Abstract
The yeast species Saccharomyces bayanus and Saccharomyces pastorianus are of industrial importance since they are involved in the production process of common beverages such as wine and lager beer; however, they contain strains whose variability has been neither fully investigated nor exploited in genetic improvement programs. We evaluated this variability by using PCR-restriction fragment length polymorphism analysis of 48 genes and partial sequences of 16. Within these two species, we identified “pure” strains containing a single type of genome and “hybrid” strains that contained portions of the genomes from the “pure” lines, as well as alleles termed “Lager” that represent a third genome commonly associated with lager brewing strains. The two pure lines represent S. uvarum and S. bayanus, the latter a novel group of strains that may be of use in strain improvement programs. Hybrid lines identified include (i) S. cerevisiae/S. bayanus/Lager, (ii) S. bayanus/S. uvarum/Lager, and (iii) S. cerevisiae/S. bayanus/S. uvarum/Lager. The genome of the lager strains may have resulted from chromosomal loss, replacement, or rearrangement within the hybrid genetic lines. This study identifies brewing strains that could be used as novel genetic sources in strain improvement programs and provides data that can be used to generate a model of how naturally occurring and industrial hybrid strains may have evolved.
The Saccharomyces sensu stricto species complex (45) contains some of the most important species for the food industry, namely, Saccharomyces cerevisiae (Meyen ex E. C. Hansen), the agent of wine, bread, ale beer, and sake fermentations; Saccharomyces bayanus (Saccardo), which is involved in wine and cider fermentations; and Saccharomyces pastorianus (E. C. Hansen), which is responsible for lager beer fermentation. In spite of their commercial importance, these yeasts still lack an unambiguous and accurate taxonomic definition (38). S. bayanus and S. pastorianus contain diverse strains, with different genetic and metabolic characteristics, that may have a hybrid origin. In their current form, both S. bayanus and S. pastorianus may be more correctly regarded as species complexes. On the basis of molecular and genetic data, Saccharomyces bayanus has been subdivided further into two groups (33, 35, 37): (i) S. bayanus var. bayanus, which is similar to the type strain CBS 380 and contains a miscellany of hybrid cultures, and (ii) S. bayanus var. uvarum (28), commonly referred to as S. uvarum, which is coincident with the formerly acknowledged species S. uvarum (Beijerink) and contains strains of nonhybrid origin, usually isolated from grapes or wine fermentations (10, 29, 30). S. uvarum is the only group found in either species in which all strains have equal and consistent characteristics (15, 34-38). Strain CBS 7001 is the most representative isolate of S. uvarum (37) and serves as a representative strain in studies of chromosomal evolution and distribution in Saccharomyces sensu stricto yeasts (12). Its genome has been fully sequenced (3, 8, 21), and auxotrophic mutants have been derived from it (39).
S. pastorianus also contains hybrid strains with diverse characteristics, including lager brewing strains, which are the most studied and industrially relevant of the allopolyploid yeasts. Lager brewing strains are thought to originate from a natural hybridization event that occurred between an S. cerevisiae strain and a non-S. cerevisiae strain, probably an S. bayanus strain (44). The existence of two types of genome in lager brewing strains has been confirmed by chromosome transfer experiments (reviewed in reference 22), Southern hybridization and sequencing analysis (5, 7, 13, 17, 18, 23, 40, 41, 46), proteome pattern analysis (20), and random amplified polymorphic DNA (RAPD) PCR (11).
The hybrid nature and the diversity of many S. bayanus and S. pastorianus strains makes it difficult to predict their metabolic abilities. It is not always possible to clearly distinguish these two species (19, 25, 27), and strains in some major culture collections are identified as “S. bayanus or S. pastorianus” (Centraal Bureau for Shimmelcultures [CBS], Utrecht, The Netherlands) or “S. bayanus/S. pastorianus” (Agricultural Research Service Culture Collection [ARS; formerly known as the Northern Regional Research Laboratory], National Center for Agricultural Utilization Research, Peoria, IL).
Comparative genomics is now being used to clarify the origin and evolution of various yeasts. Among the Saccharomyces sensu stricto strains considered in the present study, two have been completely sequenced: S. cerevisiae (14) and S. uvarum (3, 8, 21). S. cerevisiae contributes a portion of the genome of lager brewing strains. S. uvarum is a subgroup of S. bayanus, and its genome is very similar to that of other S. bayanus and, perhaps, S. pastorianus strains. We used the two genomic sequences to evaluate the genome of several S. bayanus and S. pastorianus strains by PCR-restriction fragment length polymorphism (PCR/RFLP) analysis.
The objectives of this study were (i) to evaluate the genetic diversity of isolates currently classified S. bayanus and S. pastorianus, (ii) to determine how natural hybrid yeasts within the genus Saccharomyces might occur and evolve, and (iii) to identify the non-S. cerevisiae parental strain of lager brewing strains. Our working hypothesis is that much of the diversity observed in these two species may be the result of genetic hybridization. The detailed characterization of these strains will enable more accurate predictions of their metabolic capabilities and may help to elucidate the mechanisms by which such hybrid strains are formed.
MATERIALS AND METHODS
Strains and media.
We evaluated 35 strains, including all of the cultures classified as S. bayanus and/or S. pastorianus available from the CBS and NBRC (NITE Biological Resource Center, Chiba, Japan) culture collections (Table 1). S. cerevisiae laboratory strain X2180-1A (MATa SUC2 mal mel gal2 CUP1) from the Genetic Research Resource Center, American Type Culture Collection, Manassas, VA, was used as a reference. All cultures were grown on YPD (1% yeast extract, 2% peptone, 2% glucose) at 28°C with shaking at 150 rpm and maintained on YPD supplemented with 2% agar.
TABLE 1.
List of strains and PCR/RFLP data obtained in the preliminary study
| Speciesa | Old epithet | CBSb | NBRCc | Remarks | PCR/RFLP analysis |
||
|---|---|---|---|---|---|---|---|
| FUN14 | RIP1 | HIS3 | |||||
| S. bayanus* | S. bayanus | 380 | 11022 | Type strain | Su+Sb | Su | Sb |
| S. bayanus | S. uvarum | 395 | 11025 | Type strain | Su | Su | Su |
| S. bayanus* | S. globosus | 424 | 10557 | Type strain | Sb | Su | Sb |
| S. bayanus or S. pastorianus† | 1174 | Brewing strain (Saaz) | Sc+Sb | Sc+Sb | Sb | ||
| S. pastorianus* | 1462 | 250 | Brewing strain | Sc+Su | Sc+Su | Su | |
| S. bayanus or S. pastorianus† | 1483 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus or S. pastorianus† | 1484 | Isolated from cloudy beer | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. pastorianus* | 1486 | 1961 | Brewing strain (Saaz) | Sc+Sb | Sc+Sb | Sb | |
| S. bayanus or S. pastorianus† | 1502 | Brewing strain | Su | Su | Su+Sb | ||
| S. pastorianus* | S. monacensis | 1503 | 10610 | Type strain | Sc+Sb | Sb | Sc+Sb |
| S. pastorianus* | S. carlsbergensis | 1513 | 11023 | Type strain | Sc+Sb | Sc+Sb | Sb |
| S. pastorianus* | S. pastorianus | 1538 | 11024 | Type strain | Sc+Sb | Sc+Sb | Sb |
| S. bayanus or S. pastorianus† | 1542 | Su+Sb | Su+Sb | Su+Sb | |||
| S. bayanus | 1545 | 1344 | Brewing strain | Su+Sb | Su+Sb | Su+Sb | |
| S. bayanus or S. pastorianus† | 2156 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus or S. pastorianus† | 2165 | Brewing strain | Sc | Sc | Sb | ||
| S. bayanus or S. pastorianus† | 2440 | Brewing strain (Saaz) | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus or S. pastorianus† | 2443 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus | 2946 | 573 | Su | Su | Su | ||
| S. bayanus | 3008 | 10558 | Wine strain | Sb | Su | Sb | |
| S. bayanus or S. pastorianus† | 5184 | 1343 | Brewing strain | Su+Sb | Su+Sb | Su | |
| S. bayanus or S. pastorianus† | 5792 | Brewing strain | Su | Su | Su | ||
| S. bayanus or S. pastorianus† | 5832 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus or S. pastorianus† | 6903 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus* | S. abuliensis | 7001 | S. uvarum representative | Su | Su | Su | |
| S. byanus or S. pastorianus*† | 8614 | Cider strain | Sc+Su | Sc+Su | Sc+Su | ||
| S. bayanus | 251 | Su+Sb | Su+Sb | Sb | |||
| S. bayanus | 213 | Su+Sb | Su+Sb | Sb | |||
| S. bayanus | 539 | Sb | Sb | Sb | |||
| S. bayanus | 1048 | Su+Sb | Sb | Su+Sb | |||
| S. bayanus* | 1948 | Brewing contaminant | Sb | Sb | Sb | ||
| S. pastorianus* | 2003 | Brewing strain | Sc+Sb | Sc+Sb | Sc+Sb | ||
| S. bayanus* | 2031 | Brewing strain (Pilsner) | Sb | Sb | Su+Sb | ||
| S. bayanus | 10158 | Brewing strain | Su | Su | Su | ||
| S. bayanus | 10551 | Su | Su | Su | |||
*, strains selected for further study; †, species as indicated in the CBS database at the time of purchase.
CBS (Centraal Bureau voor Shimmelcultures, Utrecht, The Netherlands) strain.
NBRC (NITE Biological Resource Center, Department of Biotechnology, National Institute for Technology and Evaluation, Chiba, Japan) strain.
PCR amplifications.
DNA fragments (500 to 1,500 bp) were amplified by PCR with two sets of primers (see Table SA2 in the supplemental material). One set of primers was based on the genomic sequence of S. cerevisiae (Saccharomyces Genome Database [http://www.yeastgenome.org/]), and the other set was based on the genomic sequence of S. uvarum strain CBS 7001 (see reference 21, in which the strain is indicated as the corresponding culture S. bayanus MCYC 623).
PCRs (25-μl reactions) contained 50 ng of genomic DNA that had been extracted according to standard procedures (6). Amplifications were performed with a GeneAmp PCR System 9700 (Applied Biosystems) and under the following conditions: 94°C for 4 min; 25 cycles of denaturing at 94°C for 30 s, annealing at the average Tm of the primers for 30 s, and elongation at 72°C for 1 min for each kilobase to be amplified; and a final elongation step at 72°C for 7 min. RFLP analysis was carried out with selected enzymes (see Table SA2 in the supplemental material) by digesting the PCR fragments overnight at 37°C and separating them on a 0.8% agarose gel.
Sequencing.
Sequence analysis was carried out by using a BigDye Terminator v3.1 Cycle Sequencing Ready Reaction (Applied Biosystems) and an ABI Prism 3100 genetic analyzer (PE Applied Biosystems). Sequences were aligned and compared to the S. uvarum genome sequence by using the tool BLAST 2 sequences (42) without the filter option (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html).
Quantitative analysis of restriction profiles.
For each strain and in each chromosome position analyzed, we evaluated the presence or absence of each of the four genomes described here (Sc, Sb, Su, and Lg), which were deduced from the corresponding restriction enzyme profiles. The types of genome could only be present or absent in each of the portions of the chromosomes analyzed, and each genome for each strain was therefore tabulated in a binary matrix (indicating 1 for present and 0 for absent) and analyzed with TREECON for Windows, version 1.3b (43). The genetic distance between isolates was calculated by using the coefficient of Nei and Li (32), and these values were used to construct a neighbor-joining (NJ) genetic distance tree. The reliability of the NJ trees was assessed by using bootstrapping with 1,000 replications.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study were deposited in the DDBJ/EMBL/GenBank database. The accession numbers are as follows: for S. bayanus strain NBRC 1948, AB196324 (LRE1), AB196325 (COQ1), AB196326 (STP22), AB196327 (STE2), AB196328 (PTR3), AB196329 (DEG1), AB196330 (HAT1), AB196331 (RRP9), AB196338 (MVP1), and AB196339 (TEX1); for S. bayanus (S. globosus) strain CBS 424, AB196332 (MRLP16) and AB196333 (DEG1); for S. pastorianus strain NBRC 2031, AB196334 (LRE1); and for S. pastorianus strain CBS 1538, AB196335 (STP22), AB196336 (CDC50), and AB196337 (PST2).
RESULTS
Strain selection.
A preliminary study was carried out with 35 strains (Table 1) classified as S. bayanus and/or S. pastorianus (45) to identify strains with the maximum number of genetic differences for further analyses.
We obtained two types of PCR fragments: (i) Sc fragments, when the primers based on the S. cerevisiae genome were used, and (ii) Su fragments, when the primers based on the S. uvarum genome were used. Since S. uvarum is a variety of S. bayanus, the primers based on the S. uvarum genome could be used to amplify the corresponding fragments from the S. bayanus strains.
The S. uvarum and S. bayanus genomes were defined by three types of RFLPs (Table 1). For FUN14-Su and HIS3-Su, the S. uvarum genome has no restriction site for BanII and RsaI, respectively, but the S. bayanus genome does. For RIP1-Su, the S. uvarum genome has a PstI restriction site that is missing in the S. bayanus genome. Six of the 35 strains tested (CBS 395, CBS 7001, CBS 2946, CBS 5792, NBRC 10158, and NBRC 10551) had exclusively Su RFLP patterns, and two (NBRC 539 and NBRC 1948) had exclusively Sb RFLP patterns. These two S. bayanus isolates differ from the current S. bayanus type strain CBS 380, which is a hybrid culture (7, 34, 35, 37).
Of the remaining 27 strains evaluated in the preliminary study, 14 had both Sc and Sb RFLPs, including the type strains of S. carlsbergensis (CBS 1513), S. monacensis (CBS 1503), and S. pastorianus (CBS 1538). Two strains had both Sc and Su RFLPs, and 11 strains had both Sb and Su RFLPs, including the type strain of S. bayanus (CBS 380) and the former type strain of S. globosus (CBS 424). A subset of 12 strains (Table 1) showing all of the possible PCR/RFLP patterns and their combinations was selected to be representative of the genetic variability in S. bayanus and S. pastorianus and used for the rest of the study.
PCR/RFLP analysis of the 12 selected strains.
We amplified three regions (centromere and both telomeres) of each of the 16 chromosomes corresponding to the S. cerevisiae chromosome set for each strain (Fig. 1). As in the preliminary study, we used one set of PCR primers based on the S. cerevisiae genome and a second set based on the S. uvarum genome, and we performed RFLP analysis on the resulting DNA fragments. During this process we identified a third RFLP pattern that we refer to as the Lager-type pattern (Lg). RFLP analyses also were made with Sc-based amplicons that could not be differentiated from the Su amplicons. From these analyses we identified “pure” strains that carry only one type of genome and three types of “hybrid” strains that carry genetic material from more than one genome.
FIG. 1.
Type of genomes deduced by PCR/RFLP or sequencing for all of the fragments obtained. Bars: □ Sc type; ▪, Sb type; ░⃞, Su type; ▥, Sb type = Su type; , Lg type. *, differentiation between Sb and Su obtained by sequencing; the difference in these cases is only of a few substitutions.
Saccharomyces bayanus and S. uvarum pure genetic lines.
We identified strains that carry only the S. uvarum genome, e.g., CBS 7001, and only the S. bayanus genome, e.g., NBRC 1948. Of the 48 DNA fragments examined, 32 could be used in RFLP analyses to distinguish S. bayanus and S. uvarum, 10 had to be sequenced to be assigned to one of the two genomes, and 6 were monomorphic (Fig. 1).
Group 1 hybrids: S. cerevisiae/S. bayanus/Lager genome.
This group is representative of lager brewing strains and includes S. pastorianus strain CBS 1538, S. carlsbergensis strain CBS 1513, S. monacensis strain CBS 1503, and lager strains CBS 1486 and NBRC 2003. Chromosomes I and X were the same for all five strains. Sc fragments were amplified from all five strains but were not recovered from the same locations for all of the strains. Saccharomyces carlsbergensis, S. monacensis, and S. pastorianus all differ from contemporary lager brewing strains, e.g., NBRC 2003 (Fig. 1). In 32 of the 48 Su amplicons analyzed, the RFLP pattern for these five strains corresponds to the Sb pattern. For the other 16 fragments, only the Lg pattern was detected. Of the 16 Lg DNA fragments, 6 were sequenced and were 89 to 94% identical to the corresponding S. uvarum sequences and 77 to 88% identical to the corresponding S. cerevisiae sequences (Table 2). The S. uvarum PCR/RFLP pattern was not detected in any of the tested chromosomes. Thus, we concluded that the S. uvarum genome was not present in lager brewing strains. Either the Sb or the Lg pattern was detected at all sites except for CDC50 on the right arm of chromosome III in S. carlsbergensis CBS 1513 and NBRC 2003 (Fig. 1), where only S. cerevisiae sequences were detected.
TABLE 2.
Percent sequence identity of six Lg fragments to the S. uvarum and S. cerevisiae nucleotide sequences
| Gene fragment | Chromosome location | Strain sequenced |
% Identity |
||
|---|---|---|---|---|---|
| Name | Size (bp) | S. uvarum | S. cerevisiae | ||
| MRPL16 | II | CBS 424 | 555 | 94 | 83 |
| LRE1 | III | NBRC 2031 | 540 | 89 | 77 |
| STP22 | III | CBS 1538 | 891 | 89 | 77 |
| CDC50 | III | CBS 1538 | 894 | 93 | 79 |
| PST2 | IV | CBS 1538 | 497 | 94 | 88 |
| DEG1 | VI | CBS 424 | 648 | 91 | 78 |
Group 2 hybrids: S. cerevisiae/S. uvarum/S. bayanus/Lager.
This group includes only two strains, CBS 1462 and CBS 8614. S. cerevisiae/S. uvarum hybrids have been isolated previously from enological substrates (29) but not from brewing habitats. CBS 1462 is the first culture of this type from a brewing context. Strain CBS 8614 was isolated from cider and has been previously described as a triple hybrid containing genetic material from S. cerevisiae, S. bayanus, and S. kudriavzevii (16, 26). In both strains we usually detected either an Sb or an Su pattern, but there were some cases in which both Sb and Su patterns were detected simultaneously (Fig. 1). CBS 8614 contains portions of three different genomes on the left arm of chromosome I (Sc/Su/Sb) and on the right arm of chromosome VIII (Sc/Su/Sb). Both CBS 8614 and CBS 1462 contain portions of three different genomes near the centromere of chromosome VI (Sc/Su/Lg). CBS 8614 has recently been reported to be a triploid (31).
Group 3 hybrids: S. bayanus/S. uvarum/Lager.
This group includes S. bayanus type strain CBS 380, S. globosus strain CBS 424, and strain NBRC 2031, which is a brewing strain designated Pilsner but does not contain the S. cerevisiae genome. Most of the strains have only a single genome represented at most of the analyzed sites. CBS 424 had only a single genome represented at most of the analyzed sites, of which the Lg pattern was the most common. NBRC 2031 had two different genomes represented at 14 of 48 loci examined and CBS 380 had two different genomes at 3 of 48 loci (Fig. 1). There was no clear pattern of association between the presence of Lg fragments and the fragments from the other genomes. When an Lg fragment was found for a site in CBS 424, then no other genome was represented at that site. An Lg fragment was detected for six fragments common to CBS 380 and CBS 424. For the Pilsner strain NBRC 2031, Lg fragments were found only if a Su or Sb fragment for the same region also was present.
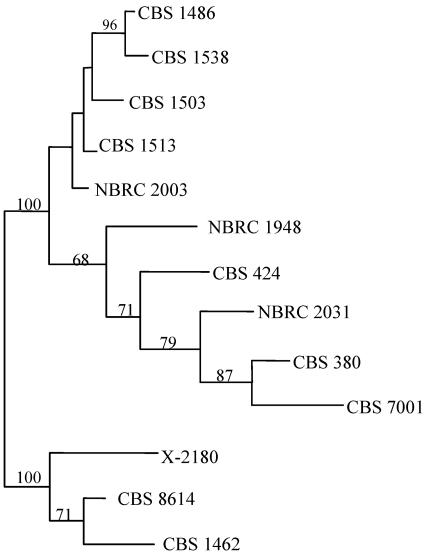
Clustering analysis.
We constructed a neighbor-joining tree based on the presence or absence of the different genome types in each strain (Fig. 2). The three hybrid lines form three distinct clusters. All strains of group 3 are between their putative parental strains, S. bayanus (strain NBRC 1948) and S. uvarum (strain CBS 7001). The separation of S. bayanus and S. uavrum is consistent with our hypothesis that these strains represent “pure” genetic lines.
FIG. 2.
Neighbor-joining tree constructed using the PCR-RFLP profiles of all of the genes analyzed. The numbers at the nodes are the bootstrap values of >50% based on 1,000 pseudoreplications.
DISCUSSION
From our results we identified two “pure” genetic lines: (i) S. uvarum, a group of yeasts already defined as a pure line with well-known characteristics and proposed as a proper species (34, 36), and (ii) S. bayanus, a grouping of strains proposed here for the first time and represented by NBRC 1948. These strains are not the same as S. uvarum and probably should be considered as members of a different species, since the differences detected by PCR/RFLP analysis reflect species level polymorphism. Further studies are now being carried out to test this hypothesis.
We identified three types of hybrid lines: (i) S. bayanus/S. uvarum/Lager, (ii) S. cerevisiae/S. bayanus/Lager, and (iii) S. cerevisiae/S. bayanus/S. uvarum/Lager. We termed “Lager” (or “Lg”) derivatives of the Su amplicons that could be identified by RFLP analysis and that were first identified from lager brewing strains. We did not identify any strains that carry exclusively Lg alleles and found such alleles at only 16 of the 48 examined loci; at these same loci the Su and Sb alleles were either identical or differed by a few base pairs. The relatedness of the Lg and the Su alleles suggests that Lg strains might be currently in the process of separating from S. uvarum.
Based on our results, we recommend that the current S. bayanus species be separated into one “hybrid” and two “pure” genetic lines: (i) S. uvarum for strains with characters analogous to CBS 7001, (ii) S. bayanus for strains with characters analogous to NBRC 1948, and (iii) the S. bayanus hybrid line for strains that include portions of the S. bayanus and S. uvarum genomes but not the S. cerevisiae genome. The currently accepted type strain of S. bayanus CBS 380 may be retained for historical purposes but should not be used as a reference in taxonomic, metabolic, or ecological studies of the Saccharomyces sensu stricto yeasts. Although this strain has been previously reported to be a hybrid, our data are not consistent with its classification as an S. cerevisiae/S. uvarum hybrid (31, 34) since we did not detect any Sc alleles in this strain at any of the 48 loci that we examined.
We propose that the S. pastorianus name be used for the multiple genetic lines that contain the S. cerevisiae genome, namely, the S. cerevisiae/S. bayanus/Lager hybrid line and the S. cerevisiae/S. bayanus/S. uvarum/Lager hybrid line. Lager brewing strains are the most studied and industrially important naturally occurring Saccharomyces hybrids. We confirmed that these yeasts contain components of an S. cerevisiae-like genome and found that the non-S. cerevisiae portions of the genome were derived from S. bayanus and/or S. uvarum. The identification of the source of the non-S. cerevisiae portion of the genome could aid the rational design and construction of novel hybrids for the brewing industry. S. bayanus strains represented by NBRC 1948 have the same PCR/RFLP banding pattern as the lager brewing strains at 32 of 48 DNA fragments examined, suggesting that these strains might be a good source of new alleles for use in strain improvement programs.
All of the lager brewing strains we examined contained genetic components from three different genomes—Sc, Sb, and Lg—but in different proportions and in different locations for each chromosome. In particular, our results confirm the finding that current lager brewing strains, e.g., NBRC 2003, contain an almost complete S. cerevisiae genome, unlike older lager brewing strains, e.g., CBS 1538, which contain a smaller portion of the S. cerevisiae genome (2, 7, 9, 31). The Su genome was not detected at any of the 48 loci examined; however, it seems to be the origin of the Lg genome. Multiple hybridization events and subsequent chromosomal loss and rearrangement, events known to occur in Saccharomyces yeasts (1, 9), are required to explain the genetic composition of lager brewing strains. The hybrid genetic line S. bayanus/S. uvarum/Lager could have originated from multiple hybridization events and is the best current candidate for the ancestor of the non-S. cerevisiae parental strain of lager brewing yeasts. The results of protein profiles of lager brewing strains are consistent with this hypothesis (20).
The hybrid S. pastorianus line (S. cerevisiae/S. bayanus/S. uvarum/Lager) could represent another intermediate in the evolution of the current lager brewing strains. Additional chromosome losses, replacements, and rearrangements, including all or portions of the S. cerevisiae and S. uvarum genomes, would still be required to yield the genome and chromosome composition found in the Lager brewing strains currently in use (4, 7, 9, 11, 24, 40, 41, 46). We recommend that synthetic hybrids for use in brewing be created by hybridizing at least one S. cerevisiae and S. bayanus NBRC 1948 type strain with an S. bayanus strain carrying different characters of interest.
Supplementary Material
Acknowledgments
We thank G. A. Garcia (Scuola Superiore Sant'Anna [Italy]) for critically reading the manuscript and for helpful discussions, R. Rossello-Mora (CSIS-UIB [Spain]) for comments on the genetic distance analysis, Y. Van de Peer (University of Ghent [Belgium]) for use of the TREECON software package, and Y. Itokui and Y. Wada (Suntory Research Center [Japan]) for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Antunovics, Z., H.-V. Nguyen, C. Gaillardin, and M. Sipiczki. 2005. Gradual genome stabilization by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 5:1141-1150. [DOI] [PubMed] [Google Scholar]
- 2.Azumi, M., and N. Goto-Yamamoto. 2001. AFLP analysis of type strains and laboratory and industrial strains of Saccharomyces sensu stricto and its applications to phenetic clustering. Yeast 18:1145-1154. [DOI] [PubMed] [Google Scholar]
- 3.Bon, E., C. Neuvegliese, S. Casaregola, F. Artiguenave, P. Wincker, M. Aigle, and P. Durrens. 2000. Genomic exploration of the hemiascomycetous yeast: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 487:37-41. [DOI] [PubMed] [Google Scholar]
- 4.Bond, U., N. Cassandra, D. Donnelly, and T. C. James. 2004. Aneuploid and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridization. Curr. Genet. 45:360-370. [DOI] [PubMed] [Google Scholar]
- 5.Borsting, C., R. Hummel, E. R. Schultz, T. M. Rose, M. B. Pedersen, J. Knudsen, and K. Kristiansen. 1997. Saccharomyces carlsbergensis contains two functional genes encoding the acyl-CoA binding protein, one similar to the ACB1 gene from S. cerevisiae and one identical to the ACB1 gene from S. monacensis. Yeast 13:1409-1421. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D., D. Dawson, and T. Steerns. 2000. Yeast DNA isolations, p. 110-111. In D. Burke, D. Dawson, and T. Steerns (ed.), Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Casaregola, S., H.-V. Nguyen, G. Lapathitis, G. Kotyk, and C. Gaillardin. 2001. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int. J. Syst. Evol. Microbiol. 51:1607-1618. [DOI] [PubMed] [Google Scholar]
- 8.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 9.De Barros Lopes, M., J. R. Bellon, N. J. Shirley, and P. F. Ganter. 2002. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1:323-331. [DOI] [PubMed] [Google Scholar]
- 10.Demuyter, C., M. Lollier, J.-L. Legras, and C. Le Jeune. 2004. Predominance of Saccharomyces uvarum during spontaneous alcoholic fermentation, for three consecutive years, in an Alsatian winery. J. Appl. Microbiol. 97:1140-1148. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Espinar, M. T., E. Barrio, and A. Querol. 2003. Analysis of the genetic variability in the species of the Saccharomyces sensu stricto complex. Yeast 20:1213-1226. [DOI] [PubMed] [Google Scholar]
- 12.Fisher, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405:451-454. [DOI] [PubMed] [Google Scholar]
- 13.Fuji, T., H. Yoshimoto, N. Nagasawa, T. Bogaki, Y. Tamai, and M. Hamachi. 1996. Nucleotide sequences of alcohol acetyltransferase genes from lager brewing yeast, Saccharomyces carlsbergensis. Yeast 12:593-598. [DOI] [PubMed] [Google Scholar]
- 14.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:563-567. [DOI] [PubMed] [Google Scholar]
- 15.Giudici, P., C. Caggia, A. Pulvirenti, and S. Rainieri. 1998. Karyotyping of Saccharomyces strains with different temperature profiles. J. Appl. Microbiol. 84:811-819. [DOI] [PubMed] [Google Scholar]
- 16.Groth, C., J. Hansen, and J. Piskur. 1999. A natural chimeric yeast containing genetic material from three species. Int. J. Syst. Bacteriol. 49:1933-1938. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J., H. Cherest, and M. C. Kielland-Brandt. 1994. Two divergent MET10 genes, one from Saccharomyces cerevisiae and one from Saccharomyces carlsbergensis encode for the α subunit of sulfite reductase and specify potential binding sites for FAD and NADPH. J. Bacteriol. 176:6050-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, J., and M. C. Kielland-Brandt. 1994. Saccharomyces carlsbergensis contains two functional MET2 alleles similar to homologues from S. cerevisiae and S. monacensis. Gene 140:33-40. [DOI] [PubMed] [Google Scholar]
- 19.Josepa, S., J. M. Guillamon, and J. Cano. 2000. PCR differentiation ofSaccharomyces cerevisiae from Saccharomyces bayanus/Saccharomyces pastorianus using specific primers. FEMS Microbiol. Lett. 193:255-259. [DOI] [PubMed] [Google Scholar]
- 20.Joubert, R., P. Brignon, C. Lehmann, C. Monribot, F. Gendre, and H. Boucherie. 2000. Two-dimensional gel analysis of the proteome of lager brewing yeasts. Yeast 16:511-522. [DOI] [PubMed] [Google Scholar]
- 21.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 22.Kielland-Brandt, M. C., T. Nilsson-Tillgren, C. Gjermansen, S. Holmberg, and M. B. Pedersen. 1995. Genetics of brewing yeasts, p. 223-254. In A. H. Rose, E. Wheals, and J. S. Harrison (ed.), The yeasts, vol. 6, 2nd ed. Academic Press, Ltd., London, England. [Google Scholar]
- 23.Kodama, Y., F. Omura, and T. Ashikari. 2001. Isolation and characterization of a gene specific to lager brewing yeast that encodes a branched-chained amino acid permease. Appl. Environ. Microbiol. 67:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart, L., S. G. Oliver, and D. Delneri. 2002. Tools for the study of genome rearrangements in laboratory and industrial yeast strains. Yeast 19:441-448. [DOI] [PubMed] [Google Scholar]
- 25.Manzano, M., L. Cocolin, B. Longo, and G. Comi. 2004. PCR-DGGE differentiation of strains of Saccharomyces sensu stricto. Antonie Leeuwenhoek 85:255-259. [DOI] [PubMed] [Google Scholar]
- 26.Masneuf, I., J. Hansen, C. Groth, J. Piskur, and D. Dubourdieu. 1998. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64:3887-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 36:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naumov, G. I. 2000. Saccharomyces bayanus var. uvarum comb. nov., a new variety established by genetic analysis. Mikrobiologyia 69:410-414. [PubMed] [Google Scholar]
- 29.Naumov, G. I., I. Masneuf, E. S. Naumova, M. Aigle, and D. Dubourdieu. 2000. Association of Saccharomyces bayanus var. uvarum with some French wines: genetic analysis of yeast populations. Res. Microbiol. 151:683-691. [DOI] [PubMed] [Google Scholar]
- 30.Naumov, G. I., E. S. Naumova, Z. Antunovics, and M. Sipiczki. 2002. Saccharomyces bayanus var. uvarum in Tokaj wine-making of Slovakia and Hungary. Appl. Microbiol. Biotechnol. 59:727-730. [DOI] [PubMed] [Google Scholar]
- 31.Naumova, E. S., G. I. Naumov, I. Masneuf-Pomarede, M. Aigle, and D. Dubourdieu. 2005. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast 22:1099-1115. [DOI] [PubMed] [Google Scholar]
- 32.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, H.-V., and C. Gaillardin. 1997. Two subgroups within the Saccharomyces bayanus species evidenced by PCR amplification and restriction polymorphism of the non-transcribed spacer 2 in the ribosomal DNA unit. Syst. Appl. Microbiol. 20:286-294. [Google Scholar]
- 34.Nguyen, H. V., and C. Gaillardin. 2005. Evolutionary relationships between the former species Saccharomyces uvarum and the hybrids Saccharomyces bayanus and Saccharomyces pastorianus; reinstatement of Saccharomyces uvarum (Beijerinck) as a distinct species. FEMS Yeast Res. 5:471-483. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, H.-V., A. Lepingle, and C. Gaillardin. 2000. Molecular typing demonstrates homogeneity of Saccharomyces uvarum strains and reveals the existence of hybrids between S. uvarum and S. cerevisiae, including the S. bayanus type strain CBS 380. Syst. Appl. Microbiol. 23:71-85. [DOI] [PubMed] [Google Scholar]
- 36.Pulvirenti, A., H.-V. Nguyen, C. Caggia, P. Giudici, S. Rainieri, and C. Zambonelli. 2000. Saccharomyces uvarum, a proper species within Saccharomyces sensu stricto. FEMS Microbiol. Lett. 192:191-196. [DOI] [PubMed] [Google Scholar]
- 37.Rainieri, S., C. Zambonelli, J. E. Hallsworth, A. Pulvirenti, and P. Giudici. 1999. Saccharomyces uvarum, a distinct group within Saccharomyces sensu stricto. FEMS Microbiol. Lett. 177:177-185. [DOI] [PubMed] [Google Scholar]
- 38.Rainieri, S., C. Zambonelli, and Y. Kaneko. 2003. Saccharomyces sensu stricto: systematics, genetic diversity and evolution. J. Biosci. Bioeng. 96:1-9. [PubMed] [Google Scholar]
- 39.Talarek, N., E. J. Louis, C. Cullin, and M. Aigle. 2004. Developing methods and strains for genetic studies in the Saccharomyces bayanus var. uvarum species. Yeast 21:1195-1203. [DOI] [PubMed] [Google Scholar]
- 40.Tamai, Y., T. Momma, H. Yoshimoto, and Y. Kaneko. 1998. Co-existence of two types of chromosomes in the bottom fermenting yeast, Saccharomyces pastorianus. Yeast 14:923-933. [DOI] [PubMed] [Google Scholar]
- 41.Tamai, Y., K. Tanaka, N. Uemoto, K. Tomizuka, and Y. Kaneko. 2000. Diversity of the HO gene encoding an endonuclease for mating-type conversion in the bottom fermenting yeast Saccharomyces pastorianus. Yeast 16:1335-1343. [DOI] [PubMed] [Google Scholar]
- 42.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 sequences: a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 43.Van de Peer, Y., and R. De Wachter. 1994. TreeCon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 44.Vaughan-Martini, A., and C. P. Kurtzman. 1985. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int. J. Syst. Bacteriol. 35:508-511. [Google Scholar]
- 45.Vaughan-Martini, A., and A. Martini. 1998. Saccharomyces Meyen ex Reess, p. 358-373. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 46.Yamagishi, H., and T. Ogata. 1999. Chromosomal structures of bottom-fermenting yeasts. Syst. Appl. Microbiol. 22:341-353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.