Abstract
A fluorescence in situ hybridization-flow cytometry (FISH/FC)-based method was optimized using artificial mixtures of pure cultures of methanotrophic bacteria. Traditional oligonucleotide probes targeting 16S rRNAs of type I (MG84/705 probe) and type II (MA450 probe) methanotrophs were labeled with fluorescein or Alexa fluor and used for FISH, followed by fluorescence-activated FC analysis and cell sorting (FACS). The method resulted in efficient separation of target cells (type I or type II methanotrophs) from the artificial mixtures. The method was then applied for detection and enrichment of type I and type II methanotroph populations from a natural sample, Lake Washington sediment. Cells were extracted from the sediment, fixed, and subjected to FISH/FC/FACS. The resulting subpopulations were analyzed by reverse transcriptase PCR surveys of 16S rRNA, pmoA (encoding a subunit of particulate methane monooxygenase), and fae (encoding formaldehyde-activating enzyme) genes. The functional gene analysis indicated specific separation of the type I and type II methanotroph populations. 16S rRNA gene analysis revealed that type I methanotrophs comprised 59% of the subpopulation separated using the type I-specific probe and that type II methanotrophs comprised 47.5% of the subpopulation separated using the type II-specific probe. Our data indicate that the FISH/FC/FACS protocol described can provide significant enrichment of microbial populations of interest from complex natural communities and that these can be used for genetic tests. We further tested the possibility of direct whole-genome amplification (WGA) from limited numbers of sorted cells, using artificial mixtures of microbes whose genome sequences are known. We demonstrated that efficient WGA can be achieved using 104 or more cells separated by 16S rRNA-specific FISH/FC/FACS, while fewer cells resulted in less specific WGA.
Methanotrophs are a physiologically specialized group of bacteria capable of utilizing methane as a sole source of carbon and energy, and they have been recognized as major players in local and global elemental cycling in aerobic environments (20). Methanotrophs have been detected in a variety of environments, and in some they represent significant fractions of total microbial communities (9, 40). Estimates of methanotroph abundance in natural samples are based on a number of complementary techniques, such as determination of methane oxidation rates (3, 9, 29, 40), determination of the fatty acid composition of the total microbial population (9, 29, 40), PCR-based (2, 14, 15, 19, 25, 26, 28), quantitative PCR-based (5, 27, 29), or reverse transcriptase PCR (RT-PCR)-based (24, 32) surveys, and direct counting using microscopy combined with fluorescence in situ hybridization (FISH) (6, 8, 10). The most widely used PCR primers for detecting type I and type II methanotrophic bacteria target rRNA genes (15, 19) or functional genes encoding particulate methane monooxygenase (pmoA) (14), methanol dehydrogenase (mxaF) (14), formaldehyde-activating enzyme (fae) (25), or the D subunit of the formyltransferase/hydrolase complex (fhcD) (25), and large databases of these genes have been compiled from a variety of environments (5, 11, 14, 16, 19, 24, 26, 28, 31, 33, 39, 40). The most widely used FISH probes target rRNA (15, 19); however, several protocols have been developed recently for mRNA-targeted (pmoA or mxaF) FISH detection (12, 34).
In recent years, applying fluorescence-activated flow cytometry (FC) to the goals of environmental microbiology has received much attention (1, 38, 42), based on the promise of fast and accurate detection of small particles, potentially translated into qualitative and quantitative detection of microbial populations in natural environments. So far, FC has been applied successfully to studies of bacterial community structure, composition, and activity in aquatic ecosystems (7, 18). The approach has also been tested for analyzing soil and sediment microbial populations as well as for viral detection (13). However, the recovery and separation of microbial cells from soil or sediment particles still remain challenging tasks (13, 35). One of the most intriguing potential applications of FC is the possibility of direct extraction of specific subpopulations from environmental samples, omitting the cultivation step, followed by genetic or even genomic characterization. The aim of the present study was to establish a protocol for FISH-FC analysis and separation of methanotroph populations, allowing for subsequent genetic and genomic analyses. We first tested the feasibility of this approach and optimized the protocol by separating specific subpopulations from artificial mixes of cultured methanotrophs. We then applied the protocol to separate methanotroph populations from a complex microbial community inhabiting the top layer of Lake Washington sediment.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Methylobacterium extorquens AM1, Methylobacillus flagellatus KT, Methylosinus trichosporium OB3b, Methylosarcina lacus LW14, Methylomonas sp. strain LW13, Methylosinus sp. strain LW2, and Methylococcus capsulatus Bath were employed in the optimization experiments. M. extorquens and M. flagellatus were grown in a previously described minimal medium (21) supplemented with 100 mM methanol. Methanotroph cultures were grown in NMS medium (44). For optimization of mmoX-targeted FISH, Methylomonas LW13 cells were grown with or without a Cu supplement, as previously described (2).
Cell extraction.
Lake Washington sediment samples were collected as described previously (26). Cells were extracted from 2 to 5 ml of the sediment. The following three methods for cell extraction were tested: (i) shaking for 20 min at 200 rpm at room temperature, (ii) vortexing using an MBB-8 apparatus (Biospec Products) for 2 min with 0.1-mm zirconia-silica beads (Biospec Products) at 5°C, and (iii) homogenization using a PRO200 homogenizer 115V (PROScientific) for 5 min at position 3 on ice. Samples were then diluted 20 to 50 times with filter-sterilized Lake Washington water (LWW) supplemented with the following: 0.1 M NaCl, 1% sucrose, and 0.1% of either Triton X or Tween 80. To eliminate sediment particles, the blended material was either filtered through 5-μm NY20 filters (Millipore) or centrifuged at 750 × g for 3 min. Cells from the resulting filtrates or supernatants were pelleted by centrifugation at 8,000 × g for 15 min and resuspended in either LWW or phosphate-buffered saline (PBS; 0.1 M Na2HPO4, 20 mM KH2PO4, 137 mM NaCl, 27 mM KCl).
The efficiency of recovery was tested by CFU counts, microscopic observations, and estimations of DNA yields. To ensure optimal CFU counts, cells were plated onto the following media prepared with LWW: 0.1× nutrient agar (BD Diagnostics), 0.1× Luria-Bertani (LB) agar (BD Diagnostics), 0.1× TGY (0.5% tryptone, 0.1% yeast extract, 0.1% glucose) agar, or MMB agar (http://www.dsmz.de/media/med628.htm). To specifically address the methanotroph extraction efficiency, cells were plated on LWW agar or on 0.1× NMS agar, and plates were incubated either under a methane-air (50:50) atmosphere or under an air atmosphere to subtract colonies growing on agar alone. Plates were incubated aerobically at 16°C, room temperature (approximately 24°C), or 30°C for up to 1 month. The maximal CFU counts were obtained on 0.1× LB agar. DNAs were extracted using an UltraClean Soil DNA kit (MO BIO Laboratories, Inc.), and their concentrations were measured spectrophotometrically (37).
Probes and conditions for FISH.
The following probes were used in this study: 16S rRNA-targeted probes MA450, MG84, and MG705 (15) and a polynucleotide mmoX-targeted probe. The polynucleotide probe was PCR amplified using the chromosome of Methylomonas sp. strain LW13 as a template and PCR primers described earlier (2) and either labeled with Alexa fluor 488-dUTP included in the PCR mix or labeled using a Fluorescein-High Prime kit (Roche). The PCR fragments were purified by ethanol precipitation and digested by HincII, HinfI, HpaII, and AluI, resulting in 30- to 60-bp fragments.
Cells of pure cultures (108 to 109) or cells extracted from the sediment (108 CFU) were harvested by centrifugation, resuspended in 1 ml of PBS, and fixed with 4% paraformaldehyde (1:3 [vol/vol]) for 8 to 12 h on ice. The fixed cells were washed with 1 ml of PBS twice and resuspended in 200 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 7.5, 20% formamide, 0.1% sodium dodecyl sulfate). Each sample was divided amongst three tubes, and the tubes were prewarmed at 50 to 58°C. One of the tubes was used as a control, the second was mixed with a specific fluorescent probe (5 ng/μl of oligonucleotide probe or 50 ng/μl of polynucleotide probe), and the third was mixed with a nonspecific probe. Cell suspensions were incubated at an appropriate hybridization temperature (50°C for the MA450 probe, 58°C for the MG84/705 pair, and 56°C for the polynucleotide probe) for 8 to 12 h. After hybridization, cells were pelleted by centrifugation (8,000 × g for 3 min), separated from the supernatant, and incubated in 500 μl of hybridization buffer for an additional 20 min, followed by incubation in 500 μl of wash buffer (20 mM Tris-HCl, pH 8.0, 0.9 mM NaCl, 0.1% sodium dodecyl sulfate) for 20 min. Cells were collected by centrifugation, resuspended in 1 to 3 ml of cold PBS, homogenized for 15 to 30 s, and used for microscopic observations, FC analysis, and cell sorting. Cells were observed using an epifluorescence microscope (PASCAL LSM 5) and a ×100 oil immersion objective, and data were analyzed using Zeiss LSM Image software.
Flow cytometry and cell sorting.
A BD LSR benchtop flow cytometer (Becton Dickinson) was used to measure the forward angle light scattering, right angle light scattering, and fluorescence of microbial cells. These parameters were acquired as pulse height signals for 10,000 events at a rate of 600 to 3,000 events per second. Subsequent analysis and cell sorting were performed using a BD FACS Vantage SE instrument. The instrument tubing was sterilized using, sequentially, 10% bleach, 3% hydrogen peroxide, 70% ethanol, and sterile PBS. Data analysis and graphics were acquired using the WinMDI 2.1 software package (http://facs.scripps.edu/software.html). Cells were collected at 1,000 events per 0.25-ml PCR tube, with a total of eight tubes per experiment.
Diagnostic RT-PCR.
RT-PCR amplifications were carried out directly with sorted cells, using a one step RT-PCR kit (QIAGEN). pmoA was amplified using the primer set A189 (5′-GGNGACTGGGACTTCTGG-3′)/A682 (5′-GAASG CNGAGAAGAASGC-3′) (23). mmoX was amplified using the primer set mmoXA (5′-ACCAAGGARCARTTCAAG-3′)/mmoXD (5′-CCGATCCAGATDCCRCCCCA-3′) (3). fae was amplified in two steps, as described previously (25), using the primer sets fae1f (5′-GTCGGCGACGGCAAYGARGTCG-3′)/fae1r (5′-GTAGTTGWANTYCTGGATCTT-3′) and fae2f (5′-GCACACATCGACCTSATCATSGG-3′)/fae2r (5′-CCAGTGRATGAAVACGCCRAC-3′). Eubacterial 16S rRNA genes were amplified using the EUB27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and EUB1492r (5′-TACGGYTACCTTGTTACGACTT-3′) primers. The resulting PCR fragments were cloned into the pCR2.1 vector using a TOPO TA kit (Invitrogen). Plasmids were purified using a QIAprep spin miniprep kit (QIAGEN). Sequencing reactions were performed using a BlueDye3.1 kit. Reaction analyses were performed by the Department of Biochemistry sequencing facility at the University of Washington, using an ABI 3700 high-throughput capillary DNA analyzer.
Whole-genome amplification.
Cells of M. capsulatus were labeled with the MG84/705 probe set and sorted from a mixed culture as described above. Pools containing desired numbers of cells were collected, and cells were pelleted by centrifugation at 10,000 × g for 10 min. DNA was amplified using a GenomePhi whole-genome amplification kit (GE Healthcare) according to the manufacturer's instructions. The amplified DNA was digested with BamHI (NEB) and randomly cloned into the pUC19 vector (NEB). Clone libraries were generated using Escherichia coli JM109 as a host, up to 50 clones from each library were verified by the presence of inserts, and these were sequenced as described above.
Phylogenetic analysis.
Sequences were aligned using the Clustal W program (41). The Phylip program package (17) was used for phylogenetic analysis.
Nucleotide sequence accession numbers.
The sequences obtained in this work were deposited with GenBank under the following accession numbers: DQ367733 to DQ367735 (16S rRNA genes), DQ367737 to DQ367742 (pmoA genes), and DQ367743 to DQ367746 (fae genes).
RESULTS AND DISCUSSION
FISH-based cell sorting with artificial mixtures of methylotrophic bacteria.
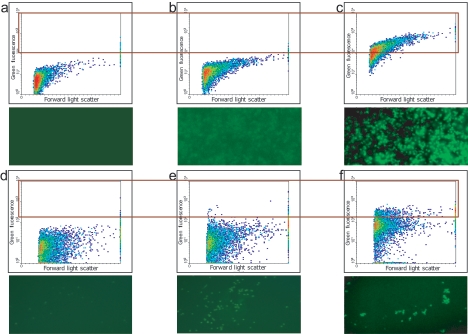
Previously designed probes for 16S rRNA-targeted methanotroph detection and standard FISH protocols (15) were first tested with laboratory strains of the methanotrophs M. capsulatus, M. lacus, Methylomonas sp. strain LW13 (type I methanotrophs), M. trichosporium, and Methylosinus sp. strain LW2 (type II methanotrophs). Microscopic observations and FC data demonstrated that cells of the first three strains were successfully labeled with the MG84/MG705 probe pair specific for type I methanotrophs (Fig. 1a to c) and that cells of the last two strains were successfully labeled with the MA450 probe specific for the Methylosinus/Methylocystis group of type II methanotrophs (data not shown). In addition, we tested a polynucleotide probe for detection of a functional gene mRNA translated from the mmoX gene previously detected in Methylomonas sp. strain LW13. This probe only resulted in a good signal with cells grown in the absence of copper, in accordance with the known expression pattern for the soluble methane monooxygenase (30). Thus, we demonstrated that not only 16S rRNA but also mRNA can be detected by FISH and FC (Fig. 1d to f).
FIG. 1.
FC (top) and fluorescence microscopy (bottom) analyses of fixed cells of M. capsulatus (a to c) and Methylomonas sp. strain LW13 (d to f). (a and d) No probe; (b) nonspecific probe (NON-EUB338); (c) type I methanotroph-specific oligonucleotide probe (MG84/705); (e and f) Methylomonas sp. strain LW13 mmoX-specific polynucleotide probe; (e) cells grown in the presence of Cu; (f) cells grown without Cu. Each plot contains 10,000 events. The boxed part of the plot indicates the events gated for sorting.
We then tested the applicability of these probes to the specific separation of cells of interest from mixed bacterial populations. We produced artificial mixtures of fixed cells of four cultures, M. capsulatus, M. trichosporium, M. extorquens, and M. flagellatus, in which target cells were present in different proportions (5 to 25% of total cells). These were hybridized with either the MG84/705 probe set (targeting M. capsulatus) or the MA450 probe (targeting M. trichosporium), followed by FC and cell sorting. Cells displaying high fluorescence were collected and analyzed by RT-PCR amplification of 16S rRNA and pmoA genes, followed by cloning and sequencing. A total of 25 randomly chosen clones from each library were analyzed. All clones in both 16S rRNA gene and pmoA libraries originating from the MG84/705-based sort belonged to the target organism, M. capsulatus. Similarly, all clones originating from the MA450-based sort belonged to the target organism, M. trichosporium. We performed similar experiments with a bacterial mixture including Methylomonas sp. strain LW13, M. extorquens, and M. flagellatus, using with the mmoX-targeted polynucleotide probe, and obtained similar results: DNAs amplified from the sorted cells belonged to Methylomonas sp. strain LW13. These results demonstrated that FISH-FC-based separation of cells from simple bacterial mixtures allowed discrimination with high efficiency between target and nontarget cells, and thus this protocol could be applied to enriching cells of interest from more complex natural populations.
Recovery of bacterial cells from Lake Washington sediment.
Effective extraction of bacterial cells from sediment is the first essential step for correct assessment of natural populations, and it remains a challenging task. We tested several types of extraction procedures for their efficiency of recovering bacterial cells from Lake Washington sediment. The shaking protocol produced the lowest cell counts (6.2 × 104 ± 1.8 × 104 CFU ml−1 plated onto 0.1× LB agar). The MBB-8 vortexing protocol resulted in higher cell counts (4.5 × 106 ± 1.2 × 106 CFU ml−1), but the best results were achieved by applying homogenization using a PRO200 homogenizer (2.8 × 108 ± 0.2 × 108 CFU ml−1). The addition of 0.1 M NaCl and dilution of treated samples (1:20 or 1:50) in LWW supplemented with 1% sucrose and 0.1% Triton X-100 further increased the extraction efficiency by 10 to 15%. The number of CFU in a methane atmosphere was 6 × 105 colonies after this extraction protocol, compared to 8 × 102 CFU when sediment dilutions were plated without extraction. Therefore, a protocol involving the homogenization of lake samples followed by dilution was applied for all cell sample preparations and resulted in extraction of approximately 3 × 108 cells from 1 ml of lake sediment, on average. Since filtration resulted in a significant decrease in cell number (data not shown), it was replaced by two steps of centrifugation, first at 750 × g for 3 min and then at 5,000 × g for 15 min. The efficiencies of these steps were followed by CFU counts and estimations of DNA recovery. The first centrifugation step resulted in the collection of sediment debris and eukaryotic organisms (algae, protists, etc.), with a small number of bacterial cells and DNA (6 × 103 CFU and 30 μg DNA per g of sample), while the second centrifugation step resulted in relatively pure bacterial cell samples (2.8 × 108 CFU and 200 μg DNA per g of sample). Flow cytometric analysis of the samples showed that 98% of the detected events stained with the DNA stain 4′,6′-diamidino-2-phenylindole (DAPI).
Separation and enrichment of type I and type II methanotrophs from Lake Washington sediment.
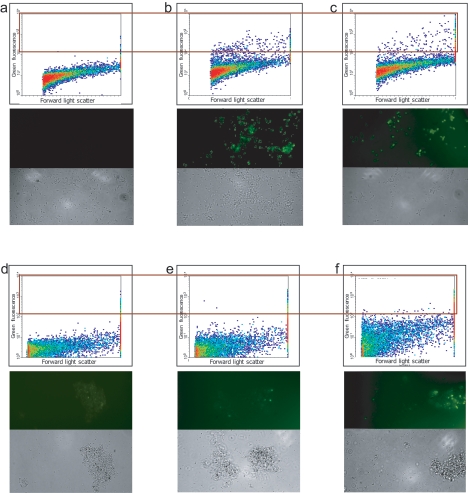
Cells extracted from the lake sediment as described above were fixed and hybridized with the group-specific probe set MG84/705 (type I methanotrophs) or MA450 (type II methanotrophs), as done for pure culture mixtures. Hybridization with the MG84/705 probe pair resulted in a larger number of positive (bright) cells than the number of cells hybridizing with the MA450 probe (Fig. 2d to f). Similarly, cell counts deduced from FC analysis correlated with previous observations on the dominant presence of type I methanotrophs (4.7% ± 1.3% of total events) at the site compared to type II methanotrophs (1.2% ± 0.4% of total events) (9). Cells were sorted as described above. To increase the efficiency of cell separation and to decrease the background, only cells displaying the highest fluorescence signals were gated (2% of cells hybridized with the MG84/705 probe and 0.2% of cells hybridized with the MA450 probe) and collected (Fig. 2). The enrichment of the target cells was tested via RT-PCR amplification of pmoA, fae, and 16S rRNA gene fragments. For each gene, a clone library was constructed, and 25 to 50 randomly chosen clones were sequenced and analyzed.
FIG. 2.
FC (top), fluorescence microscopy (middle), and phase-contrast microscopy (bottom) analyses of fixed cells of an artificial mixture of M. capsulatus, M. trichosporium, M. extorquens, and M. flagellatus (a to c) and of cells extracted from Lake Washington sediment (d to f). (a and d) No probe; (b, c, and f) type I methanotroph-specific oligonucleotide probe (MG84/705); (e) type II methanotroph-specific probe (MA450). For panels b and c, 10% and 5% of target cells (M. capsulatus), respectively, were included in the mixture. Each plot contains 10,000 events. The boxed part of the plot indicates the events gated for sorting.
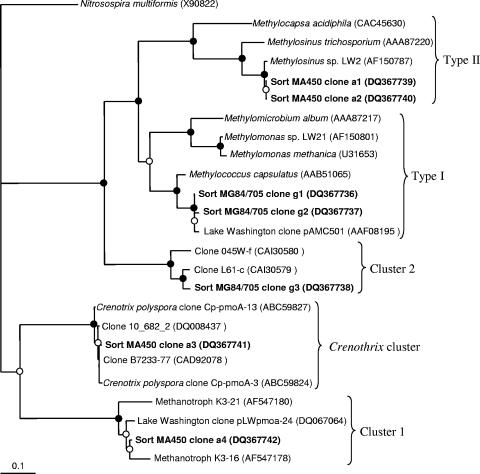
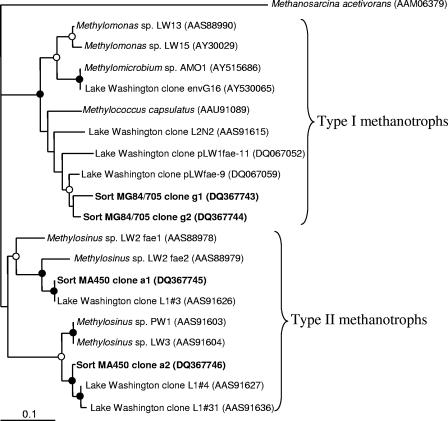
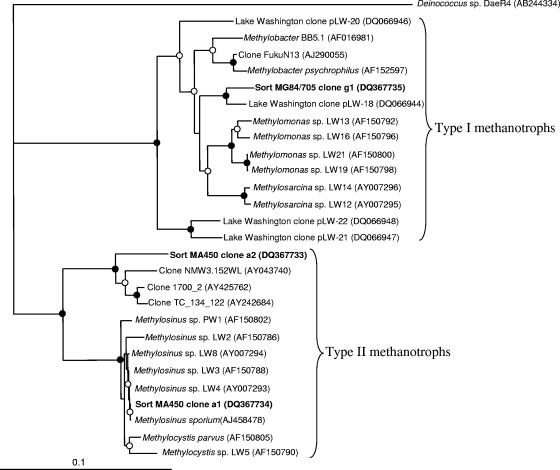
Three types of pmoA sequences were identified in the library obtained from sorted cells hybridized with the MG84/705 (type I) probes. Two of these (clones g1 and g2 [37 and 42% of total clones, respectively]) were closely related to each other and to the sequences from type I methanotrophic bacteria, while the third (g3 [21% of the total clones]) was closely related to a group of deeply branching sequences not represented so far by any cultivated organisms (Fig. 3). These sequences were recently ascribed to cluster 2 of pmoA genes (28, 36). Two types of fae sequences were amplified from the same cells and were equally represented in the library, and these were closely related to each other and to type I methanotroph sequences (Fig. 4). Eighty-six percent of 16S rRNA sequences amplified from the same cells were related to gammaproteobacterial sequences, with 59% being related to the sequences of type I methanotrophs (Fig. 5) and 27% being related to the sequences of Pseudomonas sp. The remaining 14% of sequences were related to the sequences of gram-positive bacteria.
FIG. 3.
Phylogenetic tree showing relationships of the translated PmoA sequences (160 positions) uncovered in this work to the sequences from known type I and type II methanotrophs and to the sequences of uncultivated organisms. The Nitrosospira multiformis sequence was used as an outgroup. A distance algorithm (Protdist) was employed, with 1,000 bootstrap analyses. Closed circles indicate bootstrap support of over 90%, and open circles indicate bootstrap support of over 55%.
FIG. 4.
Phylogenetic tree showing relationships of the translated Fae sequences (98 positions) uncovered in this work to the sequences from known type I and type II methanotrophs and to the sequences of uncultivated organisms. The Methanosarcina acetivorans sequence was used as an outgroup. A distance algorithm (Protdist) was employed, with 1,000 bootstrap analyses. Closed circles indicate bootstrap support of over 90%, and open circles indicate bootstrap support of over 50%.
FIG. 5.
Phylogenetic tree showing relationships of 16S rRNA gene sequences (543 to 605 positions) uncovered in this work to the sequences from known type I and type II methanotrophs and to the sequences of uncultivated organisms. A distance algorithm (DNAdist) was employed, with 1,000 bootstrap analyses. Closed circles indicate bootstrap support of over 90%, and open circles indicate bootstrap support of over 55%. The Deinococcus sequence was used as an outgroup.
Four types of pmoA sequences were identified in the library obtained from sorted cells hybridized with the MA450 (type II) probe. Clones a1 and a2 (11 and 9%, respectively, of all sequences) were closely related to each other and to pmoA genes of the Methylosinus/Methylocystis group. Clone a4 (50% of total clones) was related to the group of deeply diverging type II pmoA genes previously described for tundra soil isolates related to Methylosinus/Methylocystis (Fig. 3) (33) and recently ascribed to cluster 1 of pmoA genes (28, 36). Closely related sequences were also previously detected in the Lake Washington sediment via RT-PCR surveys (Fig. 3) (32). Clone a3 (30% of total clones) was closely related to pmoA genes of the recently described yet uncultivated filamentous methanotroph Crenothrix polyspora (39) and to a large group of sequences recovered from the Eastern Snake River Plain aquifer, designated group II unculturables (16). While C. polyspora is a gammaproteobacterium, its pmoA sequence has been noted to be more closely related to amoA sequences and to the divergent alphaproteobacterial sequences (cluster 1) (39) than to gammaproteobacterial sequences. Two types of fae sequences were identified in the library obtained from the same cells and were equally represented in the library, and both were closely related to fae genes described for type II methanotroph isolates from Lake Washington as well as to the sequences retrieved from Lake Washington via PCR surveys (Fig. 4) (26). In the 16S rRNA gene clone library obtained from the same cells, 62.5% of the sequences were related to alphaproteobacterial sequences, with 35% being most related to Methylocystis sp. sequences (approximately 98% similarity) and 12.5% being more distantly related to the sequences of alphaproteobacterial methanotrophs (Fig. 5), while 15% were related to the sequences of Sphingomonas sp. Of the 37.5% of nonalphaproteobacterial sequences, 5% were related to the sequences of Pseudomonas sp., and the rest were related to the sequences of gram-positive bacteria.
The presence of Pseudomonas-like sequences as well as sequences related to the sequences of gram-positive bacteria in both 16S rRNA clone libraries is likely due to nonspecific sorting based on natural bacterial fluorescence or due to cell adhesion. Similar problems with the specificities of FISH probes have been noted before (38). The majority of the sorted cells, however, seemed probe specific, as type I methanotroph-targeted sorting resulted in enrichment of sequences related to those of known type I methanotrophs, and type II methanotroph-targeted sorting resulted in enrichment of sequences related to those of known type II methanotrophs. The limited diversity of phylogenetic and functional genes in the analyzed PCR-based libraries likely reflects the dominant nature of the organisms in question in each subpopulation. While most of the sequences uncovered in this work were closely related to the sequences previously uncovered from the site by PCR or RT-PCR surveys, some of the sequences were novel and had not previously been detected in Lake Washington. The divergent 16S rRNA sequence distantly related to those of type II methanotrophs (clone a2) (Fig. 5) may represent a novel group of type II methanotrophs, the divergent pmoA clone (g3) may represent a divergent group of gammaproteobacterial pmoA genes (cluster 2) (Fig. 3), and another divergent pmoA clone (a3) may represent a group of pmoA genes shared by alpha- and gammaproteobacterial methanotrophs (Crenothrix cluster) (Fig. 3). While this last possibility is intriguing, additional experiments, such as double (rRNA and mRNA) FISH, are necessary to link this novel group of pmoA sequences to the phylogenetic identities of the organisms in question.
We were not able to perform FC analysis or fluorescence-activated cell sorting (FACS) using the mmoX-targeted polynucleotide probe (see above), as this probe produced a very weak fluorescent signal, consistent with the lack of or low expression of mmoX in the sediment community under ambient conditions (M. Kalyuzhnaya, unpublished observations).
Overall, the results described above suggest that the FISH/FC/FACS approach is effective in obtaining preparations of cells enriched in either type I or type II methanotrophs from lake sediment and is suitable for further genetic analysis of functional genes in this population.
WGA.
One of the attractive applications of FISH-FC-based separation of specific populations of cells could be in the genomic characterization of uncultured microbes. While the amounts of DNA isolated from sorted cells would not be sufficient for traditional shotgun sequencing, DNAs could be amplified by using commercially available enzymes for whole-genome amplification (WGA). We tested the feasibility of such an approach and determined the number of cells required to produce sufficient amounts of DNA for specific amplification, using a model methanotroph, M. capsulatus, whose genome sequence is available (43). We used an artificial mix of M. capsulatus with M. extorquens and M. flagellatus in approximately equal proportions. The genomic sequences of the two control organisms are also available (http://www.integratedgenomics.com/genomereleases.html#6 and http://genome.jgi-psf.org/draft_microbes/metfl/metfl.home.html). Cells were labeled using the MG84/705 probe and subjected to FISH/FC/FACS as described above. Pools containing 10, 102, 103, 104, or 105 of the sorted cells were used for WGA. The resulting DNAs were used to construct shotgun libraries corresponding to each pool, and up to 50 clones from each library were sequenced. Our data indicated that a direct correlation existed between the number of cells used for WGA and the specificity of WGA. In the first three pools, 80%, 50%, and 5% of the tested clones contained nonspecific DNA. This DNA seemed to be noncoding, as judged by BLAST analyses with either a nonredundant nucleotide or nonredundant protein database, and likely was a product of nonspecific (background) amplification (11, 22). The remaining sequences in these libraries carried M. capsulatus DNA, as judged from BLAST analyses. One hundred percent of tested clones in libraries corresponding to pools with 104 and 105 cells carried M. capsulatus DNA, and the hits were distributed randomly along the chromosome, with only one hot spot, in the glutamine synthetase gene (data not shown). Likely, the cause of this hot spot was a cloning bias toward the respective BamHI fragment of 0.2 kb. Our results are in agreement with the results from other groups indicating that at least 103 microbial cells or at least 1 ng of DNA is necessary for high-fidelity WGA (4, 11, 22).
Conclusion.
The FISH/FC/FACS approach for separation of microbial populations of interest from natural samples described here represents a promising tool for genetic and genomic characterization of as yet uncultivated or unculturable microbes. In this work, we demonstrated successful separation of type I and type II methanotrophs and enrichment of the desired type from a complex natural community, Lake Washington sediment, using traditional FISH probes. Future work will address the specificity of cell sorting using phylogenetic probes and will further explore the feasibility of probes targeting functional gene RNA transcripts for FISH/FC/FACS and subsequent WGA.
Acknowledgments
We acknowledge support from the Microbial Observatories Program funded by the National Science Foundation (MCB-0131957).
We also acknowledge the staff of the Cell Analysis Facility at the University of Washington for assistance with flow cytometric analysis and David Stahl for sharing his microscope facilities.
REFERENCES
- 1.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auman, A. J., and M. E. Lidstrom. 2002. Analysis of sMMO-containing type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517-524. [DOI] [PubMed] [Google Scholar]
- 3.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergen, A. W., Y. Qi, K. A. Haque, R. A. Welch, and S. J. Chanock. 2005. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC Biotechnol. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodrossy, L., N. Stralis-Pavese, M. Konrad-Koszler, A. Weilharter, T. G. Reichenauer, D. Schofer, and A. Sessitch. 2006. mRNA-based parallel detection of active methanotroph populations by use of a diagnostic microarray. Appl. Environ. Microbiol. 72:1672-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne, D. G., A. J. Holmes, N. Iversen, and J. C. Murrell. 2000. Fluorescent oligonucleotide rDNA probes for specific detection of methane oxidising bacteria. FEMS Microbiol. Ecol. 31:29-38. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, L., M. R. Landry, J. Constantinou, H. A. Nolla, S. L. Brown, H. Liu, and D. A. Caron. 1998. Response of microbial community structure to environmental forcing in the Arabian Sea. Deep-Sea Res. II 45:2301-2325. [Google Scholar]
- 8.Carini, S., N. Bano, G. LeCleir, and S. B. Joye. 2005. Aerobic methane oxidation and methanotroph community composition during seasonal stratification in Mono Lake, California (USA). Environ. Microbiol. 7:1127-1138. [DOI] [PubMed] [Google Scholar]
- 9.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 10.Dedysh, S. N., M. Derakshani, and W. Liesak. 2001. Detection and enumeration of methanotrophs in acidic sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Detter, J. S., J. M. Jett, S. M. Lucas, E. Dalin, A. R. Arellano, M. Wang, J. R. Nelson, J. Chapman, Y. Lou, D. Rokhsar, T. L. Hawkins, and P. M. Richardson. 2002. Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics 80:691-698. [DOI] [PubMed] [Google Scholar]
- 12.Dias, J. C. T., C. M. Silva, A. H. Mounteer, F. M. L. Passos, and V. R. Linardi. 2003. Molecular diversity of mesophilic and thermophilic bacteria in a membrane bioreactor determined by fluorescent in situ hybridization with mxaF- and rRNA-targeted probes. J. Basic Microbiol. 43:202-209. [DOI] [PubMed] [Google Scholar]
- 13.Duhamel, S., and S. Jacquet. 2006. Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J. Microbiol. Methods 64:316-332. [DOI] [PubMed] [Google Scholar]
- 14.Dumont, M. G., and J. C. Murrell. 2005. Community-level analysis: key genes of aerobic methane oxidation. Methods Enzymol. 397:413-427. [DOI] [PubMed] [Google Scholar]
- 15.Eller, G., S. Stubner, and P. Frenzel. 2001. Group specific 16S rRNA targeted probes for the detection of type I and type II methanotrophs by fluorescence in situ hybridisation. FEMS Microbiol. Lett. 198:91-97. [DOI] [PubMed] [Google Scholar]
- 16.Erwin, D. P., I. K. Erickson, M. E. Delwiche, F. S. Colwell, J. L. Strap, and R. L. Crawford. 2005. Diversity of oxygenase genes from methane- and ammonia-oxidizing bacteria in the Eastern Snake River Plain aquifer. Appl. Environ. Microbiol. 71:2016-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 2003. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, Mass.
- 18.Fuchs, B. M., D. Woebken, M. V. Zubkov, P. Burkill, and R. Amann. 2005. Molecular identification of picoplankton populations in contrasting water of the Arabian Sea. Aquat. Mol. Ecol. 39:145-157. [Google Scholar]
- 19.Gulledge, J., A. Ahmad, P. A. Steudler, W. J. Pomerantz, and C. M. Cavanaugh. 2001. Family- and genus-level 16S rRNA-targeted oligonucleotide probes for ecological studies of methanotrophic bacteria. Appl. Environ. Microbiol. 67:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harder, W., M. Attwood, and J. R. Quayele. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 22.Holbrook, J. F., D. Stabley, and K. Sol-Church. 2005. Exploring whole genome amplification as a DNA recovery tool for molecular genetic studies. J. Biomol. Technol. 16:125-133. [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, A. J., A. M. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 24.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalyuzhnaya, M. G., and L. Chistoserdova. 2005. Community-level analysis: genes encoding methanopterin-dependent enzymes. Methods Enzymol. 397:443-454. [DOI] [PubMed] [Google Scholar]
- 26.Kalyuzhnaya, M. G., M. E. Lidstrom, and L. Chistoserdova. 2004. Utility of environmental primers targeting ancient enzymes: methylotroph detection in Lake Washington. Microb. Ecol. 48:463-472. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi, T., K. Iwasaki, H. Nishinara, Y. Takamura, and O. Yagi. 2002. Quantitative and rapid detection of the trichloroethylene-degrading bacterium Methylocystis sp. M in groundwater by real-time PCR. Appl. Microbiol. Biotechnol. 59:731-736. [DOI] [PubMed] [Google Scholar]
- 28.Knief, C., S. Kolb, P. L. Bodelier, A. Lipski, and P. F. Dunfield. 2006. The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration. Environ. Microbiol. 8:321-333. [DOI] [PubMed] [Google Scholar]
- 29.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 695:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murrell, J. C., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 31.Nercessian, O., M. G. Kalyuzhnaya, S. B. Joye, M. E. Lidstrom, and L. Chistoserdova. 2005. Analysis of fae and fhcD genes in Mono Lake, California. Appl. Environ. Microbiol. 71:8949-8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nercessian, O., E. Noyes, M. G. Kalyuzhnaya, M. E. Lidstrom, and L. Chistoserdova. 2005. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71:6885-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacheco-Oliver, M., I. R. McDonald, D. Groleau, J. C. Murrell, and C. B. Miguez. 2002. Detection of methanotrophs with highly divergent pmoA genes from Arctic soils. FEMS Microbiol. Lett. 209:313-319. [DOI] [PubMed] [Google Scholar]
- 34.Pernthaler, A., and R. Amann. 2004. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl. Environ. Microbiol. 70:5426-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter, J., R. Pickup, and C. Edwards. 1997. Evaluation of flow cytometric methods for detection and viability assessment of bacteria from soil. Soil Biol. Biochem. 29:91-100. [Google Scholar]
- 36.Ricke, P., S. Kolb, and G. Braker. 2005. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 71:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecker, K., B. Bendinger, B. Schoning, P. H. Nielsen, J. L. Nielsen, C. Barnavi, E. R. Toenshoff, H. Daims, and M. Wagner. 2006. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. USA 103:2363-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundh, I., D. Bastviken, and L. J. Tranvik. 2005. Abundance, activity, and community structure of pelagic methane-oxidizing bacteria in temperate lakes. Appl. Environ. Microbiol. 71:6746-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, N., O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methé, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. K. Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M. Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLOS Biol. 2:1616-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]