Abstract
Polyethylene glycol dehydrogenase (PEGDH) from Sphingopyxis terrae (formerly Sphingomonas terrae) is composed of 535 amino acid residues and one flavin adenine dinucleotide per monomer protein in a homodimeric structure. Its amino acid sequence shows 28.5 to 30.5% identity with glucose oxidases from Aspergillus niger and Penicillium amagasakiense. The ADP-binding site and the signature 1 and 2 consensus sequences of glucose-methanol-choline oxidoreductases are present in PEGDH. Based on three-dimensional molecular modeling and kinetic characterization of wild-type PEGDH and mutant PEGDHs constructed by site-directed mutagenesis, residues potentially involved in catalysis and substrate binding were found in the vicinity of the flavin ring. The catalytically important active sites were assigned to His-467 and Asn-511. One disulfide bridge between Cys-379 and Cys-382 existed in PEGDH and seemed to play roles in both substrate binding and electron mediation. The Cys-297 mutant showed decreased activity, suggesting the residue's importance in both substrate binding and electron mediation, as well as Cys-379 and Cys-382. PEGDH also contains a motif of a ubiquinone-binding site, and coenzyme Q10 was utilized as an electron acceptor. Thus, we propose several important amino acid residues involved in the electron transfer pathway from the substrate to ubiquinone.
Polyethylene glycols (PEGs) are produced in large quantities totaling more than 100 million tons per year and are used extensively in many industries, including in pharmaceuticals, cosmetics, detergents, and lubricating oils. They are used in a broad spectrum of sizes and in a variety of derivatives. After use, most of them eventually show up in sewage water, since they are water soluble, but they are biodegraded by many aerobic bacteria belonging to different genera (12, 34). Several groups have also reported anaerobic degradation of PEGs and nonionic detergents that contain PEGs (13, 14). Thus, PEGs are thought to be environmentally compatible xenobiotic polymers.
Several Sphingomonas species have been reported to biodegrade PEGs, in either a single culture (e.g., Sphingomonas macrogoltabidus) (15) or a mixed culture (e.g., Sphingomonas terrae and a Rhizobium sp.) (18) (S. terrae was renamed as Sphingopyxis terrae). In all cases, it has been determined that the first step on the degradation pathway is the oxidation of the terminal alcohol groups with a dye-linked dehydrogenase (16) called PEG dehydrogenase (PEGDH). Purification and characterization of PEGDH from S. terrae were reported by Kawai et al. (17, 19), who suggested that the enzyme is bound to the membrane and recognizes a broad range of primary aliphatic and aromatic alcohols as substrates. The gene encoding the enzyme (pegA) was cloned, sequenced, and expressed in Escherichia coli (29). The deduced amino acid sequence suggested that PEGDH did not contain a signal peptide sequence in spite of its membrane-bound characteristics. A comparison with known sequences showed that PEGDH belongs to a group of glucose-methanol-choline (GMC) flavoprotein oxidoreductases (3), a group of enzymes having noncovalently bound flavin adenine dinucleotide (FAD) as a cofactor. Membrane-bound, dye-linked bacterial alcohol dehydrogenases (ADHs) (ADH of Pseudomonas oleovorans [32] and 4-nitrobenzyl ADH of a Pseudomonas species [10]) had high levels of similarity to PEGDH (46% and 35% identity, respectively). These three ADHs shared the characteristic FAD-binding signatures and the same proposed catalytic residues in the GMC group, but no signal peptides (29). ADH of Pseudomonas oleovorans and 4-nitrobenzyl ADH of a Pseudomonas species were involved in the catabolism of alkanes and 4-nitrobenzyl alcohol (and benzyl alcohol), respectively, just as PEGDH is involved in the catabolism of PEG. The fact that benzyl alcohol was a better substrate for PEGDH than PEG might support the notion that these catabolic ADHs have evolved from the same origin, sharing the same protein structures. In addition, PEGDH is similar to both oxidases and dehydrogenases in the GMC group, and its sequence is thought to be a hybrid of these two enzymes. This fact evoked an interest in the catalytic mechanisms of this novel type of dehydrogenase. Except for PEGDH, this family of dehydrogenases has scarcely been characterized with respect to conserved signatures and active-site residues.
Flavoenzymes are known to have a variety of folding topologies, but their catalytic apparatuses have some recurrent features (7). Flavoenzymes that catalyze dehydrogenation reactions share a few invariant features in the hydrogen bond interaction between their protein and flavin constituents. Similarly, the positioning of the reactive part of the substrate with respect to the cofactor is generally conserved. Modulation of substrate and cofactor reactivities and the exact positioning of the substrate in the active site are key elements in these enzymes' modes of action. Among GMC oxidoreductases, glucose oxidase (GOX) has been crystallized and well characterized and is often used as a model structure for the group (35). PEGDH shows 30.5% identity with GOX from Aspergillus niger and 28.5% identity with GOX from Penicillium amagasakiense. Using GOX from A. niger as the template, we performed three-dimensional (3D) modeling of PEGDH and analyzed the kinetic parameters of wild-type and mutant enzymes. Mutation was introduced into His-467 and Asn-511, corresponding to His-538 and His-581 in the catalytic site of GOX, and into four cysteine residues found in the catalytic domain of PEGDH (PEGDH included eight cysteine residues). Previous data (17, 29) strongly suggested that cysteine residues play a part in the activity.
In this paper, we show the molecular functions of the amino acid residues found in the active site and illustrate a predicted electron transfer pathway of PEGDH based on the 3D modeling.
MATERIALS AND METHODS
Materials, bacterial strains, and plasmids.
Restriction enzymes, T4 DNA ligase, and Ex Taq DNA polymerase were purchased from Takara Bio (Kyoto, Japan). The primers used for site-directed mutagenesis are listed in Table 1. The bacterial strains used in this study were derivatives of E. coli. Their relevant genotypes and plasmids are listed in Table 1. Rotenone, coenzyme Q10 (CoQ10), and other reagents were obtained from commercial sources.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and description | Reference or source |
|---|---|---|
| Strains | ||
| DH 5α | deoR endA1 gyrA96 hsdR17(rk− mk+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argF)U169 φ80lacZ ΔM15 F− λ− | Takara Bio |
| BL21 (DE3)(pLysS) | E. coli B; F−dcm ompT hsdS(rB− mB−) ga λ(DE3) [pLysS Camr] | Takara Bio |
| Plasmids | ||
| pET23d | Ampr His tag | Novagen |
| pPEGDHEX | Ampr His tag peg A wild type | 29 |
| EX-N1 5′-AGCCATGGACAAATTCGACTTTGTCGTGG-3′ | ||
| EX-C1 5′-CCTCGAGGGTGTAAGATTCGGCTTCATC-3′ | ||
| pPEGDHEX-H467Aa | Ampr His tag pegA mutant H467A (CAT→GCT) | This work |
| EX-N1 and 5′-GGTACCAACCGGUAGCUATAGATCGT-3′ | ||
| EX-C1 and 5′-CCGGTTGGTACCTGCAAGATG-3′ | ||
| pPEGDHEX-N511Ha | Ampr His tag pegA mutant N511H (AAC→CAC) | This work |
| EX-N1 and 5′-CCCGATACAATTGAAGGCATGA-3′ | ||
| EX-C1 and 5′-CTTCAATTGTATCGGGACACACAAAT-3′ | ||
| pPEGDHEX-C472Aa | Ampr His-tag peg A mutant C472A (TGC→GCC) | This work |
| EX-N1 and 5′-GGTACCAACCGGATG-3′ | ||
| EX-C1 and 5′-GTTGGTACCGCCAAGATGGGTCGG-3′ | ||
| pPEGDHEX-C379Aa | Ampr His-tag peg A mutant C379A (TGC→GCC) | This work |
| EX-N1 and 5′-GGACACACACGTGGCCGCTAAATCCGCCGC-3′ | ||
| EX-C1 and 5′-TAGCTGCCACGTGTGTGTCC-3′ | ||
| pPEGDHEX-C382Aa | Ampr His-tag peg A mutant C382A (TGT→GCC) | This work |
| EX-N1 and 5′-GGACACACACGTGGCAGCT-3′ | ||
| EX-C1 and 5′-CGGATTTAGCGCCCACGTGTGTGTCC-3′ | ||
| pPEGDHEX-C297Aa | Ampr His-tag peg A mutant C297A (TGT→GCC) | This work |
| 5′-ATTTCUGCCUTTAATGTACCAA-3′ | ||
| 5′-ATTAAGGCGAAATCTACATG-3′ |
Obtained by site-directed mutagenesis. Sequences introduced for site-directed mutagenesis are underlined.
Mutagenesis.
Six amino acids in PEGDH (His-467, Asn-511, Cys-297, Cys-379, Cys-382, and Cys-472) were selected as candidates for site-directed mutagenesis, which was performed by the method of Crameri and Stemmer (4), except for Cys-297, for which we used the method for the QuikChange II XL Site-Directed Mutagenesis Kit (Funakoshi, Tokyo, Japan). Five of these amino acids were changed to alanine to lose activity, and Asn-511 was changed to histidine for known replaceability by histidine for the activity. Using PCR and the primers listed in Table 1, we separately amplified the 5′ and 3′ regions of the gene divided by the mutation site. DNA manipulations were carried out as described by Sambrook et al. (26). The amplified fragments were cloned in pGEM-T Easy Vector (Promega, Madison, WI) and digested by the restriction enzymes whose recognition sites were introduced by the primers. Two fragments of each gene were ligated and inserted between the NcoI and XhoI sites of pET23d. The resulting plasmids, pPEGDHEX-H467A, pPEGDHEX-N511H, pPEGDHEX-C379A, pPEGDHEX-C382A, and pPEGDHEX-C472A, were introduced into E. coli BL21(DE3)(pLysS). A sixth plasmid, pPEGDHEX-C297A, in which Cys-297 was mutated to alanine, was constructed by using the primers listed in Table 1. Their DNA sequences were confirmed using an ABI310 DNA sequencer (Applied Biosystems, Foster City, CA). Expression of PEGDH mutants was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting (22) using a polyclonal antibody raised against PEGDH purified from E. coli(pPEGDH) (29).
Purification of PEGDHs.
E. coli BL21(DE3)(pLysS)(pPEGDH) was precultured in 4 ml of 2× yeast extract-tryptone medium (1.0% yeast extract-1.6% tryptone-0.5% NaCL) medium containing 50 μg/ml ampicillin and 1% glucose at 37°C overnight. The medium was then transferred into 100 ml of LB medium containing 50 μg/ml ampicillin and 1% glucose in a 500-ml shaking flask. The culture was shaken at 150 rpm until the optical density at 600 nm reached 0.6 at 37°C. Then, isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. The cells were cultivated for an additional 3 h at 25°C. The culture was centrifuged, and the cells were suspended in 50 mM Tris-HCl (pH 8.8) containing 500 mM NaCl, 10% ethylene glycol, and 0.2% n-dodecyl maltoside (buffer A) (29). The cells were disrupted by sonication, and cell debris was removed by centrifugation at 40,000 × g for 20 min at 4°C. The supernatant was applied to a Ni-nitrilotriacetic acid agarose column (QIAGEN, Valencia, CA) equilibrated with buffer A. The column was washed with the same buffer, and the bound protein was eluted with buffer A containing 100 mM imidazole. Mutant PEGDHs were purified in the same way as the wild-type PEGDH. All PEGDHs were used as histidine-tagged proteins without any further purification. Approximately 3 mg of the wild-type PEGDH was obtained from 2 g (wet weight) cells cultivated in 800 ml medium. The yield of purification for each PEGDH was approximately 90%.
Enzyme assay.
PEGDH activity was measured using PEG 6000, benzyl alcohol, PEG 1000, tetraethylene glycol (TEG), and 1-hexanol as substrates. The wild-type and mutant PEGDHs (0.4 μM each) were incubated in 0.1 M Tris-HCl (pH 9.0) containing 0.2 M KCl in a total volume of 1 ml, as described previously (29). PEGDH activity was examined by measuring the reduction of 2,6-dichloroindophenol (DCIP; ɛ = 19,000) (27) at 600 nm with a UV-1600 spectrophotometer (Shimadzu, Kyoto, Japan). The enzyme activity was also measured by the reduction of CoQ10 (molar extinction coefficient [ɛ] = 14,500) (23) at 270 nm. CoQ10 was suspended in 20% dodecyl maltoside at a concentration of 200 μM and dissolved by being heated at 50°C for 10 min in the dark. After confirming the absorbance of the supernatant, an appropriate amount of it was used for the reaction. To estimate the inhibitory effect of rotenone on the enzyme activity, the absorption change of CoQ10 at 270 nm was measured in 0.1 M Tris-HCl (pH 9.0) containing 0.2 M KCl, 0 to 500 μM rotenone, 0.2 μM PEGDH, 5 mM PEG 6000, and 0.4 μM CoQ10. The protein concentration was determined using a Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as a standard. Apparent Km values were estimated on the basis of Lineweaver-Burk plots. The precision of absorbance was 0.001. The deviation for Km values was ±0.03 for all wild-type and mutant PEGDHs, but kcat values for wild-type and C472A and other mutant PEGDHs were in the range of ±0.2 and ±0.04, respectively. The absorption spectra of the wild-type and mutant enzymes were measured with the spectrophotometer mentioned above. The absorbance was scanned with a 1-cm-path-length quartz cell containing 250 μl solution, which contained approximately 4 μM of PEGDH, 0.1 M Tris-HCl buffer (pH 9.0), and 0.2 M KCl. Stopped-flow analysis was carried out with a UV-3000 Shimadzu spectrophotometer equipped with a rapid-mixing attachment (model RMA-1A). The precision of absorbance was 0.0001. The reactions were initiated by rapid mixing of 100 μl of each of two solutions, one containing 2 μM purified PEGDH and the other containing 0.2 M Tris-HCl (pH 9.0), 0.4 M KCl, and 10 mM PEG 6000. The absorption change of FAD at 412 nm was monitored for 2.0 s at 30°C with a 1-cm-path-length quartz cell. The data are shown as average values of two or three independent experiments, each performed in triplicate.
Measurement of free sulfhydryl groups in the wild and mutant PEGDHs.
The reaction mixture contained 0.1 M sodium phosphate buffer (pH 7.26), 1 mM EDTA, 5 mM 5,5′-dithio-bis 2-nitrobenzoic acid, and an appropriate amount of the purified PEGDHs. After incubation at room temperature for 2 h, the absorption of the thio-nitrobenzoic acid produced was measured at 412 nm (5). Glutathione (reduced form) was used as a standard.
Modeling of 3D structure.
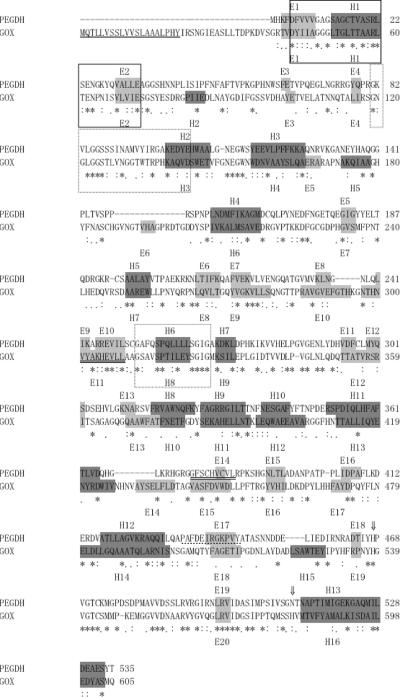
There are two structurally defined homologues of PEGDH: GOX (EC 1.1.3.4) from A. niger (Protein Data Bank code, 1cf3; 30.5% identity with PEGDH) and GOX (EC 1.1.3.4) from P. amagasakiense (Protein Data Bank code, 1gpe; 28.5% identity with PEGDH) (35). Two GOXs have sequence identity of 68.5% with each other. Both are homodimers that contain FAD and some number of N-acetylglucosamine and mannose moieties. According to the SCOP (http://scop.mrc-lmb.cam.ac.uk/scop/) taxonomy, the N-terminal domain and the end of the C-terminal domain (1cf3: 25 to 346 and 543 to 605) belong to the 3.3.1 “FAD/NAD(P)-binding domain” (core, three layers, β/β/α; central parallel beta-sheet of five strands, order 32145; top antiparallel beta-sheet of three strands, meander), and the C-terminal domain (347 to 542) belongs to the 4.15.1 “FAD-linked reductases, C-terminal domain” (alpha-plus-beta sandwich). MODELLER release 6v2 (developed by A. Sali [25]; http://salilab.org/modeller/modeller.html) was used for modeling. We chose 1cf3 as the template structure. Because the software does not have energy parameters for FAD, an FAD molecule was modeled using the “block” residue option under the restraint that it conformed to the template structure. PSI-BLAST (1) was used to align PEGDH and GOX (Fig. 1). Benzyl alcohol was placed in the enzyme as a model substrate. Fraaije and Mattevi (7) reported that flavoprotein enzymes share common features in flavin and substrate orientation. Following their observation, the orientation of benzyl alcohol to the FAD was decided (see Fig. 6). The 3D structure of PEGDH predicted by homology modeling is shown in Fig. 2.
FIG. 1.
Sequence alignment of PEGDH from S. terrae and GOX from A. niger. Gaps, indicated by dashes, were introduced into the sequences to maximize homology. Below the alignment, identical amino acids are marked by asterisks. Similar amino acids marked by colons or periods, according to their similarities. ⇓, active-site residues; box, FAD/NAD(P)-binding Rossmann fold; dashed boxes, GMC oxidoreductase signatures (3); underline, signal peptide sequence; double underline, quinone-binding motif; dotted underline, membrane-anchoring motif; α-helices and β-strands are shown as gray background with E and black background with H, respectively.
FIG. 6.
Proposed electron transfer pathway from flavin via three cysteines to ubiquinone embedded in the cytoplasm membrane. UQ, ubiquinone.
FIG. 2.
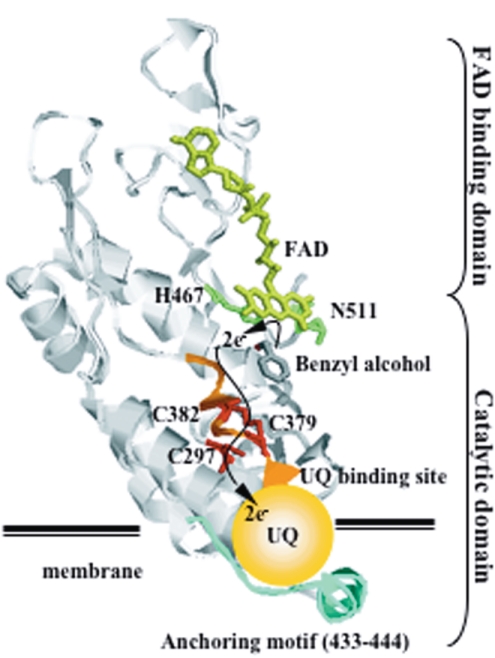
Predicted 3D structure of PEGDH by homology modeling. Yellow, FAD; green, active-site residues; pink, substrate-binding residues; red-orange, three cysteine residues; orange, ubiquinone-binding site; and dark blue, membrane-anchoring motif.
RESULTS
Purification of mutant PEGDHs.
The mutant PEGDHs were successfully constructed and purified by Ni-nitrilotriacetic acid affinity chromatography. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that the molecular weights of mutant proteins were identical to that of the wild-type PEGDH. The stabilities and expression levels of mutant PEGDH proteins were checked by Western blot analysis (data not shown). The analysis revealed that mutation did not change the higher structure of the protein and that the expression levels of mutant PEGDHs were 70 to 90% of that of the wild type.
Roles of H467A, N511H, and C472A.
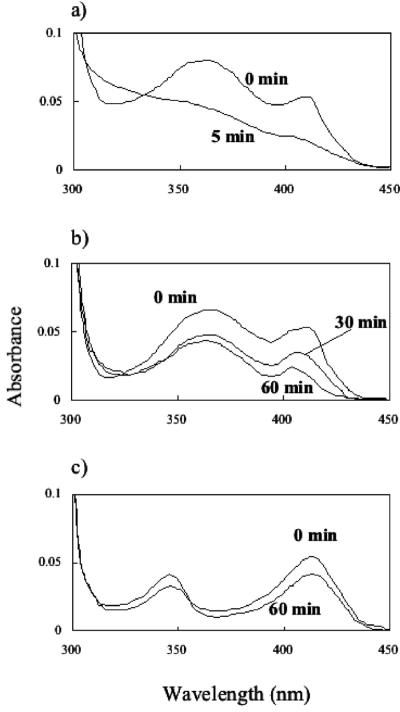
Mutations were introduced into His-467 and Asn-511, which were expected to be catalytic residues according to the comparison with the GMC oxidoreductase family enzymes (3). As Cys-472 was conserved in the GMC oxidoreductase, a mutation was also introduced in this residue. To compare the flavin spectra of the wild-type and mutant PEGDHs, the absorption spectra of the purified PEGDHs were determined. The wild-type and mutant PEGDHs showed two absorption peaks at 356 and 412 nm, except H467A and N511H, each of which showed a shifted peak at 346 nm, and the absorbance of H467A and N511H with the 346 nm peak was lower than that of the wild type, as shown in Fig. 3. The absorption ratios (A280/A412) were almost the same between the wild type and mutants. Based on the calculation using a ɛ of 11,300 for FAD at 412 nm, each subunit of the wild-type and mutant PEGDHs contained 1 FAD molecule. Thus, every mutant existed as a holoenzyme.
FIG. 3.
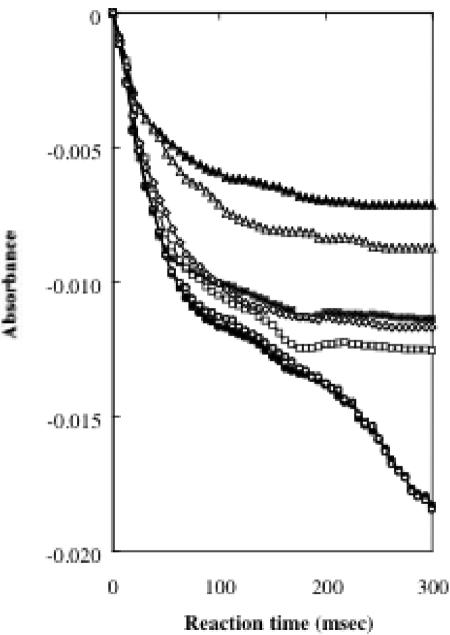
Absorption spectra of wild- type and mutant PEGDHs. The absorption spectra were measured with and without PEG 6000. Flakes of PEG 6000 were added at a final concentration of 5 mM and maintained at 30°C. (a) Wild type and C472A. (b) C297A, C379A, and C382A. (c) H467A and N511H.
To examine the effects of mutations on enzyme kinetics, the steady-state apparent kinetic parameters were determined using PEG 6000, PEG 1000, TEG, 1-hexanol, and benzyl alcohol as substrates (Table 2). H467A and N511H showed no significant change in their affinities toward substrates, except for benzyl alcohol. The apparent kcat values decreased to approximately 1/50 to 1/90 with H467A and to approximately 1/20 to 1/30 with N511H compared with the wild type. H467A and N511H showed higher apparent Km values and lower apparent kcat values for benzyl alcohol than those of the wild type. Catalytic efficiencies (kcat/Km) for benzyl alcohol of H467A and N511H were less than 0.5% of that of the wild type. kcat was always higher with N511H than with H467A, but the value with N511H was only 3 to 5% of the wild type. Although N511H is equivalent to catalytic active sites in GOX, Asn-511 could not be replaced with His in PEGDH. Detectable activity (3 to 5% of that of the wild type) might be due to the pH used for the reaction (pH 9.0), as dissociation constants of two protons and three protons of His are around 6.0 and 9.2, respectively. These results showed that His-467 and Asn-511 are directly involved in the enzymatic activity. The apparent Km and kcat values of C472A showed no differences from those of the wild type, suggesting that Cys-472 does not play an important role in catalytic activity, although the cysteine residue at this position is well conserved among dehydrogenases of GMC oxidoreductases and GOX.
TABLE 2.
Kinetic properties of purified wild-type and mutant PEGDHs
| Protein | PEG 6000
|
PEG 1000
|
TEG
|
1-Hexanol
|
Benzyl alcohol
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) | |
| Wild type | 2.4 | 12 | 5.0 | 1.1 | 15 | 13 | 6.7 | 7.6 | 1.1 | 0.45 | 11 | 24 | 0.11 | 18 | 170 |
| H467A | 2.4 | 0.15 | 0.06 | 1.1 | 0.17 | 0.15 | 6.8 | 0.15 | 0.02 | 0.44 | 0.20 | 0.56 | 0.81 | 0.29 | 0.36 |
| N511H | 2.3 | 0.36 | 0.16 | 1.2 | 0.46 | 0.38 | 6.9 | 0.37 | 0.05 | 0.45 | 0.58 | 1.3 | 0.84 | 0.67 | 0.80 |
| C472A | 2.3 | 11 | 5.0 | 1.2 | 15 | 13 | 6.9 | 7.6 | 1.1 | 0.44 | 11 | 25 | 0.10 | 18 | 180 |
| C379A | 2.5 | 0.61 | 0.25 | 1.1 | 0.62 | 0.57 | 8.3 | 0.62 | 0.08 | 1.7 | 1.4 | 0.84 | 1.4 | 1.4 | 1.0 |
| C382A | 2.5 | 0.80 | 0.32 | 1.0 | 0.82 | 0.82 | 8.1 | 0.82 | 0.10 | 1.7 | 1.8 | 1.1 | 1.2 | 1.8 | 1.5 |
| C297A | 2.5 | 0.86 | 0.34 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1.1 | 2.1 | 1.9 |
The proton is thought to be transferred from substrate to FAD by dehydrogenation in the catalytic domain. To confirm the residues' roles in dehydrogenation, the velocities of the initial step in the enzymatic reaction (the flavin reduction rate) in the wild-type and mutant enzymes were compared. A stopped-flow experiment was performed by monitoring (0 to 3.0 s) the absorption change at 412 nm, as shown in Fig. 4. The wild type and C472A showed the same rate (100%). In contrast, values for H467A and N511H were decreased to approximately 40 and 50%, respectively, suggesting that His-467 and Asn-511 should play catalytically important roles in the active site.
FIG. 4.
Kinetic traces of reactions by wild-type and mutant PEGDHs. Absorption at 412 nm was monitored after adding 5 mM PEG 6000. Closed circle, wild type; open circle, C472A; closed triangle, H467A; open triangle, N511H; closed square, C379A; open square, C382A; and open diamond, C297A.
Roles of cysteine residues in PEGDH activity.
In the previous study, enzymatic activity was inhibited by p-chloromercuribenzoic acid and heavy metals (17, 29), suggesting that a thiol group was involved in PEGDH activity. The amino acid sequence of PEGDH contains eight cysteine residues (29). Four cysteines (Cys-16, -194, -252, and -472) were located in the FAD-binding domain, and others (Cys-165, -297, -379, and -382) were located in the catalytic domain (Fig. 2). Since an S-S bridge helps to stabilize the protein structure in GOX (35), we measured the number of S-S bridges in PEGDH and identified the position of an S-S bridge by constructing several mutant enzymes, such as C297A, C379A, and C382A (Table 3). The wild type contained six free thiol groups, indicating the presence of one S-S bridge in the subunit. The number of free thiol groups decreased to five with C297A but increased to seven with C379A and C382A. Thus, we can conclude that an S-S bridge exists between Cys-379 and Cys-382, the position of which is different from that in GOX and which seems to play a different role in PEGDH than in GOX.
TABLE 3.
Measurement of free thiol groups in wild-type and mutant PEGDHs
| PEGDH | Free thiol groups/ monomer proteina |
|---|---|
| Wild type | 5.82 ± 0.10 |
| C472A | 4.97 ± 0.15 |
| C297A | 5.07 ± 0.05 |
| C379A | 7.16 ± 0.07 |
| C382A | 7.20 ± 0.04 |
The values of free thiol groups were measured as described in Materials and Methods.
C297A, C379A, and C382A showed FAD absorption spectra similar to those of the wild type and C472A (Fig. 3), suggesting that holoenzymes were formed in these mutants. Stopped-flow analysis showed that three cysteines are equivalently involved in the activity, because three mutants showed approximately the same behavior (Fig. 4). In all mutants, the apparent Km values for PEG 6000 and PEG 1000 were almost the same as in the wild type, but the apparent Km values for TEG, 1-hexanol, and benzyl alcohol were higher than that of the wild type (Table 2). The apparent kcat values decreased to approximately 1/8 to 1/25 with C379A and to approximately 1/6 to 1/18 with C382A and C297A compared with the wild type. Since considerable activity (4 to 15% of the wild-type activity left) was retained in every mutant PEGDH, these residues are likely involved in some way in binding or catalysis, although they are probably not directly involved in dehydrogenation.
Electron acceptor.
A previous study suggested that coenzyme Q could be an acceptor for PEGDH in vitro (19). As sphingomonads in general contain ubiquinone 10 (CoQ10) (28), the activity of the cloned enzyme was confirmed with CoQ10, as shown in Table 4. The apparent Km value of CoQ10 (11 μM) was lower than that of DCIP (30 μM), and the apparent kcat value of CoQ10 (0.33 s−1) was lower than that of DCIP (9.7 s−1). As rotenone is known as a specific inhibitor for the proton-pumping NADH-ubiquinone oxidoreductase (complex I; EC 1.65.3) and is thought to act on the quinone binding site, we measured the I50 values of rotenone (the concentrations of rotenone that caused 50% inhibition) (6, 24). The I50 values toward CoQ10 and DCIP were 0.24 μM and 0.28 μM, respectively, suggesting that the concentration of the inhibitor did not depend on the type of electron acceptor (CoQ10 and DCIP).
TABLE 4.
Comparison of kinetic parameters of the wild-type PEGDH for CoQ10 and DCIP
| Electron acceptor | Km (μM) | kcat (s−1) | I50a (μM) |
|---|---|---|---|
| CoQ10 | 11 | 0.33 | 0.24 |
| DCIP | 30 | 9.7 | 0.28 |
Proposed 3D structure.
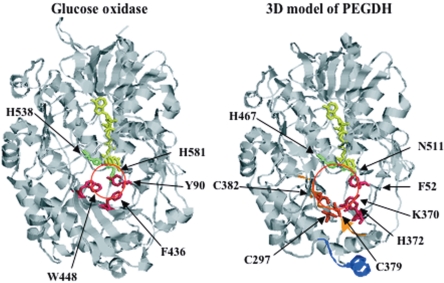
The sequence identities between PEGDH and GOX were not very high, but the alignment of PEGDH and GOX covered almost all of the protein length (Fig. 1). PEGDH has fewer predicted helices and strands (13 and 19, respectively) than GOX (16 α-helices and 20 β-strands). The ADP-binding motif (5 to 34) and the signature 1 and 2 consensus sequences (81 to 104 and 253 to 267, respectively) of GMC oxidoreductases (3) were also present. The secondary structure (β1 strand-α1 helix-β2 strand-α2 helix-β3 strand) and the sequence motif (V/IXGXGX1-2GXXGXXXG/A) of the FAD/NAD(P)-binding Rossmann fold (20) were conserved in both enzymes, although the α2-helix of GOX is apparently absent in PEGDH. Both enzymes have one tightly bound FAD per monomer protein. The differences between them are mainly in the N-terminal region (a signal peptide in GOX consisting of approximately 40 amino acids). Four amino acid regions of GOX (188 to 201, 292 to 296, 427 to 434, and 522 to 525) are absent in PEGDH, and the carbohydrate content in GOX is also absent in PEGDH. Sequences in the regions around FAD (the FAD-binding domain) and the active center (the substrate-binding domain) were well conserved. Residues potentially involved in catalysis (His-467 and Asn-511) were identified in the vicinity of the flavin ring (Fig. 2).
The reacting CH group in benzyl alcohol was placed at the re site (7), so that the distance from the CH group to the N-5 atom in FAD was 3.5 Å and the angle N-10-N-5-CH was 105°. We assumed that the benzene ring of the substrate is parallel to the FAD ring and that the hydroxyl group of the substrate is on the opposite side from the FAD ring. The orientation of benzyl alcohol in the active site seems to be stabilized by hydrophobic contacts with Lys-370, His-372, and, to a lesser extent, Phe-52 and FAD, as in the case of GOX (Phe-436, Trp-448, Tyr-90, and FAD, respectively) (35). Since the position of Trp-448 of GOX corresponded to Cys-379 and Cys-382 rather than His-372, three cysteine residues might play the same role as Trp-448, forming a bigger cavity in the active site (approximately 13 Å) than that of GOX (approximately 9 Å), as shown in Fig. 2. In the present model complex (Fig. 2), the hydroxyl group of benzyl alcohol sits virtually equidistant between His-467 and Asn-511, but these residues were not catalytic sites. The expanded cavity in the active site of PEGDH might have resulted in different roles of His-467 and Asn-511 and different catalytic sites.
DISCUSSION
In PEGDH, His-467 and Asn-511 corresponded to the positions of His-538 and His-581 in GOX. The reduction rate of FAD decreased remarkably but retained 1 to 5% of the wild-type activity in these two mutant enzymes, suggesting that both residues are catalytically important active residues but not catalytic sites. Two histidines (His-538 and His-581) exist as hydrogen bond donors in the active site of GOX (35). The former is well conserved in all members of the GMC oxidoreductase family (3). The latter, however, is not conserved in any members of the family (His-581 is replaced by Asn), although both histidines may share an overall structural similarity and similar reaction mechanisms (35). As shown in Table 2, the apparent Km values of H467A and N511H did not change toward any of the substrates except benzyl alcohol, but apparent kcat values decreased by 2 orders of magnitude compared with the wild-type enzyme. This suggested that His-467 and Asn-511 are crucial for catalysis in PEGDH. This, in turn, would be explained by the difference in hydrophobicity between PEG and benzyl alcohol. The stronger affinity of His-467 versus Asn-511 to hydrophobic substrates could cause an inappropriate positioning of benzyl alcohol in N511H. Fraaije et al. (8) reported that the spectral characteristics at shorter wavelengths were drastically changed by a mutation in the histidine residue on a flavoenzyme, as found in H467A and N511H. These data indicate that His-467 and Asn-511 are important active residues of PEGDH. N511H is equivalent to amino acids found in the active site of GOX, but activity was almost completely lost. This result suggested that Asn-511 plays an important catalytic role in the activity of PEGDH and that the catalytic mechanism in PEGDH might be different from that in GOX.
The absorption spectra of C297A, C379A, and C382A were the same as those of the wild type and C472A (Fig. 3), but the reduction rates of flavin were decreased in the three mutants. The DCIP-dependent enzymatic activities of these mutants also decreased for all the substrates, and affinities to the substrates decreased toward 1-hexanol and benzyl alcohol. These results suggested that Cys-297, Cys-379, and Cys-382 are important for activity and substrate binding. The calculation of the number of free thiol groups suggested that Cys-379 and Cys-382 formed a disulfide bridge. The disulfide bridge was reported to be important for intramolecular electron transfer to ubiquinone in PQQ-dependent methanol dehydrogenase (2), and a C-X-X-C motif of DsbB in E. coli was essential for respiration-coupled oxidation (21). An essential role of a disulfide bond between two adjacent cysteines for electron transfer from PQQH2 to heme c was strongly suggested for ADH IIB from Pseudomonas putida and ADH from Comamonas testosteroni (31). These results suggest that cysteine residues including a disulfide bond might be involved in electron transfer with PEGDH. The fact that the Km value for CoQ10 was lower than that of DCIP suggests that CoQ10 would be an indigenous electron acceptor that binds to the quinone-binding site on PEGDH. The I50 value by rotenone did not differ between CoQ10 and DCIP, suggesting that electrons pass only along the quinone-binding site and are transferred from that site to the electron acceptor. As the disulfide bridge is located on the quinone-binding site and Cys-297 is located close to it, the electrons were possibly transferred to CoQ10 via these three cysteine residues.
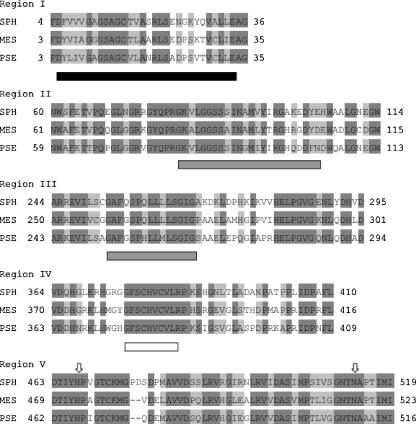
The 3D model structure showed that the cavity of the active center of PEGDH is wider than that of GOX, as suggested in Fig. 2. A large substrate, such as PEG, might be able to enter this cavity. The disulfide bridge between Cys-379 and Cys-382, as well as Cys-297 itself, is also located in the cavity of the active center on 3D structural images. The reduction of affinity for small substrates, such as TEG, 1-hexanol, and benzyl alcohol, on cysteine mutants indicated that three cysteines might play a role in substrate binding. Although the disruption of the disulfide bond in methanol dehydrogenase completely inactivated its activity (2), C379A, C382A, and C297A kept 4 to 15% of the activity of the wild type. Two cysteine residues seem to be enough for PEG binding (the same Km value as the wild type for each mutant) but inappropriate for binding of TEG, l-hexanol, and benzyl alcohol (a higher Km value than the wild type for each mutant) on the quinone-binding site, which might result in electron flow to an electron acceptor through the quinone-binding site, although a disulfide bond is lost. Two homologues of PEGDH from S. terrae have been found. These homologues are PEGDH of Mesorhizobium loti (49% identity) (11) and ADH of Pseudomonas putida (49% identity), which may recognize a PEG chain of octylphenol polyethoxylate as a substrate (accession no. AB100375). The amino acid alignment of these three dehydrogenases showed that the FAD/NAD(P)-binding Rossmann fold, the GMC oxidoreductase signatures 1 and 2 (3), the quinone-binding motif, catalytic residues (His and Asn), and the amino acid residues in the vicinity of the catalytic residues were highly conserved (Fig. 5). The quinone-binding motif was completely conserved, and a Cys-X-X-Cys motif was also found within this motif. These five regions were located in the internal structure of the protein, suggesting that these regions were directly involved in enzymatic activity. The enzymatic activity of C297A was the same as that of C379A and that of C382A, thus demonstrating the significance of Cys-297 in PEGDH, but Cys-297 was not conserved in PEGDH from M. loti or in ADH from P. putida.
FIG. 5.
Comparison of regions I, II, III, IV, and V among S. terrae PEGDH, M. loti PEGDH and P. putida ADH. SPH, S. terrae PEGDH; MES, M. loti PEGDH; and PSE, P. putida ADH. Black box, FAD/NAD(P)-binding Rossmann fold; gray boxes, GMC oxidoreductase signatures (3); white box, quinone-binding motif; arrows, His and Asn in the active site. Identical amino acids and similar amino acids are shown as dark-gray background and gray background, respectively.
In our previous study, the PEGDH from S. terrae seemed to be located in the membrane fraction, and detergents solubilized the PEGDH from the membrane (17, 19). The predicted amino acid sequence, however, did not show any continuous hydrophobic region on its hydropathy plot and did not have a membrane transsegment compartment (29). On the other hand, a putative membrane-anchoring motif (433 to 444) was reported by Hagerhall and Hederstedt (9), Szeto et al. (30), and Vergeres and Waskell (33). This motif was identified in the amino acid sequence of PEGDH; it exists on the surface of PEGDH and is close to the quinone-binding site. Since CoQ10 served as an electron acceptor, the disposition of the motif would be beneficial to the enzyme's ability to transfer electrons to the quinone molecule embedded in the membrane, as shown in Fig. 6.
From these results, we propose the following catalytic mechanism for PEGDH (Fig. 6). When a substrate approaches the active site from the interface of each monomeric protein, it is stabilized at the re site (7) by binding to hydrophobic or aromatic amino acids, FAD, and three cysteine residues. The hydrogen atom is subtracted from the —CH2OH group on the substrate by unknown catalytic sites. His-467 and Asn-511 in the active site are deeply related to the catalysis, and an electron is finally delivered from the quinone-binding site to the ubiquinone embedded in the cytoplasm membrane, as shown in Fig. 6. To the best of our knowledge, this is the first report on the reaction mechanism of a dehydrogenase belonging to the GMC oxidoreductase family. It also should be noted that the quinone-binding motif and the C-X-X-C motif on it were well conserved among sequences homologous to PEGDH.
Acknowledgments
This work was supported by a grant-in-aid (no. 14560068) to F.K. for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
We are grateful to J. A. Duine for discussions at the start of this work.
REFERENCES
- 1.Altchul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake, C. C., M. Ghosh, K. Harlos, A. Avezoux, and C. Anthony. 1994. The active site of methanol dehydrogenase contains a disulphide bridge between adjacent cysteine residues. Nat. Struct. Biol. 1:102-105. [DOI] [PubMed] [Google Scholar]
- 3.Cavener, D. R. 1992. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 223:811-814. [DOI] [PubMed] [Google Scholar]
- 4.Crameri, A., and W. P. Stemmer. 1995. Combinatorial multiple cassette mutagenesis creates all the permutations of mutant and wild-type sequences. BioTechniques 18:194-196. [PubMed] [Google Scholar]
- 5.Dowd, S. R., E. A. Pratt, Z. Y. Sun, and C. Ho. 1995. Nature and environment of the sulfhydryls of membrane-associated d-lactate dehydrogenase of Escherichia coli. Biochim. Biophys. Acta 1252:278-283. [DOI] [PubMed] [Google Scholar]
- 6.Fisher, N., and P. R. Rich. 2000. A motif for quinone binding sites in respiratory and photosynthetic systems. J. Mol. Biol. 296:1153-1162. [DOI] [PubMed] [Google Scholar]
- 7.Fraaije, M. W., and A. Mattevi. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25:126-132. [DOI] [PubMed] [Google Scholar]
- 8.Fraaije, M. W., R. H. H. van den Heuvel, W. J. H. van Berkel, and A. Mattevi. 1999. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J. Biol. Chem. 274:35514-35520. [DOI] [PubMed] [Google Scholar]
- 9.Hagerhall, C., and L. Hederstedt. 1996. A structural model for the membrane-integral of succinate: quinone oxidoreductases. FEBS Lett. 389:25-31. [DOI] [PubMed] [Google Scholar]
- 10.James, K. D., M. A. Hughes, and P. A. Williams. 2000. Cloning and expression of nynD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol. 182:3136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesama, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, F. 1995. Breakdown of plastics and polymers by microorganisms. Adv. Biochem. Eng. 52:151-194. [DOI] [PubMed] [Google Scholar]
- 13.Kawai, F. 2002. Microbial degradation of polyethers. Appl. Microbiol. Biotechnol. 58:30-38. [DOI] [PubMed] [Google Scholar]
- 14.Kawai, F. 2002. Biodegradation of polyethers (polyethylene glycol, polypropylene glycol, polytetramethylene glycol, and others), p. 267-298. In A. Steinbüchel (ed.), Biopolymers, vol. 9. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 15.Kawai, F., and S. Enokibara. 1996. Role of novel dye-linked dehydrogenases in the metabolism of polyethylene glycol by pure cultures of Sphingomonas sp. N6. FEMS Microbiol. Lett. 141:45-50. [Google Scholar]
- 16.Kawai, F., T. Kimura, Y. Tani, and H. Yamada. 1984. Involvement of a polyethylene glycol (PEG)-oxidizing enzyme in the bacterial metabolism of PEG. Agric. Biol. Chem. 48:1349-1351. [Google Scholar]
- 17.Kawai, F., T. Kimura, Y. Tani, H. Yamada, and M. Kurachi. 1980. Purification and characterization of polyethylene glycol dehydrogenase involved in the bacterial metabolism of polyethylene glycol. Appl. Environ. Microbiol. 40:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai, F., and H. Yamanaka. 1986. Biodegradation of polyethylene glycol by symbiotic mixed culture (obligate mutualism). Arch. Microbiol. 146:125-129. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, F., H. Yamanaka, M. Ameyama, E. Shinagawa, K. Matsushita, and O. Adachi. 1985. Identification of the prosthetic group and further characterization of a novel enzyme, polyethylene glycol dehydrogenase. Agric. Biol. Chem. 49:1071-1076. [Google Scholar]
- 20.Kleiger, G., and D. Eisenberg. 2002. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H · O hydrogen bonds and Van der Waals interactions. J. Mol. Biol. 323:69-76. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, T., Y. Takahashi, and K. Ito. 2001. Identification of a segment of DsbB essential for its respiration-coupled oxidation. Mol. Microbiol. 39:158-165. [DOI] [PubMed] [Google Scholar]
- 22.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 23.Morton, R. A., U. Gloor, O. Schindler, G. M. Wilson, L. H. Chopard-dit-Jean, F. W. Hemming, O. Isler, W. M. F. Leat, J. F. Pennock, R. Ruegg, U. Schwieter, and O. Wiss. 1958. Die Structur des Ubichinons aus Schweinherzen. Helv. Chim. Acta 4:2343-2357. [Google Scholar]
- 24.Prieur, I., J. Lunardi, and A. Dupuis. 2001. Evidence for a quinone binding site close to the interface between NUOD and NUOB subunits of complex I. Biochim. Biophys. Acta 1504:173-178. [DOI] [PubMed] [Google Scholar]
- 25.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 1339-1341. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schilke, B., C. Voisine, H. Beinert, and E. Craig. 1999. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Society for Industrial Microbiology. 1999. Journal of industrial microbiology and biotechnology, vol. 23, no. 4/5. Special issue for sphingomonads. Springer, Berlin, Germany.
- 29.Sugimoto, M., M. Tanabe, M. Hataya, S. Enokibara, J. A. Duine, and F. Kawai. 2001. The first step in polyethylene glycol degradation by sphingomonads proceeds via a flavoprotein alcohol dehydrogenase containing flavin adenine dinucleotide. J. Bacteriol. 183:6694-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szeto, T. H., S. L Rowland, L. I. Rothfield, and G. F. King. 2002. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 99:15693-15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyama, H., F. Mathews, O. Adachi, and K. Matsushita. 2004. Quinohemoprotein alcohol dehydrogenase: structure, function, and physiology. Arch. Biochem. Biophys. 428:10-21. [DOI] [PubMed] [Google Scholar]
- 32.van Beilen, J. B., G. Eggink, H. Enequist, R. Bos, and B. Witholt. 1992. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol. Microbiol. 6:3121-3136. [DOI] [PubMed] [Google Scholar]
- 33.Vergeres, G., and L. Waskell. 1992. Expression of cytochorome b5 in yeast and characterization of mutants of the membrane-anchoring domain. J. Biol. Chem. 267:12583-12591. [PubMed] [Google Scholar]
- 34.White, G. F., N. J. Russell, and E. C. Tidswell. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60:216-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlfahrt, G., S. Witt, J. Hendle, D. Schomburg, H. M. Kalisz, and H.-J. Hecht. 1999. 1.8 and 1.9 Å resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr. D55:969-977. [DOI] [PubMed] [Google Scholar]