Abstract
We studied the effects of a bacterium (Pseudomonas chlororaphis) and a bactivorous protozoan (Uronema sp.) on transformations of labile dissolved organic carbon (DOC). In 36-day time series experiments, bacteria were grown on glucose both with and without protozoa. We measured bulk organic carbon pools and used electrospray ionization mass spectrometry to characterize dissolved organic matter on a molecular level. Bacteria rapidly utilized glucose, depleting it to nondetectable levels and producing new DOC compounds of higher molecular weight within 2 days. Some of these new compounds, representing 3 to 5% of the initial glucose-C, were refractory and persisted for over a month. Other new compounds were produced and subsequently used by bacteria during the lag and exponential growth phases, pointing to a dynamic cycling of organic compounds. Grazers caused a temporary spike in the DOC concentration consisting of labile compounds subsequently utilized by the bacteria. Grazing did not increase the complexity of the DOC pool already established by the bacteria but did continually decrease the particulate organic carbon pool and expedited the conversion of glucose-C to CO2. After 36 days, 29% of initial glucose-C remained in pure bacteria cultures, while only 6% remained in cultures where a grazer was present. In this study the bacteria were the primary shapers of the complex DOC continuum, suggesting higher trophic levels possibly have less of an impact on the qualitative composition of DOC than previously assumed.
Dissolved organic matter (DOM) comprises the largest, yet least-characterized reservoir of reduced organic carbon in aquatic systems, estimated at 700 × 1015 g C (11). DOM is important in the carbon and nitrogen cycles, the scavenging and solubilization of trace contaminants, and biogeochemical cycles of other elements (3, 14). Heterotrophic bacteria process and reprocess some of this DOM (2), channeling about one-half of oceanic primary production through the microbial loop (8).
The role of bacteria in the rate and extent of DOM mineralization and their production of (semi)refractory DOM have received less attention. Some studies indicate that bacteria produce refractory DOM that is resistant to further utilization (5, 12, 37, 39). Ogawa et al. (26) showed that a natural inoculum of marine bacteria (and undoubtedly nanoflagellates and viruses) growing on labile compounds (glucose and glutamate) produce new DOM compounds that appear to be refractory for at least a year. It was not known if a single strain of bacteria could produce similar refractory material. Bacterioplankton can also be a source of photoreactive C-DOM that is refractory to a natural bacteria assemblage following photochemical alteration (18). What kinds and how many different compounds make up the refractory DOM pool are largely unknown.
In aquatic ecosystems, bacteria are consumed by protozoa and other zooplankton, which in turn release DOM as colloidal matter (17, 40) and macromolecular organic complexes (24). A substantial portion (>50%) of primary and bacterial production can be consumed by a single class of protozoa, the Ciliata (10). Therefore, ciliates can act as trophic links, nutrient regenerators, and DOM producers (38)—roles often overlooked in traditional food webs. Little is known about the effects of additional trophic levels on the production and composition of refractory DOM. Nagata and Kirchman (24) suggested that the release of DOM by protozoa is potentially important in aquatic food webs and nutrient cycles. Kujawinski et al. (19) used electrospray ionization (ESI) Fourier transform ion cyclotron resonance mass spectrometry (MS) to identify 80 new DOM compounds produced when a protozoan grazed on bacteria.
Here, we analyzed the effects on DOM dynamics of a pure bacteria culture alone and when grazed on by a ciliate. We used conventional, bulk analysis to examine how trophic structure influenced dissolved organic carbon (DOC) transformations. We used ESI-MS for molecular-level characterization of compounds produced during bacterial growth and grazing. ESI-MS has been used for identification and quantification of specific compounds (13, 22, 28, 29). Terrestrial and marine DOM pools have been characterized by ESI-MS (15, 16, 20, 21). ESI-MS has also been used to gain insights into changes in DOM due to protozoan grazing (19), for characterization of DOM in rainwater (32), and to discriminate between possible refractory and labile DOM compounds in freshwater samples (30).
MATERIALS AND METHODS
Experimental setup.
Production and dynamics of DOM were examined using glucose as the sole utilizable carbon source (10 mM C initial concentration). There were three treatments: sterile control, bacteria only, and bacteria plus ciliates. Experiments took place in 125-ml flasks (baked at 500°C for 4 h before use) containing 25 ml sterile, low-carbon (62.8 μM C from nitrilotriacetic acid, 0.8 μM C from Na2EDTA) modified mineral basal solution (MBS). In the modified MBS, ammonia and phosphate concentrations were 5 times lower, with a carbonate buffer concentration 50 times lower and metal concentrations 100 times lower.
All flasks (except the sterile controls) were initially inoculated with the bacterium Pseudomonas chlororaphis. Initial samples were taken directly after inoculation (day 0). Half of the remaining flasks were inoculated with the bactivorous protozoan Uronema sp. after 3 days, when the bacteria had reached stationary growth. Flasks were capped with aluminum foil to maintain sterility but permit air exchange. All cultures were incubated in the dark on a shaker table at 50 rpm and 20°C. Triplicate sacrificial flasks were used for each time point to prevent contamination from repeated sampling of the same flasks.
Bacterial strain.
We used a single freshwater heterotrophic bacterial strain, Pseudomonas chlororaphis (ATCC 17418), a gamma proteobacterium (34). Flasks were inoculated with 10 μl of triple-washed cells harvested from cultures at the end of the log growth phase. At each time point, abundance was estimated by measuring the optical density (1-cm path length) at 550 nm of a 1-ml sample.
Grazer.
We used a freshwater ciliate, Uronema sp. (Scuticociliatida), as the bactivorous protozoan grazer. This ciliate is about 20 μm in its longest dimension. Prior to the experiment, ciliates were maintained in culture using protozoan pellets (Carolina Biological Supply) as a nutrient source. Before being introduced into P. chlororaphis cultures, ciliates were filtered through a polyester mesh (41-μm openings) to remove detritus. Ciliates in the <41-μm fraction were triple rinsed with low-carbon MBS and then concentrated onto a 5-μm Nucleopore filter and resuspended at a final density of 1,600 cells/ml. Flasks were inoculated with 25 μl of concentrated ciliate suspension (∼40 cells). At each time point, abundance was determined by direct counts of a 1-ml sample using a Sedgewick Rafter counting cell.
Bulk chemical analysis.
At each time point, samples were taken from each flask for particulate organic carbon (POC), particulate nitrogen (PN), DOC, total dissolved nitrogen (TDN), and dissolved inorganic nitrogen (DIN).
For the POC and PN analysis, 23 ml of sample was filtered through a precombusted (500°C for 4 h) 25-mm-diameter Whatman GF/F filter. The filter was dried and analyzed using a Carlo Erba Instruments NA 1500 series 2 elemental analyzer (23). Results for POC and PN were corrected based on filters processed from the sterile controls. The filtrate containing the dissolved fraction was transferred to acid-washed capped polypropylene vials and stored frozen (−20°C) in the dark until needed for all other analyses.
DOC and TDN were measured using a Shimadzu 5000 high-temperature combustion analyzer for DOC (4, 33) with an inline NOx detector (Antek model 7000 total N analyzer) for TDN analysis (31). DIN was measured using a Lachat, Inc., auto analyzer (ammonium; QuikChem 31-107-06-1-A; nitrate plus nitrite, QuikChem 31-107-04-1-A). DON was determined as the difference between TDN and DIN.
Compound-level analyses.
All compound-level analyses were performed using an Agilent 1100 liquid chromatography (LC) mass spectrometer equipped with an ESI source and a quadrupole mass selective detector (30, 32). An autosampler introduced 20 μl of individual sample into the LC mobile phase that went directly to the ESI source (no LC column was used). The mobile phase was 50:50 (vol/vol) methanol-0.05% formic acid in deionized water (pH 3.5) with a flow rate of 0.220 ml min−1. Samples were analyzed in both the positive and negative ionization mode of the ESI source. The flow of the drying gas (N2 at 350°C) in the source was 10 ml/min (24 lb/in2 gauge). The ions were introduced in the mass spectrometer through a capillary (3,000 V); the fragmentor voltage was 40 V. The quadrupole temperature was 99°C, and the quadrupole voltage was kept at the default voltage of 0 V for both ionization modes. The scanned mass range was 50 to 1,000 m/z.
For each DOM sample, six replicate injections were made to facilitate a robust interpretation of the data. The raw mass spectra were recorded on Agilent software (Chemstation version A.7.01) and then imported into Access (Microsoft) for instrument blank correction and statistical analysis as described by Seitzinger et al. (30, 32) and then into Excel (Microsoft) for data interpretation. To determine the background spectra due to the medium and bacteria inoculation, samples collected immediately after bacteria were added to the medium (day 0 in the experiments) were analyzed in positive and negative ionization ESI modes. These background spectra were used to correct the subsequent time series ESI spectra. Glucose ions were identified in the background spectra (day 0), based on glucose standards in a medium matrix, to track the glucose utilization in subsequent uncorrected time series ESI spectra.
ESI is a soft ionization technique, and fragmentation of the molecular ions does not occur under standard instrument conditions (22). Studies using ESI coupled with high-resolution MS (Fourier-transformed ion cyclotron resonance MS) on terrestrial DOM and humic and fulvic acids also concluded that fragmentation was not a factor in the analysis (20, 36). Previous analyses of DOM from aquatic systems have shown that DOM compounds are mostly detected as singly charged ions (20, 35) with humic and fulvic material and with C18-extracted marine DOM (16). In addition, organic compound standards were detected in a singly charged state (unpublished data). Therefore, in the current study we assumed that the detected mass-to-charge ratios (m/z) were a good proxy for the molecular weight of the compounds.
Interpretation of ESI-MS data requires caution regarding the limitations indicated above, as well as the potential detection of several different compounds within the 1-unit mass resolution of the ESI-MS used in this study. Different compounds have different ion efficiencies resulting in different ion abundances in the ESI-MS spectra unrelated to their individual concentration in solution. Ion abundance cannot be used as a direct measure of concentration. Also, some DOM compounds in a sample may not be effectively ionized by the ESI technique, due to the polarity of the compound and the sample matrix, and would therefore not appear in the mass spectrum (20).
RESULTS
Bulk DOC, POC, and population dynamics: bacteria-only experiment.
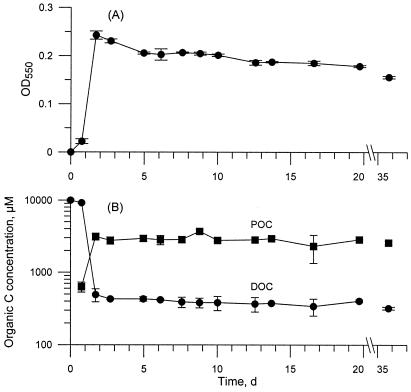
Glucose was rapidly utilized during the exponential growth phase of the bacteria as indicated by changes in optical density and DOC concentration (Fig. 1). The initial decrease in DOC concentration from 10 mM to around 440 μM coincided with the approximate 10-fold increase in optical density by day 2. The DOC concentration decreased slightly after the initial steep DOC decline but stayed about 250 μM higher than the medium background (67 μM) for the remainder of the experiment.
FIG. 1.
Bacteria-only experiment. (A) Average ± SD optical density over time. (B) Concentration of organic carbon in dissolved and particulate phases. Note the axis break between 20 and 35 days and the logarithmic scale for carbon concentration.
The experiment results were divided into three stages based on the bacterial abundance and the DOC trends: the lag phase, where the bacterial abundance was low and the DOC was high (day 1; stage I); the exponential growth phase, where the bacterial abundance increased to a maximum and a steep decrease in DOC was observed (day 2; stage II); and the stationary growth phase, where a slight decrease in both bacterial abundance and DOC concentration occurred (days 3 to 36; stage III). If first-order kinetics were assumed for the utilization of DOC, the decay constant during day 1 (stage I) was 0.028 per day before increasing to 3.1 ± 0.1 per day in stage II. In stage III the decay constant decreased to 0.013 ± 0.002 per day, a 250-fold reduction.
The POC concentration rapidly increased to 3.1 mM by day 2 and then remained constant during the rest of the experiment (Fig. 1). The C/N ratio of the particulate matter was quite uniform throughout the experiment (data not shown), with an overall mean value of 4.5 (standard deviation [SD], 0.9).
The dissolved nitrogen concentration remained high throughout the experiment (data not shown). The ammonium concentration on day 1 was 3,150 μM, which compared well with the expected medium concentration of 3,000 μM. Ammonium decreased to about 2,800 μM on day 2 and remained there for the remainder of the experiment. Dissolved N was present essentially exclusively as ammonium; DON was below detection, and nitrite plus nitrate were about 1 μM.
(ii) Bacteria plus bactivorous protozoan experiment.
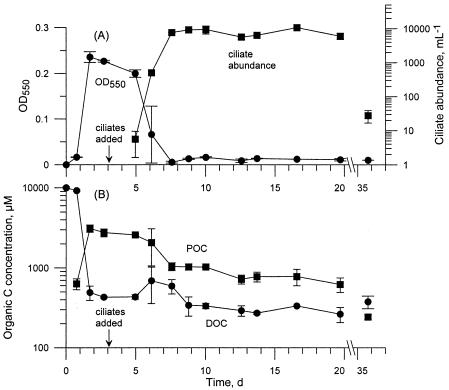
Following the introduction of Uronema just after day 3 samples were taken, the optical density of the cultures decreased from 0.2 on day 5 to 0.006 on day 8. Uronema abundance increased from 6 to 5,080 per ml in the same time period (Fig. 2). From day 8 onwards, the optical density of the remaining cultures was only slightly above the sterile control values. Ciliate abundance varied cyclically from day 8 through day 20, from highs of 9,000 to 10,600 per ml to lows of 5,600 to 6,000 per ml (Fig. 2). Ciliates at these cell densities contribute negligibly to optical density (D. F. Gruber, S. J. Tuorto, and G. L. Taghon, unpublished data). Ciliate abundance declined sharply to 28 per ml between day 20 and day 36.
FIG. 2.
Bacteria plus bactivorous protozoan experiment. (A) Average ± SD optical density and ciliate abundance over time. (B) Concentration of organic carbon in dissolved and particulate phases. Note the axis break between 20 and 35 days and the logarithmic scales for ciliate abundance and carbon concentration.
DOM was released during the increase in ciliate and decrease in bacteria abundance between day 5 and day 8 (Fig. 2). The maximum DOC increase was between day 5 and day 6, from 430 to 690 μM. During this period, Uronema increased from 8 to 500 per ml, while the OD decreased from 0.2 to 0.07. After these transient changes, the DOC concentration decreased to an average value of 315 μM for the remainder of the experiment, a level slightly lower, by 53 μM, than the DOC concentration for the bacteria-only experiment during the same time period.
POC concentration steadily decreased following Uronema addition, from 3,100 μM at the end of the exponential growth phase of P. chlororaphis, to values of 620 to 1,000 μM during the interval of greatest Uronema abundance (days 8 to 20), to a final value of 240 μM on day 36 (Fig. 2). Once Uronema was established in the cultures, the C/N ratio of the particulate matter gradually increased from its pre-Uronema value of 4.5 to 6.2 on day 36 (data not shown).
The dissolved nitrogen concentration again remained high throughout the experiment (data not shown). The ammonium concentration reached its lowest level of 2,200 μM on day 5 and then gradually increased to 2,800 μM by day 20. DON was below detection, and the nitrate plus nitrate concentration was less than 1 μM.
ESI-MS characterization. (i) Characterization of DOM produced by bacteria.
The loss of glucose ions to an undetectable level in both positive and negative ESI spectra after day 2 also indicates that glucose was rapidly utilized during the exponential growth phase of the bacteria.
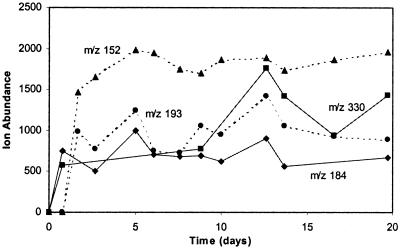
The ESI spectrum changed from day 0 to day 1 and again from day 1 to day 2; it then remained relatively constant. New m/z values were detected at day 1 in both positive (118 masses) and negative (100 masses) ionization mode that were not there on day 0. Most of these new detected masses (80 in positive mode and 81 in negative mode) did not occur beyond day 1. The remaining new m/z values identified at day 1 were present throughout the experiment. On day 2 additional new masses (45 in positive mode and 27 in negative) were detected, and all of these remained present throughout the experiment (Fig. 3).
FIG. 3.
Examples of new m/z values and ion abundance values identified in positive mode on day 1 (solid line) and on day 2 (dotted line) that remained present for the duration of the experiment. Note that ion abundance is not a direct measure of concentration because ion abundance varies per compound in the ESI-MS technique.
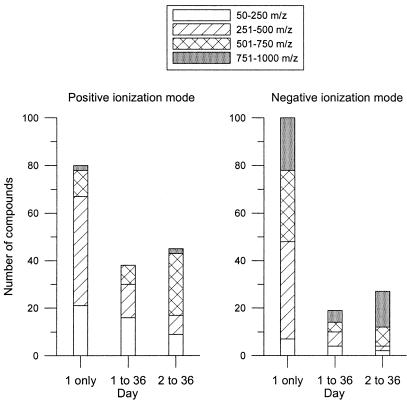
After identifying the new m/z values in the ESI spectra from the different growth stages, the molecular weight distribution of these masses was examined. The m/z values were grouped into four categories (50 to 250, 251 to 500, 501 to 750, and 751 to 1,000 m/z) to simplify the interpretation (Fig. 4). In positive ionization mode, 84% of the masses detected on day 1 only had an m/z lower than 500 (Fig. 4). This distribution is comparable to masses detected on day 1 that remained present through day 36 (Fig. 4). In contrast with day 1 results, only 38% of the new masses detected on day 2 had an m/z less than 500 (Fig. 4).
FIG. 4.
Molecular weight distribution for the bacteria-only experiment of newly detected masses at day 1 only, masses first detected on day 1 and present for the remainder of the experiment, and masses first detected on day 2 and present for the remainder of the experiment. (The newly detected masses were present in background-corrected spectra; the mass distribution was therefore not influenced by glucose ions or any other ions present in the initial medium.)
There was a shift towards higher m/z values for compounds detected in negative ionization mode compared to positive mode (Fig. 4). In negative ionization mode, about 50% of the m/z values in the day 1-only and day 1 through day 36 distributions fell in the lower-than-500 m/z categories (Fig. 4). This change is mostly due to the increase in the number of masses in the 751-to-1,000 m/z category. For the day 2 through day 36 distribution, 85% of the masses were greater than 500 m/z, with the majority greater than 750 m/z (Fig. 4).
(ii) Characterization of DOM following bactivorous protozoan addition.
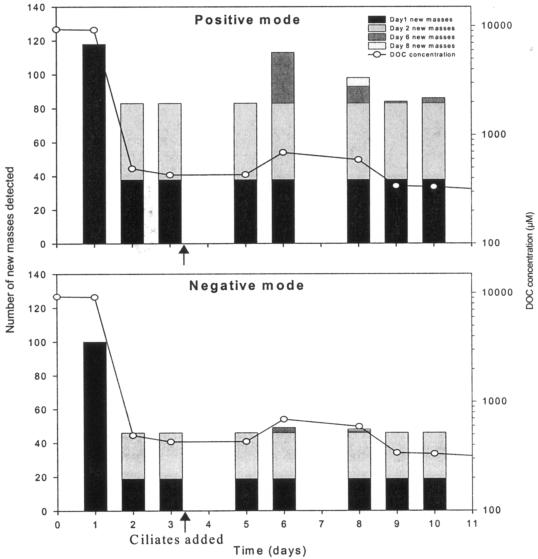
Additional new compounds were detected 3 days after Uronema addition that were not present in the bacteria-only treatment (Fig. 5). No new m/z values due to Uronema inoculation were detected in either ionization mode on day 5, 48 h after inoculation. For subsequent samples, most new masses were identified in the positive ionization mode. At day 6, 30 new masses were detected, all with a mass of <400 m/z. In the day 8 sample, 15 new compounds with masses of <400 m/z were identified. For day 9, when the transient DOC pulse decreased to the same DOC concentration as for the bacteria-only treatment, only one new compound was detected. Of the new m/z identified in the positive ionization mode on day 6, day 8, and day 9, 10 occurred on day 6 and day 8 and 1 compound remained in all three samples (Fig. 5). Thus, a total of 35 new compounds were detected in the grazer treatment in addition to the 132 compounds already present in the bacteria-only spectra, an increase of ∼27%.
FIG. 5.
New masses detected in time point samples with positive and negative mode of ESI-MS. This figure is a composite of results from the bacteria-only and bacteria plus bactivorous protozoan experiments. For comparison, the dissolved organic carbon concentrations of the bacteria plus bactivorous protozoan time points are included.
In the negative mode, only three new compounds were detected on day 6, and two of those had an m/z of >400. Two new compounds were identified on day 8. In the subsequent sample after the DOC increase due to the ciliate addition (day 9), no new masses were detected. A total of four new compounds were identified in day 6 and day 8 samples, of which one was present both days (Fig. 5).
DISCUSSION
The conversion of DOM to bacterial biomass has been studied in many natural aquatic and laboratory systems and has resulted in many published values for yield or growth efficiency (9, 27). The rate and extent of the transformation of labile DOM to refractory compounds is less studied but is at the heart of the issue of carbon storage in biologically unavailable pools. In this study we found that a pure culture of bacteria rapidly produced a complex pool of DOM from a simple labile compound. Many of the new DOM compounds (49 to 68%) were themselves used within 1 day, while the remainder formed a small refractory DOM pool (3% of the initial carbon). The addition of a bactivorous protozoan increased the extent of organic carbon remineralization from 71% to 94% but had no effect on the complexity of the DOM pool.
Changes in bulk organic carbon and nitrogen pools.
The results of our bacteria-only experiment were generally similar to those of Ogawa et al. (26), although there were substantial differences in experimental conditions. For example, they used glucose as a labile DOM source, but at much lower initial concentration than we did (208 μM C versus 10,000 μM C). They also used a natural, marine bacterial assemblage as inoculum, while we used an isolated, freshwater strain. In both experiments, glucose was undetectable within 2 days. After 2 days in their experiment, bacteria converted 7% of the initial glucose carbon to POC and 15% to other forms of DOC, and the remaining 78% was respired. After 2 days in our experiment, P. chlororaphis converted 31% of glucose carbon to POC and 5% to other forms of DOC, and the remaining 64% was presumably lost as CO2. DOC continued to decrease in Ogawa et al.'s experiment (26) and after 365 days represented 5% of the initial glucose carbon. They concluded this DOC was refractory to further bacterial transformation. In our experiment, DOC decreased only slightly over days 2 to 36, reaching a final level of 3% of the initial glucose carbon.
Introduction of a small population of a bactivorous protozoan (Uronema sp.) after the bacteria had reached stationary growth changed the distribution of carbon among the particulate, dissolved, and gaseous pools. The Uronema population rapidly increased (Fig. 2A). Although we did not directly measure the grazing rate, the size of the transient increase in DOC between days 5 and 6 (Fig. 2B) was consistent with published data on ciliate grazing rates. Assuming a bacterial carbon content of 5.8 × 10−9 μmol C/cell (70 fg C/cell [7]) and assuming between 20 and 88% of bacterial carbon is released by ciliate grazing (39), then the population of 500 ciliates/ml would have had to graze at a rate of 4,300 to 18,000 bacteria/ciliate/h to account for the 260 μM increase in DOC (also assuming no bacterial uptake of the released DOC). While high, this estimated range of grazing rates is within values reported in other studies (41).
The main effect of grazing on the bulk carbon pools was to increase the amount of glucose-C converted to CO2. The POC concentration steadily declined from day 5 to day 20 at an average of 130 μM C/day (Fig. 2B). DOC concentration also declined over this time interval, although at a slower rate (average of 11 μM C/day). We assume that any differences between the amounts of carbon originally added as glucose and measured as DOC or POC represent carbon respired as CO2 and lost from the system. The mass balance for nitrogen, an element that should not have been lost from the system, was excellent. We accounted for 94% of the added NH4-N as particulate or dissolved nitrogen throughout the incubation period.
The most likely explanation for the increase in carbon conversion to CO2 is the well-known inefficiency of energy and carbon transfer between trophic levels. Typically, 10 to 20% of food energy is transferred between successive trophic levels (25). On day 20, after the population of Uronema had been at 5,600 to 10,600 per ml for 13 days (Fig. 2A), the percentage of glucose-C present as POC was only 22% of the bacteria-only value. By the end of the experiment there was 2,600 μM POC in the bacteria-only flasks and only 9% of that, 240 μM POC, in the flasks with bacteria plus bactivorous protozoan. Thus, our results are quite consistent with a 10 to 20% trophic transfer efficiency.
A puzzling result is the continued, albeit slight, decline in DOC concentration in the grazer treatment. After the transient pulse had subsided by day 9, the DOC concentration continued to decline from 340 μM to 260 μM by day 20 (Fig. 2B), while DOC was constant at 370 μM in the bacteria-only treatment (Fig. 1B). If the residual DOC present after P. chlororaphis had depleted all the glucose were refractory, then we would expect similar residual levels of DOC in both treatments. One possibility is uptake of DOC by Uronema. Protozoa can actively take up low-molecular-weight (LMW) compounds across the cell membrane and larger dissolved compounds by pinocytosis (10). While such DOC uptake may be insufficient to meet the cell's metabolic needs, it represents a sink for DOC. The increase in DOC to a level comparable to the bacteria-only treatment by day 36 (Fig. 2B), after the Uronema population had declined precipitously (Fig. 2A), is consistent with this explanation.
The presence of Uronema led to an increase in the C/N ratio of the particulate matter. The C/N ratio of the bacteria-only treatment was invariant at 4.5, indicating that N was not limiting. Protozoa have higher C/N ratios than their bacterial prey (6), and so grazing can result in a release of the excess nitrogen from the particulate to the dissolved phase. The average NH4 concentration in the grazer treatment was 2,650 μM, compared to 2,250 μM in the bacteria-only treatment, consistent with this explanation.
Formation of DOM compounds during bacterial growth.
P. chlororaphis changed the DOC pool from a single labile compound (glucose) to a more complex pool. We detected ∼200 labile and refractory compounds (masses) in this study. Detection of masses in the day 1 sample (118 in positive and 100 in negative ESI mode) that were not present on day 0 indicates production or release of compounds associated with the bacterial utilization of glucose. Of these new masses, a majority no longer appeared later in the experiment (Fig. 4), which could indicate complete remineralization of these labile compounds, incorporation into biomass by P. chlororaphis during the exponential growth phase, or release as new compounds on day 2. The utilization of newly released compounds suggests a dynamic cycling of organic compounds by P. chlororaphis during the lag and exponential growth phases. Increasing the sampling frequency during these phases could give additional insight into production and consumption of DOM compounds by P. chlororaphis.
A number of new masses were identified in the day 1 and day 2 samples that remained present for the remainder of the experiment (Fig. 4). These new masses appeared to be compounds that were refractory to utilization by P. chlororaphis for the duration of the experiment. These refractory compounds were likely responsible for the relatively constant DOC concentration from day 2 through day 36. A larger percentage of masses detected in negative ESI mode exhibited higher molecular weights (m/z values) compared to those detected in positive mode (Fig. 4). This indicates that P. chlororaphis produced more high-molecular-weight (HMW) compounds with acidic functional groups (e.g., carboxyl and amino groups) than compounds with basic functional groups (e.g., hydroxyl and amine groups) during day 1 and day 2.
Earlier studies indicated that bacteria can transform labile compounds to compounds that are resistant to further mineralization. This transformation resulted in an increase of the average molecular weight of the initial substrate DOC pool (5, 12). Molecular weight was determined in these studies by size exclusion/gel filtration, which can only be used to infer a molecular weight range, based on elution time. In our study we were able to observe a shift to higher molecular weight with a mass unit resolution within the 50 to 1,000 m/z bin (the LMW-DOM pool). It is possible that the labile compounds released on day 1 were taken up, transformed into new compounds, and released on day 2 as refractory compounds. This scenario is supported by the shift towards higher molecular weights for masses detected in day 1 and day 2 samples (Fig. 4). However, the mechanisms responsible for this “transformation” are still poorly understood. Analysis of the elemental composition of the formed compounds using high-resolution mass spectrometry might help to elucidate differences in elemental composition as well as the number of functional groups (15, 19, 36). Kujawinski et al. (19) found in their study of biological DOM that the most common mass difference could be explained by the addition of multiples of -CH2CH2O, although the underlying chemistry was not clear.
Impact of bactivorous protozoan grazers on DOM.
The addition of a grazer increased the complexity of the DOM pool as established by P. chlororaphis, but only temporarily. The DOC and ESI results from the Uronema experiment (Fig. 2 and 5) point to an initial release of DOM compounds that were utilized within 3 days. The compounds that were released due to grazing were mostly low-molecular-weight compounds (<400 m/z). These compounds appeared, like the compounds detected on day 1 only in the P. chlororaphis experiment, to be utilized by P. chlororaphis. Thus, the addition of Uronema did not change the composition or the molecular weight distribution of the DOM pool over a long time period. The utilization of these compounds could be due to a low availability of more labile organic compounds at this time point in the experiment. Taylor et al. (39) also suggested that protozoan grazing enhanced DOC release and that the DOC released was composed of primarily low-molecular-weight compounds that were reused relatively quickly.
Most previous studies investigating DOM produced by bacteria have looked at shifts in ranges of molecular weight or overall changes in some functional groups (e.g., amino acids). With ESI-MS, we could see production and subsequent utilization of compounds in time series experiments. The next step is to focus on the elemental composition and structure of these compounds and find clues for why these compounds are labile or refractory. The similar molecular weight distributions of labile and refractory compounds within the <1,000 m/z bin identified in this study suggest that molecular weight is not the determining factor. The size-reactivity continuum model (1) suggests an overall low bioavailability (reactivity) of low-molecular-weight compounds. The degradation of reactive HMW DOM (>1,000 MW) by bacteria leads to the formation of more refractory LMW DOM (<1,000 MW) in this model. Implied in this approach is that, overall, refractory LMW DOM is older than reactive HMW DOM. In our study we found that, within the LMW DOM pool, bacteria released DOM with decreased reactivity (starting from labile glucose), even though this DOM was recently produced. The results from this study are still consistent with the size-reactivity model (1), because the refractory compounds are still part of the LMW pool, but with the addition that the refractory compounds can be formed within the LMW pool and are not necessarily all breakdown products from HMW compounds. With tools like ESI-MS, the overall dynamics of individual compounds within the LMW DOM pool can be investigated.
An important conclusion of this study is that the addition of a higher trophic level (i.e., grazer) to a single-bacteria system has a substantial effect on the rate and extent at which carbon remineralization occurs, but it has a minimal impact on the quantity and quality of DOC. It has been hypothesized (24) that ciliate grazers may also shape DOC as they release compounds that are unable to be reused by bacteria. Our experiment suggests that bacteria could be the primary shapers of DOC and play the prominent role in converting labile organic carbon to refractory. Therefore, while the bacteria-ciliate interaction is crucial for determining the fate of particulate organic carbon, it possibly has a smaller influence on dissolved organic carbon in aquatic systems.
Acknowledgments
This work was supported by Biocomplexity grant OCE0120453 from the National Science Foundation.
We thank Peter Morin for kindly providing the Uronema sp. used in this study and Dave Caron for additional ciliate cultures and advice on isolation and culturing methods. Georgina Spyres participated in preliminary experiments, and Ron Lauck performed some of the chemical analyses. Jim Ammerman and Kay Bidle provided useful dialogue on bacteria physiology.
REFERENCES
- 1.Amon, R. M. W., and R. Benner. 1996. Bacterial utilization of different size classes of dissolved organic matter. Limnol. Oceanogr. 41:41-51. [Google Scholar]
- 2.Azam, F., and R. E. Hodson. 1977. Size distribution and activity of marine microheterotrophs. Limnol. Oceanogr. 22:492-501. [Google Scholar]
- 3.Baines, S. B., and M. L. Pace. 1991. The production of dissolved organic matter by phytoplankton and its importance to bacteria: patterns across marine and freshwater systems. Limnol. Oceanogr. 36:1078-1090. [Google Scholar]
- 4.Benner, R., and M. Strom. 1993. A critical evaluation of the analytical blank associated with DOC measurements by high-temperature catalytic oxidation. Mar. Chem. 41:153-160. [Google Scholar]
- 5.Brophy, J. E., and D. J. Carlson. 1989. Production of biologically refractory dissolved organic carbon by natural seawater microbial populations. Deep-Sea Res. 36:497-507. [Google Scholar]
- 6.Caron, D. A. 1991. Evolving role of protozoa in aquatic nutrient cycles, p. 387-414. In P. C. Reid, C. M. Turley, and P. H. Burkill (ed.), Protozoa and their role in marine processes, vol. 25. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 7.Caron, D. A., E. L. Lim, G. Miceli, J. B. Waterbury, and F. W. Valois. 1991. Grazing and utilization of chroococcoid cyanobacteria and heterotrophic bacteria by protozoa in laboratory cultures and a coastal plankton community. Mar. Ecol. Prog. Ser. 76:205-217. [Google Scholar]
- 8.Cole, J. J., S. Findlay, and M. L. Pace. 1988. Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43:1-10. [Google Scholar]
- 9.del Giorgio, P. A., and J. J. Cole. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Systematics 29:503-541. [Google Scholar]
- 10.Fenchel, T. 1987. Ecology of protozoa. Science Tech Publishers, Madison, Wis.
- 11.Hedges, J. I., G. Eglinton, P. G. Hatcher, D. L. Kirchman, C. Arnosti, S. Derenne, R. P. Evershed, I. Kogel-Knabner, J. W. de Leeuw, R. Littke, W. Michaelis, and J. Rullkotter. 2000. The molecularly-uncharacterized component of nonliving organic matter in natural environments. Org. Geochem. 31:945-958. [Google Scholar]
- 12.Heissenberger, A., and G. J. Herndl. 1994. Formation of high molecular weight material by free-living marine bacteria. Mar. Ecol. Prog. Ser. 111:129-135. [Google Scholar]
- 13.Hua, Y., W. Lu, M. S. Henry, R. H. Pierce, and R. B. Cole. 1996. On-line liquid chromatography-electrospray ionization mass spectrometry for determination of the brevetoxin profile in natural “red tide” algae blooms. J. Chromatogr. A 750:115-125. [Google Scholar]
- 14.Jumars, P. A., D. L. Penry, J. A. Baross, M. J. Perry, and B. W. Frost. 1989. Closing the microbial loop: dissolved carbon pathway to heterotrophic bacteria from incomplete ingestion, digestion and sbsorption in animals. Deep-Sea Res. 36:483-495. [Google Scholar]
- 15.Kim, S., A. J. Simpson, E. B. Kujawinski, M. A. Freitas, and P. G. Hatcher. 2003. High resolution electrospray ionization mass spectrometry and 2D solution NMR for the analysis of DOM extracted by C-18 solid phase disk. Org. Geochem. 34:1325-1335. [Google Scholar]
- 16.Koch, B. P., M. R. Witt, R. Engbrodt, T. Dittmar, and G. Kattner. 2005. Molecular formulae of marine and terrigenous dissolved organic matter detected by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Geochim. Cosmochim. Acta 69:3299-3308. [Google Scholar]
- 17.Koike, I., S. Hara, K. Terauchi, and K. Kogure. 1990. Role of sub-micrometre particles in the ocean. Nature 345:242-244. [Google Scholar]
- 18.Kramer, G. D., and G. J. Herndl. 2004. Photo- and bioreactivity of chromophoric dissolved organic matter produced by marine bacterioplankton. Aquat. Microb. Ecol. 36:239-246. [Google Scholar]
- 19.Kujawinski, E. B., R. Del Vecchio, N. V. Blough, G. C. Klein, and A. G. Marshall. 2004. Probing molecular-level transformations of dissolved organic matter: insights on photochemical degradation and protozoan modification of DOM from electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 92:23-37. [Google Scholar]
- 20.Kujawinski, E. B., M. A. Freitas, X. Zang, P. G. Hatcher, K. B. Green-Church, and R. B. Jones. 2002. The application of electrospray ionization mass spectrometry (ESI MS) to the structural characterization of natural organic matter. Org. Geochem. 33:171-180. [Google Scholar]
- 21.Leenheer, J. A., C. E. Rostad, P. M. Gates, E. T. Furlong, and I. Ferrer. 2001. Molecular resolution and fragmentation of fulvic acid by electrospray ionization/multistage tandem mass spectrometry. Anal. Chem. 73:1461-1471. [DOI] [PubMed] [Google Scholar]
- 22.Loo, J. A., and R. R. Ogorzalek Loo. 1997. Electrospray ionization mass spectrometry of peptides and proteins, p. 385-419. In R. B. Cole (ed.), Electrospray ionization mass spectrometry, fundamentals, instrumentation, and application. John Wiley & Sons, Inc., New York, N.Y.
- 23.Mortlock, R. A., and P. N. Froelich. 1989. A simple method for the rapid determination of biogenic opal in pelagic marine sediments. Deep-Sea Res. 36:1415-1426. [Google Scholar]
- 24.Nagata, T., and D. L. Kirchman. 1992. Release of macromolecular organic complexes by heterotrophic marine flagellates. Mar. Ecol. Prog. Ser. 83:233-240. [Google Scholar]
- 25.Odum, E. P. 1971. Fundamentals of ecology, 3rd ed. W. B. Saunders, Philadelphia, Pa.
- 26.Ogawa, H., Y. Amagai, I. Koike, K. Kaiser, and R. Benner. 2001. Production of refractory dissolved organic matter by bacteria. Science 292:917-920. [DOI] [PubMed] [Google Scholar]
- 27.Payne, W. J. 1970. Energy yields and growth of heterotrophs. Annu. Rev. Microbiol. 24:17-52. [DOI] [PubMed] [Google Scholar]
- 28.Poon, G. K. 1997. Drug metabolism and pharmacokinetics, p. 499-525. In R. B. Cole (ed.), Electrospray ionization mass spectrometry, fundamentals, instrumentation, and application. John Wiley & Sons, Inc., New York, N.Y.
- 29.Saito, K., T. Toyo'oka, T. Fukushima, M. Kato, O. Shirota, and Y. Goda. 2004. Determination of psilocin in magic mushrooms and rat plasma by liquid chromatography with fluorimetry and electrospray ionization mass spectrometry. Anal. Chim. Acta 527:149-156. [Google Scholar]
- 30.Seitzinger, S. P., H. Hartnett, R. Lauck, M. Mazurek, T. Minegishi, G. Spyres, and R. Styles. 2005. Molecular-level chemical characterization and bioavailability of dissolved organic matter in stream water using electrospray-ionization mass spectrometry. Limnol. Oceanogr. 50:1-12. [Google Scholar]
- 31.Seitzinger, S. P., and R. W. Sanders. 1997. Contribution of dissolved organic nitrogen from rivers to estuarine eutrophication. Mar. Ecol. Prog. Ser. 159:1-12. [Google Scholar]
- 32.Seitzinger, S. P., R. M. Styles, R. Lauck, and M. A. Mazurek. 2003. Atmospheric pressure mass spectrometry: a new analytical chemical characterization method for dissolved organic matter in rainwater. Environ. Sci. Technol. 37:131-137. [DOI] [PubMed] [Google Scholar]
- 33.Sharp, J. H., R. Benner, L. Bennett, C. A. Carlson, R. Dow, and S. E. Fitzwater. 1993. Re-evaluation of high temperature combustion and chemical oxidation measurements of dissolved organic carbon in seawater. Limnol. Oceanogr. 38:1774-1782. [Google Scholar]
- 34.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 35.Stenson, A. C., W. M. Landing, A. G. Marshall, and W. T. Cooper. 2002. Ionization and fragmentation of humic substances in electrospray ionization Fourier transform-ion cyclotron resonance mass spectrometry. Anal. Chem. 74:4397-4409. [DOI] [PubMed] [Google Scholar]
- 36.Stenson, A. C., A. G. Marshall, and W. T. Cooper. 2003. Exact masses and chemical formulas of individual Suwannee River fulvic acids from ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectra. Anal. Chem. 75:1275-1284. [DOI] [PubMed] [Google Scholar]
- 37.Stoderegger, K., and G. J. Herndl. 1998. Production and release of bacterial capsular material and its subsequent utilization by marine bacterioplankton. Limnol. Oceanogr. 43:877-884. [Google Scholar]
- 38.Strom, S. L., R. Benner, S. Ziegler, and M. J. Dagg. 1997. Planktonic grazers are a potentially important source of marine dissolved organic carbon. Limnol. Oceanogr. 42:1364-1374. [Google Scholar]
- 39.Taylor, G. T., R. Iturriaga, and C. W. Sullivan. 1985. Interactions of bactivorous grazers and heterotrophic bacteria with dissolved organic matter. Mar. Ecol. Prog. Ser. 23:129-141. [Google Scholar]
- 40.Tranvik, L. 1994. Colloidal and dissolved organic-matter excreted by a mixotrophic flagellate during bacterivory and autotrophy. Appl. Environ. Microbiol. 60:1884-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zubkov, M. V., and M. A. Sleigh. 1995. Ingestion and assimilation by marine protists fed on bacteria labeled with radioactive thymidine and leucine estimated without separating predator and prey. Microb. Ecol. 30:157-170. [DOI] [PubMed] [Google Scholar]