the fluorescent siderophore produced by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 53:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cody, Y. S., and D. C. Gross. 1987. Outer membrane protein mediating iron uptake via pyoverdinpss, the fluorescent siderophore produced by Pseudomonas syringae pv. syringae. J. Bacteriol. 169:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 20.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacher. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 24.Drechsel, H., H. Stephan, H. Lotz, H. Haag, H. Zähner, K. Hantke, and G. Jung. 1995. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs. Ann. 1995:1727-1733. [Google Scholar]
- 25.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J.-F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Médigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 26.Duffy, B. K., and G. Défago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 29.Gaille, C., C. Reimmann, and D. Haas. 2003. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J. Biol. Chem. 278:16893-16898. [DOI] [PubMed] [Google Scholar]
- 30.Gardan, L., H. Shafik, S. Belouin, R. Broch, F. Grimont, and P. A. D. Grimont. 1999. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49:469-478. [DOI] [PubMed] [Google Scholar]
- 31.Gehring, A. M., E. DeMoll, J. D. Fetherson, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 32.Geoffroy, V. A., J. D. Fetherston, and R. D. Perry. 2000. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 68:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag, H., K. Hankte, H. Drechsel, I. Stojiljkovic, G. Jung, and H. Zähner. 1993. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol. 139:2159-2165. [DOI] [PubMed] [Google Scholar]
- 34.Hartwig, R. C., and R. H. Loeppert. 1993. Evaluation of soil iron, p. 465-482. In L. L. Barton and B. C. Hemming (ed.), Iron chelation in plants and soil microorganisms. Academic Press, San Diego, Calif.
- 35.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae--a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. DeBoy, A. S. Durkin, M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stall. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, J. R., T. T. O'Bryan, M. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jülich, M., K. Taraz, H. Budzikiewicz, V. Geoffroy, J.-M. Meyer, and L. Gardan. 2001. The structure of the pyoverdin isolated from various Pseudomonas syringae pathovars. Z. Naturforsch. Sect. C. 56:687-694. [DOI] [PubMed] [Google Scholar]
- 41.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 43.Kobayashi, S., H. Nakai, Y. Ikenishi, W. Y. Sun, M. Ozaki, Y. Hayase, and R. Takeda. 1998. Micacocidin A, B and C, novel antimycoplasma agents from Pseudomonas sp. II. Structure elucidation. J. Antibiot. 51:328-332. [DOI] [PubMed] [Google Scholar]
- 44.Koczura, R., and A. Kaznowski. 2003. The Yersinia high-pathogenicity island and iron-uptake systems in clinical isolates of Escherichia coli. J. Med. Microbiol. 52:637-642. [DOI] [PubMed] [Google Scholar]
- 45.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefranc Nègre, V., S. Bonacorsi, S. Schubert, P. Bidet, X. Nassif, and E. Bingen. 2004. The siderophore receptor iroN, but not the high-pathogenicity island or the hemin receptor chuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 72:1216-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 48.Loper, J. E., and. M. D. Henkels. 1997. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl. Environ. Microbiol. 63:99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marques, A. S. D. A., R. Corbière, L. Gardan, C. Tourte, C. Manceau, J. D. Taylor, and R. Samson. 2000. Multiphasic approach for the identification of the different classification levels of Pseudomonas savastanoi pv. phaseolicola. Eur. J. Plant Pathol. 106:715-734. [Google Scholar]
- 50.Martins dos Santos, V. A. P., S. Heim, E. R. B. Moore, M. Strätz, and K. N. Timmis. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264-1286. [DOI] [PubMed] [Google Scholar]
- 51.Maurhofer, M., C. Reimmann, P. Schmidli-Sacherer, S. Heeb, D. Haas, and G. Défago. 1998. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678-684. [DOI] [PubMed] [Google Scholar]
- 52.Mercado-Blanco, J., K. M. G. M. van der Drift, P. E. Olsson, J. E. Thomas-Oates, L. C. van Loon, and P. A. H. M. Bakker. 2001. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer, J.-M., V. A. Geoffroy, N. Baida, L. Gardan, D. Izard, P. Lemanceau, W. Achouak, and N. J. Palleroni. 2002. Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads. Appl. Environ. Microbiol. 68:2745-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller, D. A., L. Luo, N. Hillson, T. A. Keating, and C. T. Walsh. 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9:333-334. [DOI] [PubMed] [Google Scholar]
- 55.Mokracka, J., R. Koczura, and A. Kaznowski. 2004. Yersiniabactin and other siderophores produced by clinical isolates of Enterobacter spp. and Citrobacter spp. FEMS Immunol. Med. Microbiol. 40:51-55. [DOI] [PubMed] [Google Scholar]
- 56.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 57.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oelschlaeger, T. A., D. Zhang, S. Schubert, E. Carniel, W. Rabsch, H. Karch, and J. Hacker. 2003. The high-pathogenicity island is absent in human pathogens of Salmonella enterica subspecies I but present in isolates of subspecies III and VI. J. Bacteriol. 185:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okeke, I. N., I. C. A. Scaletsky, E. H. Soars, L. R. Macfarlane, and A. G. Torres. 2004. Molecular epidemiology of the iron utilization genes of enteroaggregative Escherichia coli. J. Clin. Microbiol. 42:36-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tárraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 61.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelludat, C., D. Brem, and J. Heesemann. 2003. Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J. Bacteriol. 185:5648-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181-1190. [DOI] [PubMed] [Google Scholar]
- 64.Perry, R. D. 2004. Yersinia, p. 219-240. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. American Society for Microbiology, Washington, D.C.
- 65.Quadri, L. E. N., J. Sello, T. A. Keatin, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 66.Rakin, A., P. Urbitsch, and J. Heesemann. 1995. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J. Bacteriol. 177:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 68.Reimmann, C., H. M. Patel, C. T. Walsh, and D. Haas. 2004. PchC thioesterase optimizes nonribosomal biosynthesis of the peptide siderophore pyochelin in Pseudomonas aeruginosa J. Bacteriol. 186:6367-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heeseman. 1998. Prevalence of the “high-pathogenicity island” of yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schubert, S., S. Cuenca, D. Fischer, and J. Heesemann. 2000. High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J. Infect. Dis. 182:1268-1271. [DOI] [PubMed] [Google Scholar]
- 72.Schubert, S., B. Picard, S. Gouriou, J. Heesemann, and E. Denamur. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70:5335-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schubert, S., S. Dufke, J. Sorsa, and J. Heeseman. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51:837-848. [DOI] [PubMed] [Google Scholar]
- 74.Schwyn, B., and J. B. Neilands. 1986. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 75.Serino, L., C. Reimmann, H. Baur, M. Beyeler, P. Visca, and D. Haas. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249:217-228. [DOI] [PubMed] [Google Scholar]
- 76.Serino, L., C. Reimmann, P. Visca, M. Beyeler, V. D. Chiesa, and D. Haas. 1997. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 179:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simon, E. H., and I. Tessman. 1963. Thymidine-requiring mutants of phage T4. Proc. Natl. Acad. Sci. USA 50:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 79.Smith, E. E., E. H. Sims, D. H. Spencer, R. Kaul, and M. V. Olson. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 187:2138-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, Y. Han, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Wang, J. Yu, H. Yang, J. Wang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 81.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 82.Völksch, B., and H. Weingart. 1997. Comparison of ethylene-producing Pseudomonas syringae strains isolated from kudzu (Pueraria lobata) with Pseudomonas syringae pv. phaseolicola and Pseudomonas syringae pv. glycinea. Eur. J. Plant Pathol. 103:795-802. [Google Scholar]
- 83.Walsh, C. T., and C. G. Marshall. 2004. Siderophore biosynthesis in bacteria, p. 18-37. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. American Society for Microbiology, Washington, D.C.
- 84.Welch, R. A., V. Burland, G. Plunkett, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]