Abstract
Environmental pollution by phosphorus from animal waste is a major problem in agriculture because simple-stomached animals, such as swine, poultry, and fish, cannot digest phosphorus (as phytate) present in plant feeds. To alleviate this problem, a phytase from Aspergillus niger PhyA is widely used as a feed additive to hydrolyze phytate-phosphorus. However, it has the lowest relative activity at the pH of the stomach (3.5), where the hydrolysis occurs. Our objective was to shift the pH optima of PhyA to match the stomach condition by substituting amino acids in the substrate-binding site with different charges and polarities. Based on the crystal structure of PhyA, we prepared 21 single or multiple mutants at Q50, K91, K94, E228, D262, K300, and K301 and expressed them in Pichia pastoris yeast. The wild-type (WT) PhyA showed the unique bihump, two-pH-optima profile, whereas 17 mutants lost one pH optimum or shifted the pH optimum from pH 5.5 to the more acidic side. The mutant E228K exhibited the best overall changes, with a shift of pH optimum to 3.8 and 266% greater (P < 0.05) hydrolysis of soy phytate at pH 3.5 than the WT enzyme. The improved efficacy of the enzyme was confirmed in an animal feed trial and was characterized by biochemical analysis of the purified mutant enzymes. In conclusion, it is feasible to improve the function of PhyA phytase under stomach pH conditions by rational protein engineering.
Phytases are phosphohydrolytic enzymes that initiate the stepwise removal of phosphate from phytate (myo-inositol hexakisphosphate), the major form of phosphorus in plant foods or feeds (28). Simple-stomached animals, such as swine, poultry, fish, and preruminant calves, do not have phytases in their gastrointestinal tracts and cannot digest phytate-phosphorus. Consequently, there are three major problems (1, 7, 41). First, the unutilized feed phosphorus ends up in manure, causing serious environmental pollution in areas of intensive animal husbandry. Second, it is necessary to supplement inorganic phosphorus in diets for swine, poultry, and fish to meet their nutrient requirement for phosphorus. This represents a significant expense in animal feeding. Third, inorganic phosphorus is nonrenewable, and the easily accessible inorganic phosphorus on earth may be exhausted in 80 years at the current rate of extraction (1). In addition, phytate can chelate trace elements, such as iron and zinc, causing widespread deficiencies of these nutrients in populations whose staple foods are legume based (18, 26).
Low-phytate plants (33) and phytase-transgenic plants (15, 40) or animals (7) have been developed to cope with the nutritional and environmental problems associated with phytate-phosphorus. Although these approaches represent remarkable scientific advances, they are limited in practical application. In contrast, microbial phytases have become a widely accepted and highly effective tool for animal industry to improve feed phytate-phosphorus bioavailability to animals and to comply with environmental laws restricting phosphorus excretion in animal waste. Numerous animal experiments have shown that adding phytase to feed at 500 to 1,000 units kg−1 may replace inorganic-phosphorus supplements for pigs and poultry and reduce their phosphorus excretion by approximately 50% (2, 13, 14).
Aspergillus niger PhyA is the first commercialized phytase, and it occupies a good share of the world market with a $500 million potential (1). Although the enzyme has good catalytic efficacy (14), its unique pH profile precludes it from full function in the stomachs of animals, where dietary phytate-phosphorus is hydrolyzed (38). Specifically, it has two pH optima, 5 to 5.5 (100%) and 2.5 (60%), and a trough in activity at pH 3.5 (22, 38). The stomach pH in many species is around 3.5 (42), which happens to be the lowest activity point in the pH profile of PhyA (22). Thus, a relatively high level of phytase supplementation is required in animal diets for adequate hydrolysis of phytate-phosphorus (13, 14).
Limited success has been achieved in altering the pH profiles of xylanase, α-amylase, glucoamylase, glycosidase, subtilisin, pepsin, and chymotrypsin by protein engineering (4, 5, 6, 9, 19, 23, 32, 37). Approaches used include replacing the amino acid residues in or near the active site, changing the pKa value of the catalytic amino acid residue acting as a general base catalyst (4, 5, 6, 9, 19, 23, 37), and modifying the enzyme surface charge (32, 36). However, all of these efforts have been almost exclusively focused on improvement of the in vitro functions of the enzymes. The feasibility of shifting the optimal pH of phytase or any given enzyme to match the stomach conditions has not been determined. Strategically, the effect of modification of amino acid residues involved in substrate binding on the enzyme’s pH profile has not been well studied. Based on the kinetics and structure of PhyA and other phytases (10, 11, 16, 24, 25), amino acid residues in the α-domain (K91, K94, E228, D262, K300, and K301) are involved in substrate binding (11, 21). As the pH profile of a given enzyme is generally determined by the ionization of the catalytic groups and is affected by various interactions involved in their microenvironments of enzyme and substrate, the unique activity drop of PhyA at pH 3.5 may be due to the interaction of these acidic and basic amino acids comprising the substrate specificity site (22). While the clustering of K91, K94, K300, K301, and other basic amino acids (R81, H82, R85, K165, and H361) (11, 21) around the active site of PhyA presumably creates a relatively favorable electrostatic environment for binding the highly negatively charged substrate phytate, it remains to be determined how alterations of biochemical properties in each of these amino acid positions actually affect the pH-activity profile and substrate affinity of PhyA. In addition, Q50 is located in the active site and has been shown to affect pH-activity profiles in other phytases (35). Therefore, the objective of the present study was to determine the impact of changing these seven selected amino acid residues into substitutes with different charges, polarities, and/or side chain lengths on the pH-activity profile of PhyA.
MATERIALS AND METHODS
Construction of phyA mutants.
Plasmid pYPP1, containing the A. niger NRRL3135 phyA gene cloned into a Saccharomyces cerevisiae expression vector, pYES2, was employed to generate the mutations (8). A total of 9 single and 12 multiple mutants at positions Q50, K91, K94, E228, D262, K300, and K301 (Fig. 1) were prepared by introducing different charges, polarities, or side chain lengths (Table 1). Oligonucleotides were synthesized to generate site-specific mutations at substrate binding sites described earlier (22). The phyA mutants in pYPP1 were constructed using the Gene Editor In Vitro Site-Directed Mutagenesis System (Promega, Madison, WI) (22). The coding region of the pYPP1 mutant construct was amplified by PCR using two primers (upstream, 5′CGG AAT TCC TGG CAG TCC CCG3′; downstream, 5′GCT CTA GAC TAA GCA AAA CAC TCC3′) and inserted into a constitutive expressing vector, pGAPZαA (Invitrogen, San Diego, CA), at EcoRI and XbaI sites. The gene was led by a signal peptide α-factor and was under the control of the GAP promoter. The desired mutations in the selected transformants were confirmed by DNA sequencing.
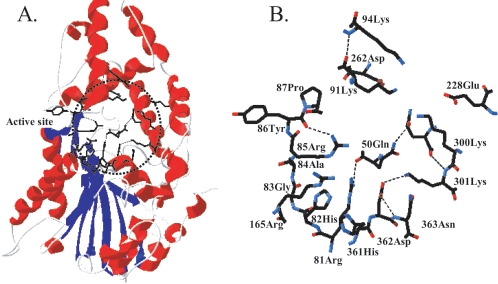
FIG. 1.
(A) The region of the mutated amino acids (dotted circle) in the three-dimensional structure of A. niger PhyA. (B) The seven mutated amino acids (Q50, K91, K94, E228, D262, K300, and K301) and related amino acids in the circled region of panel A. The structure is based on Kostrewa et al. (10). The α-helices are shown in red and the β-sheets in blue in panel A; dotted lines represent hydrogen bonds in panel B.
TABLE 1.
pH optima and activity ratios of PhyA mutants at different pH pairs
| Mutant | Optimal pHa
|
Ratio of activityb
|
||
|---|---|---|---|---|
| First | Second | pH 3.5/5.5 | pH 3.5/2.5 | |
| WT | 5-5.5 | 2.5 (67) | 0.38 ± 0.01 | 0.55 ± 0.03 |
| Q50L | 5.0 | 0.68 ± 0.01* | 1.51 ± 0.03* | |
| Q50P | 5.5 | 3.0 (61) | 0.50 ± 0.01* | 0.94 ± 0.02* |
| K91A | 5-5.5 | 2.5 (60) | 0.33 ± 0.00* | 0.55 ± 0.00 |
| K91E | 5-5.5 | 0.46 ± 0.03* | 1.76 ± 0.11* | |
| K94E | 5-5.5 | 2.0 (69) | 0.34 ± 0.01* | 0.56 ± 0.01 |
| E228Q | 5-5.5 | 2.5 (66) | 0.34 ± 0.01* | 0.52 ± 0.02 |
| E228K | 3.8 | 3.0 (83) | 1.72 ± 0.01* | 1.08 ± 0.01* |
| D262H | 5.0 | 0.87 ± 0.02* | 1.51 ± 0.03* | |
| K301E | 2.5 | 4-4.5 (93) | 1.40 ± 0.01* | 0.75 ± 0.02* |
| E228K-K300D | 5.5 | 0.52 ± 0.01* | 1.07 ± 0.03* | |
| E228K-K300T | NA | NA | NA | |
| E228K-K300R | 4.0 | 3.0 (81) | 1.44 ± 0.23* | 0.91 ± 0.14* |
| E228K-K300R-K301E | 4.0 | 4.00 ± 0.05* | 1.11 ± 0.02* | |
| E228K-K300T-K301E | 3.0-4.0 | 2.25 ± 0.03* | 1.06 ± 0.05* | |
| E228K-K300D-K301E | 4.0 | 3.0 (95) | 2.28 ± 0.03* | 0.93 ± 0.03* |
| K94E-E228K | 4.5 | 3.0 (78) | 0.69 ± 0.04* | 0.79 ± 0.04* |
| K94E-E228K-K301E | 2.5-4.5 | 1.38 ± 0.07* | 0.81 ± 0.04* | |
| K94E-K300E-K301E | 3.0-4.0 | 12.68 ± 0.14* | 1.07 ± 0.01* | |
| K300E-K301E | 3-4.5 | 6.62 ± 0.11* | 1.11 ± 0.03* | |
| K91A-E228Q-K300E | 4.5-5 | 0.70 ± 0.05* | 1.53 ± 0.11* | |
| K94A-E228A-D262A-K300D | 5.5 | 0.55 ± 0.02* | 0.94 ± 0.04* | |
Relative activity at the second optimal pH, as at the first optimal pH, is expressed as a percentage in parentheses.
The ratios of phytase activity at different pH pairs are expressed as mean ± standard error (n = 3 to 6). The buffer conditions were as follows: pH 5.5, 0.2 M citrate; pH 3.5, 0.2 M glycine-HCl; pH 2.5, 0.2 M glycine-HCl. An asterisk indicates a difference (P < 0.05) from the WT. NA, not applicable.
Transformation, phytase expression, and protein purification.
The pGAP vector containing the phyA mutant gene (10 μg) was linearized by the restriction enzyme BamHI and transformed into Pichia pastoris X33 by electroporation using an ECM 600 Electro Cell Manipulator (Gentronics, Inc., BTX Instrument Division, San Diego, CA). The transformed cells were plated in YPD agar (1% yeast extract, 2% peptone, and 2% dextrose) plus zeocin (100 μg ml−1) and incubated at 30°C for 3 days. Single colonies of the transformants were inoculated into YPD media and incubated at 30°C for 2 days for phytase expression. Phytase activity in the culture supernatant was measured to select the best transformants with high phytase expression. The expressed enzymes in the medium supernatant were sequentially purified by ultrafiltration (molecular weight cutoff, 30,000) and DEAE-cellulose column (10 mM Tris-HCl buffer, pH 7.4, with an NaCl linear gradient) and Sephadex G-100 gel exclusion column (50 mM Tris-HCl buffer containing 0.15 M NaCl, pH 7.4) chromatography (8). The purified phytase was eluted as a single peak and used for further analysis.
Phytase activity, protein, and pH profile assays.
Phytase activity was measured using sodium phytate as the substrate. One phytase unit was defined as the amount of activity that released 1 μmol of inorganic phosphorus from sodium phytate per minute at pH 5.5 and 37°C. The enzyme was diluted in 0.2 M citrate buffer, pH 5.5, and an equal volume of substrate solution containing 1% sodium phytate (Sigma) was added. After incubation of the sample for 15 min at 37°C, the reaction was stopped by addition of an equal volume of 15% trichloroacetic acid. The released inorganic phosphorus was determined as previously described (30). The total protein concentration in the samples was determined by the method of Lowry et al. (17). Samples of the purified proteins were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a Mini-Protean II cell (Bio-Rad Laboratories, Hercules, CA) (12) and Western blot analysis (8).
The initial pH profiles of the expressed phytases were determined using the following buffers (at pH 0.5 intervals): 0.2 M glycine-HCl for pH 2 to 3.5, 0.2 M citrate for pH 4 to 6.5, and 0.2 M Tris-HCl for pH 7 to 8.5. The exact optimal pH and the pH profile of the overall best mutant phytase were compared with those of the wild-type (WT) PhyA using 0.2 M glycine-HCl for pH 2.0, 2.5, 3.0, 3.2, 3.4, 3.5, 3.6, 3.8, and 4.0 and 0.2 M citrate for pH 3.5, 3.6, 3.8, 4.0, 4.2, 4.5, 5.0, 5.5, 6.0, and 6.5. Purified enzymes were diluted with each buffer of different pH to give an activity of 0.1 U ml−1 and mixed with 1% sodium phytase dissolved in the same buffer at an equal volume (0.5 ml each) to start the hydrolysis reaction. The released inorganic P and the relative phytase activity were determined as described above.
Kinetics and phytate hydrolysis profile.
The Michaelis-Menten parameters, Vmax and Km, were determined by incubation of the phytase enzyme (100 mU at pH 5.5 and 3.5, respectively) with sodium phytate substrate at 37°C in 0.2 M glycine-HCl buffer, pH 3.5, and in 0.2 M citrate buffer, pH 5.5. A total of 10 different substrate concentrations ranging from 0.1 to 10 Km were used, and the reactions were sampled at 0, 1, 2, 3, 5, and 10 min. Initial velocities were calculated from linear regions of the hydrolysis curve and plotted against the inorganic-phosphate concentration. Linear transformation was achieved using Lineweaver-Burk plots to estimate Vmax and Km parameters (3, 39). The hydrolysis profiles of sodium phytate by the phytase samples were analyzed by high-performance liquid chromatography (HPLC) (Dionex Liquid Chromatograph System DX600; Dionex Corp., Sunnyvale, CA). Phytase was incubated at 37°C in an assay mixture containing 0.2 M citrate buffer (pH 5.5) or glycine-HCl buffer (pH 3.5) and 10 mM sodium phytate. The reaction was stopped after 2, 5, 10, 15, 20, 30, 40, 50, 60, 75, and 90 min by adding an equal volume of 15% trichloroacetic acid solution, and 25 μl of 1:10-diluted samples or standards was analyzed using an AS11 ion-exchange column (Dionex Corp., Sunnyvale, CA) and a flow rate of 1 ml min−1 with a gradient of 26 to 70 mM NaOH and detected by a conductivity detector (Dionex ED50).
Hydrolysis of phytate-phosphorus in soybean meal.
The effectiveness in releasing phytate phosphorus from soybean meal was measured by incubating the feed sample with phytase (a 250-U kg−1 sample) in 0.2 M citrate buffer, pH 5.5, and in 0.2 M glycine-HCl, pH 3.5 and 2.5, respectively, at 37°C for 1 h. One gram of soybean meal was dissolved in 9 ml buffer and incubated at 37°C for 20 min with shaking. Then, 1 ml of prewarmed diluted enzyme was added to start the hydrolysis reaction. After 1 h of incubation at 37°C with shaking, the reaction was stopped by the addition of an equal volume of 15% trichloroacetic acid. The released inorganic phosphorus was determined as previously described (31).
Animal feed trial.
The Institutional Animal Care and Use Committee of Cornell University approved the protocol for the animal experiments. After being fed a low-phosphorus corn-soybean meal basal diet (34), 16 weanling pigs (28 days old) were fed the same basal diet supplemented with E228K (n = 8) or WT PhyA phytase (n = 8) at 250 U kg of feed−1 for 35 days. The growth performance, plasma inorganic-phosphorus concentrations, and plasma alkaline phosphatase activities of individual pigs were determined weekly as previously described (34).
Statistical analysis.
Data were analyzed by SAS (release 6.04; SAS Institute, Cary, NC), and the Bonferroni t test was used to compare mean differences. Significance was set at a P value of <0.05.
RESULTS
Shifts of pH optima and profile in PhyA mutants.
Transformants of all mutants except the double mutant E228K-K300T expressed extracellular phytase activity in the medium supernatants. The impacts of all mutations on the pH optima and the relative activity ratios at pH 3.5 versus at pH 5.5 or pH 2.5 (the two pH optima of the WT enzyme) are summarized in Table 1. The WT PhyA showed the unique bihump two-pH-optima profile, whereas 12 mutants lost one pH optimum and 14 mutants had the pH optimum shifted from pH 5.5 to the more acidic side. Compared with the WT enzyme, 15 mutants exhibited higher (P < 0.05) activities at pH 3.5 than at pH 5.5 or 2.5.
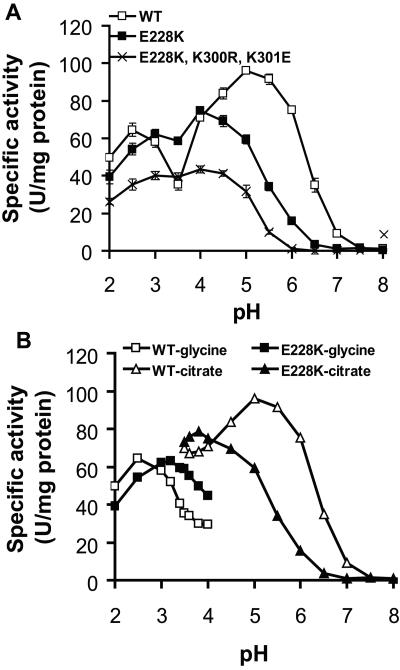
The initial pH profile assay indicated that E228K showed the best overall catalytic improvement among all single mutants (Fig. 2A). The optimal pH was shifted to 4, and the activity trough at pH 3.5 was essentially eliminated. This resulted in a 1.7-fold-higher phytase activity at pH 3.5 than the WT, which is about 38% of the activity found at pH 5.5. The mutant had similar activities at pH 3.5 and 2.5, while the WT had only 55% activity at pH 3.5 compared to the activity level at pH 2.5. Further pH profile analysis, with smaller intervals adjacent to critical points and overlapping ranges between two different buffers, showed pH 3.8 and 5 as the optimal pHs for E228K and the WT PhyA, respectively (Fig. 2B). When the glycine-HCl buffer was used, E228K displayed higher specific activity than the WT PhyA between pH 3.0 and 4.0. The opposite was true when the citrate buffer was used between pH 3.5 and 6.5. Among the multiple mutants, TK10 (E228K-K300R-K301E) showed the most favorable shift of optimal pH to 3 to 4 and a large increase in the phytase activity ratio at pH 3.5 over pH 5.5 to 4.00 ± 0.05, with slightly more activity at pH 3.5 than at 2.5 (ratio, 1.11 ± 0.02) (Fig. 2A and Table 1). Two other mutants (K94E-K300E-K301E and K300E-K301E) showed greater improvement in relative activity at pH 3.5 (Table 1) but had low specific activity or yield (data not shown). The molecular size, glycosylation, reactivity to polyclonal antibody against the purified PhyA protein from A. niger, optimal temperature, and heat stability of these two and other mutants were not significantly different from those of the WT (data not shown).
FIG. 2.
Comparisons of pH profiles of the WT, the single mutant E228K, and the triple mutant E228K-K300R-K301E. In both panels, values are expressed as the mean specific activity ± standard error (n = 3) (for panel B, standard errors were too small to be seen) of the purified enzymes at each pH, and phytase activity was determined using sodium phytate dissolved in the designated assay buffer. (A) pH 2.0 to 3.5, 0.2 M glycine-HCl; pH 4.0 to 6.5, 0.2 M citrate; and pH 7.0 to 8.0, 0.2 M Tris-HCl. (B) pH 2.0 to 4.0, 0.2 M glycine-HCl; pH 3.5 to 6.5, 0.2 M citrate. Overlapping pH points between the two buffer systems and smaller intervals were used to compare the buffer effect and the exact optimal pH of the testing phytases.
In vitro and in vivo functional assays.
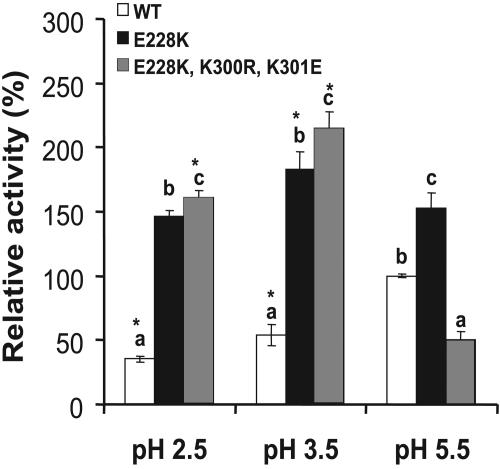
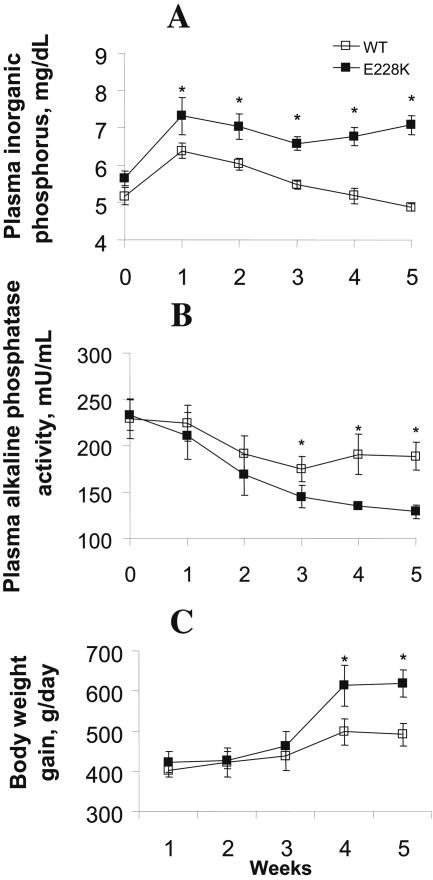
Because soybean meal is the major source of phytate in animal diets, we used it to compare the hydrolysis efficacies of the native phytate substrate by mutants E228K and TK10 with that of the WT. Given the same activity as determined at pH 5.5 using sodium phytate as the substrate, mutant E228K released 313, 238, and 53% more (P < 0.05) inorganic phosphorus from soy phytate at pH 2.5, 3.5, and 5.5, respectively, than the WT (Fig. 3). Meanwhile, the TK10 mutant showed an improved (P < 0.05) efficiency at pH 2.5 and 3.5 but decreased efficiency at pH 5.5 compared to the WT. When the WT and E228K enzymes were added to the low-phosphorus corn-soy basal diet for weanling pigs at 250 U kg−1 and fed to these pigs for 5 weeks, the mutant (E228K) phytase resulted in 13% higher (451 versus 509 g; P < 0.05) average daily body weight gain and 15 to 30% higher (P < 0.05) plasma inorganic-phosphorus concentration at weeks 2 to 5 than the WT enzyme (Fig. 4). Pigs fed E228K mutant phytase also had lower (P < 0.05) plasma alkaline phosphatase activities than those fed the WT at weeks 3 to 5, indicating less bone phosphorus resorption.
FIG. 3.
Efficacies of phytate-phosphorus hydrolysis in soybean meal by the single mutant E228K and the triple mutant E228K-K300R-K301E at pH 2.5 (0.2 M glycine-HCl), 3.5 (0.2 M glycine-HCl), and 5.5 (0.2 M citrate) at 37°C compared with that of the WT. The hydrolysis rates were calculated as the percentage of the WT phytase at pH 5.5, and the values are means ± standard errors (n = 3). An asterisk indicates a difference (P < 0.05) between pH 5.5 and other pH values within each enzyme. Different letters indicate differences (P < 0.05) between the WT and the mutants within each pH point.
FIG. 4.
Effects of supplemental E228K and WT PhyA phytases at 250 U kg−1 of a low-phosphorus corn-soybean meal diet for weanling pigs on plasma inorganic-phosphorus concentrations (A), plasma alkaline phosphatase activity (B), and body weight gain (C). The values are means ± standard errors (n = 8 for each group). An asterisk indicates a difference (P < 0.05) between treatment groups.
Kinetics characterization and hydrolysis profiles of the mutant phytase.
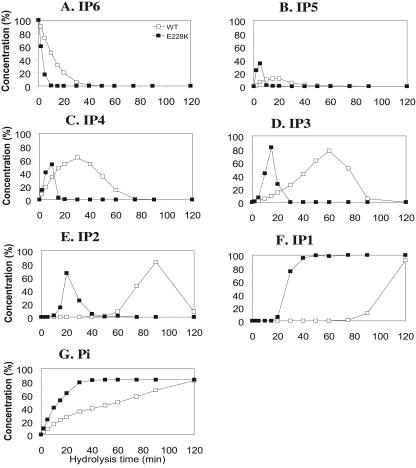
To elucidate the enzymatic basis for the improved efficacy of E228K over the WT in soy phytate hydrolysis and animal feed, we determined their kinetic parameters, Vmax and Km, for the hydrolysis of sodium phytate at pH 5.5 and 3.5 (Table 2). Compared to the WT, the Km values of E228K for sodium phytate were 79 and 30% lower (P < 0.05) at pH 5.5 and 3.5, respectively. This resulted in 70 and 160% greater overall catalytic efficiency (kcat/Km) at these two pH levels for E228K phytase. The HPLC analysis of the phytate hydrolysis profile at pH 3.5 (Fig. 5) had indicated that, although both the WT and E228K hydrolyzed sodium phytate to inositol 2-monophosphate and inorganic phosphate as the final products, the process took 120 min for the WT phytase but only 40 min for the mutant enzyme (Fig. 5). A delayed or much slower hydrolysis of inositol tetraphosphate and inositol triphosphate in the WT compared with E228K was particularly evident. There were also improvements in phytate hydrolysis by the E228K mutant phytase at pH 5.5 over the WT (data not shown). In addition, the E228K mutant enzyme had higher catalytic activity to smaller and synthetic substrates, such as p-nitrophenyl phosphate, phospho(enol)pyruvate, and fructose-6-monophosphate than the WT phytase (data not shown).
TABLE 2.
Comparison of kinetics of the WT and E228K mutant PhyA phytasesa
| pH | Phytase | Km (μM) | Vmax (μmol m−1 mg−1) | kcat (m−1) | kcat/Km (m−1 μM−1) |
|---|---|---|---|---|---|
| 5.5 | WT | 177.3 ± 22.4 | 94.7 ± 5.4 | 7,385.3 ± 421.1 | 41.64 |
| E228K | 38.6 ± 13.2* | 34.5 ± 3.9* | 2,690.2 ± 304.1* | 69.71* | |
| 3.5 | WT | 321.9 ± 37.9 | 33.8 ± 5.4 | 2,638.4 ± 421.1 | 8.20 |
| E228K | 227.8 ± 18.5* | 62.1 ± 3.9* | 4,844.3 ± 304.1* | 21.27* |
Enzymatic reactions (n = 3) were conducted at 37°C in 0.2 M glycine-HCl buffer for pH 3.5 and in 0.2 M citrate buffer for pH 5.5 using various sodium phytate concentrations (0.1 to 10 Km) and 100 mU phytase (purified) per ml reaction mixture. An asterisk indicates a difference (P < 0.05) from the WT.
FIG. 5.
Time courses of phytate hydrolysis by the WT and mutant E228K PhyAs and the subsequent appearance and disappearance of the intermediate or end products from the hydrolysis. The values are expressed as the concentration relative to the possible complete (maximal) hydrolysis. The enzymes (2 U ml−1) were incubated with 1% sodium phytate in 0.2 M glycine-HCl buffer, pH 3.5, for the designated times, and the hydrolytic metabolites were analyzed by HPLC. (A) Inositol hexaphosphate (IP6); (B) inositol pentaphosphate (IP5); (C) inositol tetraphosphate (IP4); (D) inositol triphosphate (IP3); (E) inositol diphosphate (IP2); (F) inositol monophosphate (IP1); (G) inorganic phosphate (Pi).
DISCUSSION
The most striking success in this study was in proving the feasibility of shifting the pH profile of a practically important enzyme, phytase, to enhance its catalytic efficiency in the digestive tract of a major food-producing species (swine). Among the 21 mutants, E228K showed the greatest overall improvement. The single amino acid alteration resulted in shifting the optimal pH from 5.5 to 3.8 with a significant rise in the relative activity at pH 3.5 compared with the WT. More importantly, this mutant was more efficient than the WT in releasing phytate-phosphorus from plant feed in vitro or in the gastrointestinal tracts of pigs. Two possible mechanisms may be used to explain these desirable changes. First, the substitution of a positively charged lysine at the 228 residue might affect the pKa of the acid/base catalyst (D362) indirectly, resulting in a shift of the optimal pH. Theoretically, introducing positive charges near the acid/base catalyst lowers the pKa value and shifts the optimal pH to a more acidic range (24). Second, the substitution of a positively charged lysine for the negatively charged glutamic acid may create a more favorable environment for the binding of the highly negatively charged substrate, phytate, by electrostatic attraction. Altogether, the substrate binding affinity at the acidic pH range for this mutant was improved markedly, as confirmed by the reduced Km of phytate at pH 3.5. Conversely, replacing E228 with a neutral amino acid (E228Q) negated the benefit derived from inserting lysine in the same position. However, it was somewhat surprising that the mutant also had improved hydrolysis of soy phytate at pH 5.5 compared to the WT. At pH 5.5, E228 might be protonated and interact with phytate by an H bond only, while the positively charged 228K could bind phytate by both an H bond and electrostatic interactions. This increased substrate affinity was reflected in the reduction in the Km values. Because phytate concentrations in the soybean meal (0.39% or 0.35 mM in the solution) and the swine diet (0.27% or 0.49 mM in the stomach) were lower than the substrate concentrations of sodium phytate (1%; 9 mM) used in standard laboratory assays for phytase activity, a lower Km could help the enzyme function under those conditions. Although the average pH in the stomachs of pigs is around 3.5 (42), part of newly ingested digesta may remain at somewhat higher pH levels before fully mixing with gastric fluids (29). Thus, there is an extra benefit for E228K in having a relatively strong catalytic efficiency at pH 5.5, along with the other favorable changes. Apparently, the greater efficacy of E228K in animal feed was attributable to the matching of the optimal pH to the stomach conditions, the improved substrate affinity and hydrolysis, and a net gain instead of a loss of efficiency in hydrolyzing soy phytate at pH 5.5 when the overall pH profile was shifted to the more acidic side. In addition, the higher specific activity of E228K than the WT PhyA in glycine-HCl buffer between pH 3.0 and 4.0 would allow the variant to perform better hydrolysis of phytate in the stomach, where the pH is around 3.5 and the gastric acid is predominately HCl (42).
Notably, E228K had better substrate specificity for low-molecular-weight phosphatase substrates, such as inositol di-, tri, and tetraphosphates, than the WT. Although without a structural model of the PhyA substrate (10) it is difficult for us to explain why the mutation in E228K resulted in a particular ability to hydrolyze these intermediate metabolites, the shorter time (40 versus 120 min) needed for hydrolysis of phytate to inositol monophosphate and inorganic phosphate by E228K than by the WT certainly helped the release of phytate in the digesta, which had limited transition time in the stomach. It is remarkable that the mutation in E228K made it not only a better phytase, but also a stronger acid phosphatase for several substrates. However, this observation does not support the notion (11) that the catalytic function of PhyA phytase depends on the extent of phosphorylation of myo-inositol.
Our second important finding deals with altering of pH versus the activity profile by mutating key residues in the substrate-binding domain. Previously, we showed that the replacement of negatively charged amino acids at the 300th amino acid residue (300E-300D) resulted in the optimal pH shift from 2.5 to 3 or 3.5 (22). Likewise, replacement of Q50L, K91E, or E262H in the present study abolished the pH 2.5 optimum. The residues K94 and E228 may also be involved in the activity at pH 2.5, as the replacement of K94E and E228K caused a shift of the activity peak at pH 2.5 to 2.0 or 3. Meanwhile, K301, the most conserved amino acid in the known phytase proteins (22), seems to be important for the catalytic activity and the substrate binding affinity at pH 5.5. The replacement of lysine with glutamic acid resulted in negative changes in both of the parameters (data not shown), probably due to the undesirable electrostatic repulsion between the enzyme and the substrate and between 301E and 362D (acid/base catalyst) (data not shown). Mutations of Q50L, Q50P, and D262H narrowed the pH optimum at 5 to 5.5 to a single point of pH 5 or 5.5. As mentioned above, the replacement of E228K led to a shift of the pH optimum to 3.8 and greater activity at pH 3.5. However, that change could be affected by combined mutations at residue 300: a positively charged residue 300R rendered the same pH profile as E228K, and a negatively charged residue 300D resulted in a single optimal pH at pH 5.5. When three single mutations, K91A, E228Q, and K300E, were combined, their interaction resulted in a single optimal pH at 4.5 to 5, although each single mutant had no or little effect on the pH profile. Accumulation of negative charges in the substrate binding site, such as K94E, K300E, and K301E, caused a dramatic activity decrease at pH 5.5, and the activity drop seemed to be proportional to the number of negative charges. In summary, the negatively charged amino acids in the substrate binding site had a negative effect on the catalytic efficiency at a pH around 5.5, probably due to undesirable electrostatic repulsion between the enzyme and the substrate. This supports the hypothesis of Kostrewa et al. (11) about the effect of the charge distribution within the active site on the pH profiles of A. niger acid phosphatase and phytase. However, the effects of charges at low pH depend on their location and interaction with neighboring amino acids, because those negatively charged amino acids are protonated and have no net charge (6). In addition, the local environment of the charged amino acids also affects the pKa of the acid/base catalyst, resulting in a shift in the optimal pH. The residues (E228, K300, and K301) reside close to the acid/base catalyst (D362); therefore, they could affect its pKa and the pH profile of the enzyme (23).
The triple mutant E228K-K300R-K301E showed a very impressive pH profile shift and more efficient hydrolysis of soy phytate at pH 2.5 and 3.5 than the WT. However, its specific activity was lower than that of the WT at all pH levels except pH 3.5. Thus, the catalytic improvement at pH 2.5 or 3.5 was associated with a much-reduced efficiency at pH 5.5. There was no net gain, but an actual loss of overall efficiency. This scenario teaches us the need for both in vivo and in vitro functional assays to evaluate genetically modified or engineered enzymes.
In summary, our research has clearly demonstrated that rational protein engineering can be applied to improve the pH-activity profile of phytase for optimal function in the gastrointestinal tracts of animals. The desirable shift in the optimal pH and the pH profile, along with an improved substrate affinity, in the overall best mutant was produced by the substitution of a positively charged lysine for the negatively charged glutamic acid at the 228th residue (E228K). Development of this phytase variant will enhance the field application of phytase for improving utilization of phytate-phosphorus in animal feeds, reducing the need for inorganic-phosphorus supplementation and phosphorus and preserving the nonrenewable inorganic-phosphorus deposits. Because of the great similarities between human and pig digestive systems and nutrient metabolisms (20), our results may be able to assist in the remediation of phytate-associated mineral deficiency in humans (27).
Acknowledgments
We thank Laurence Heller and Jeremy Weaver for technical assistance.
This project was developed in part under the auspices of the Cornell University Center for Biotechnology.
REFERENCES
- 1.Abelson, P. H. 1999. A potential phosphate crisis. Science 283:2015. [DOI] [PubMed] [Google Scholar]
- 2.Augspurger, N. R., D. M. Webel, X. G. Lei, and D. H. Baker. 2003. Efficacy of an E. coli phytase expressed in yeast for releasing phytate-bound phosphorus in young chicks and pigs. J. Anim. Sci. 81:474-483. [DOI] [PubMed] [Google Scholar]
- 3.Bisswanger, H. 2002. Enzyme kinetics, principles and methods, p. 51-74. Willey-VCH, Weinheim, Germany.
- 4.Cottrell, T. J., L. J. Harris, T. Tanaka, and R. Y. Yada. 1995. The sole lysine residue in porcine pepsin works as a key residue for catalysis and conformational flexibility. J. Biol. Chem. 270:19974-19978. [DOI] [PubMed] [Google Scholar]
- 5.Fang, T. Y., and C. Ford. 1998. Protein engineering of Aspergillus awamori glucoamylase to increase its pH optimum. Protein Eng. 11:383-388. [DOI] [PubMed] [Google Scholar]
- 6.Fushinobu, S., K. Ito, M. Konno, T. Wakagi, and H. Matsuzawa. 1998. Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH. Protein Eng. 11:1121-1128. [DOI] [PubMed] [Google Scholar]
- 7.Golovan, S. P., R. G. Meidinger, A. Ajakaiye, M. Cottrill, M. Z. Wiederkehr, D. J. Barney, C. Plante, J. W. Pollard, M. Z. Fan, M. A. Hayes, J. Laursen, J. P. Hjorth, R. R. Hacker, J. P. Phillips, and C. W. Forsberg. 2001. Pigs expressing salivary phytase produce low-phosphorous manure. Nat. Biotechnol. 19:741-745. [DOI] [PubMed] [Google Scholar]
- 8.Han, Y. M., D. B. Wilson, and X. G. Lei. 1999. Expression of an Aspergillus niger phytase gene (phyA) in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 65:1915-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi, M. D., G. Sidhu, I. Pot, G. D. Brayer, S. G. Withers, and L. P. McIntosh. 2000. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 299:255-279. [DOI] [PubMed] [Google Scholar]
- 10.Kostrewa, D., F. Grüninger-Leitch, A. D'Arcy, C. Broger, D. Mitchell, and A. P. G. M. van Loon. 1997. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat. Struct. Biol. 4:185-190. [DOI] [PubMed] [Google Scholar]
- 11.Kostrewa, D., M. Wyss, A. D'Arcy, and A. P. G. M. van Loon. 1999. Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 Å resolution. J. Mol. Biol. 288:965-974. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lei, X. G., P. K. Ku, E. R. Miller, and M. T. Yokoyama. 1993. Supplementing corn-soybean meal diets with microbial phytase linearly improves phytate phosphorus utilization by weanling pigs. J. Anim. Sci. 71:3359-3367. [DOI] [PubMed] [Google Scholar]
- 14.Lei, X. G., and C. H. Stahl. 2001. Biotechnological development of effective phytases for mineral nutrition and environmental protection. Appl. Microbiol. Biotechnol. 57:474-481. [DOI] [PubMed] [Google Scholar]
- 15.Li, C., E. Hegeman, R. W. Hanlon, G. H. Lacy, M. D. Denbow, and E. A. Grabau. 1997. Secretion of active recombinant phytase from soybean cell-suspension cultures. Plant Physiol. 114:1103-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, D., S. Golovan, C. W. Forsberg, and J. Jia. 2000. Crystal structure of Escherichia coli phytase and its complex with phytate. Nat. Struct. Biol. 72:108-113. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosebrough, A. L. Farrr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Maga, J. A. 1982. Phytate: its chemistry, occurrence, food interactions, nutritional significance, and methods of analysis. J. Agric. Food Chem. 30:1-9. [Google Scholar]
- 19.Mantafounis, D., and J. Pitts. 1990. Protein engineering of chymosin: modification of the optimum pH of enzyme catalysis. Protein Eng. 3:605-609. [DOI] [PubMed] [Google Scholar]
- 20.Miller, E. R., and D. E. Ullrey. 1987. The pig as a model for human nutrition. Annu. Rev. Nutr. 7:361-382. [DOI] [PubMed] [Google Scholar]
- 21.Mullaney, E. J., C. B. Daly, and A. H. J. Ullah. 2000. Advances in phytase research. Adv. Appl. Microbiol. 47:157-199. [DOI] [PubMed] [Google Scholar]
- 22.Mullaney, E. J., C. B. Daly, T. Kim, J. M. Porres, X. G. Lei, K. Sethumadhavan, and A. H. J. Ullah. 2002. Site-directed mutagenesis of Aspergillus niger NRRL 3135 phytase at residue 300 to enhance catalysis at pH 4.0. Biochem. Biophys. Res. Commun. 277:1016-1020. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, J. E., T. V. Borchert, and G. Vriend. 2001. The determinants of α-amylase pH-activity profiles. Protein Eng. 14:505-512. [DOI] [PubMed] [Google Scholar]
- 24.Ostanin, K., E. H. Harms, P. E. Stevis, R. Kuciel, M. M. Zhou, and R. L. van Etten. 1992. Overexpression, site-directed mutagenesis, and mechanism of Escherichia coli acid phosphatase. J. Biol. Chem. 267:22830-22836. [PubMed] [Google Scholar]
- 25.Ostanin, K., and R. L. van Etten. 1993. Asp (304) of Escherichia coli acid phosphatase is involved in leaving group protonation. J. Biol. Chem. 268:20778-20784. [PubMed] [Google Scholar]
- 26.Pallauf, J., and G. Rimbach. 1997. Nutritional significance of phytic acid and phytase. Arch. Anim. Nutr. 50:301-319. [DOI] [PubMed] [Google Scholar]
- 27.Porres, J. M., P. Etcheverry, D. D. Miller, and X. G. Lei. 2001. Phytase and citric acid supplementation in whole wheat bread improves phytate-phosphorus release and iron dialyzability. J. Food Sci. 66:614-619. [Google Scholar]
- 28.Reddy, N. R., S. K. Sathe, and D. K. Salukhe. 1982. Phytates in legumes and cereals. Adv. Food Res. 28:1-92. [DOI] [PubMed] [Google Scholar]
- 29.Rice, J. P., R. S. Pleasant, and J. S. Radcliffe. 2002. The effect of citric acid, phytase, and their interaction on gastric pH, and Ca, P, and dry matter digestibilities, p. 36-42. Purdue University Swine Research Report. Purdue University, West Lafayette, Ind.
- 30.Rodriguez, E., E. J. Mullaney, and X. G. Lei. 2000. Expression of the Aspergillus fumigatus phytase gene in Pichia pastoris and characterization of the recombinant enzyme. Biochem. Biophys. Res. Commun. 268:373-378. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, E., Z. A. Wood, P. A. Karplus, and X. G. Lei. 2000. Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch. Biochem. Biophys. 382:105-112. [DOI] [PubMed] [Google Scholar]
- 32.Russell, A. J., P. G. Thomas, and A. R. Fersht. 1987. Electrostatic effects on modification of charged groups in the active site cleft of subtilisin by protein engineering. J. Mol. Biol. 193:803-813. [DOI] [PubMed] [Google Scholar]
- 33.Spencer, J. D., G. L. Allee, and T. E. Sauber. 2000. Phosphorus bioavailability and digestibility of normal and genetically modified low-phytate corn for pigs. J. Anim. Sci. 78:675-681. [DOI] [PubMed] [Google Scholar]
- 34.Stahl, C. H., K. R. Roneker, W. G. Pond, and X. G. Lei. 2004. Effects of combining three fungal phytases with a bacterial phytase on plasma phosphorus status of weanling pigs fed a corn-soy diet. J. Anim. Sci. 82:1725-1731. [DOI] [PubMed] [Google Scholar]
- 35.Tomschy, A., M. Tessier, M. Wyss, R. Brugger, C. Broger, L. Schnoebelen, A. P. G. M. van Loon, and L. Pasamontes. 2000. Optimization of the catalytic properties of Aspergillus fumigatus phytase based on the three-dimensional structure. Protein Sci. 9:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomschy, A., R. Brugger, M. Lehmann, A. Svendsen, K. Vogel, D. Kostrewa, S. F. Lessen, D. Burger, A. Kronenberger, A. P. G. M. van Loon, L. Pasamontes, and M. Wyss. 2002. Engineering of phytase for improved activity at low pH. Appl. Environ. Microbiol. 68:1907-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turunen, O., M. Vuorio, F. Fenel, and M. Leisola. 2002. Engineering of multiple arginines into the Ser/Thr surface of Trichoderma reesei endo-1,4-β-xylanase II increases the thermotolerance and shifts the pH optimum towards alkaline pH. Protein Eng. 15:141-145. [DOI] [PubMed] [Google Scholar]
- 38.Ullah, A. H. J., and D. M. Gibson. 1987. Extracellular phytase (E. C. 3. 1. 3. 8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep. Biochem. 17:63-91. [DOI] [PubMed] [Google Scholar]
- 39.Ullah, A. H. J., and B. Q. Phillipy. 1994. Substrate selectivity in Aspergillus ficuum phytase and acid phosphatases using myo-inositol phosphates. J. Agric. Food Chem. 42:423-425. [Google Scholar]
- 40.Ullah, A. H. J., K. Sethumadhavan, E. J. Mullaney, T. Ziegelhoffer, and S. Austin-Phillips. 2002. Cloned and expressed fungal phyA gene in alfalfa produces a stable phytase. Biochem. Biophys. Res. Commun. 290:1343-1348. [DOI] [PubMed] [Google Scholar]
- 41.Wodzinski, R. J., and A. H. J. Ullah. 1996. Phytase. Adv. Appl. Microbiol. 42:263-302. [DOI] [PubMed] [Google Scholar]
- 42.Yi, Z., and E. T. Kornegay. 1996. Sites of phytase activity in the gastrointestinal tract of young pigs. Anim. Feed. Sci. Technol. 61:361-368. [Google Scholar]