Abstract
A novel bacterial strain that was isolated from an Italian soil and was designated SOSP1-21T forms branched mycelia in solid and liquid media and has a filamentous morphology similar to that of some genera belonging to the Actinobacteria. Electron microscopy showed that this organism has a grape-like appearance, resulting from interlacing of spores originating from sporophoric hyphae. Ten strains that are morphologically related to SOSP1-21T were recovered from soil. Phylogenetic analyses of 16S rRNA gene segments confirmed the relatedness of these strains to SOSP1-21T and indicated that the newly isolated strains form separate clades in a deeply branching lineage. The closest matches for the 16S rRNA sequences of all the strains (around 79% identity) were matches with representatives of the Chloroflexi, although the affiliation with this division was not supported by high bootstrap values. The strains are mesophilic aerobic heterotrophs and are also capable of growing under microaerophilic conditions. They all stain gram positive. Strain SOSP1-21T contains ornithine, alanine, glutamic acid, serine, and glycine as the peptidoglycan amino acids. In addition, an unusual level of C16:1 2OH (30%) was found in the cellular fatty acids. The G+C content of SOSP1-21T genomic DNA is 53.9%, and MK-9(H2) was the only menaquinone detected. All these data suggest that SOSP1-21T and the related strains may constitute a new division of filamentous, spore-forming, gram-positive bacteria. We propose the name Ktedobacter racemifer gen. nov., sp. nov. for strain SOSP1-21T.
Our understanding of the diversity and complexity of bacteria in natural environments has dramatically changed in the past two decades. When comparative analysis of 16SrRNA genes was first introduced, Woese and colleagues delineated 11 major lineages (45). Eighteen years later, about 50 bacterial phyla are recognized, and about one-half of them do not contain cultivated representatives yet (19, 36). In most cases, the delineation of a candidate phylum through analysis of 16S rRNA sequences has preceded the successful isolation of cultivated representatives. It has long been recognized that culture-dependent sampling of natural environments, performed on a limited scale and under a limited number of conditions, leads to isolation of bacteria that perform best under laboratory conditions (19). However, a similar bias might also exist in PCR-based culture-independent approaches, in which the genes that are most abundant and amplifiable might surface at high rates (22). This view is corroborated by analyzing 16S rRNA databases through rarefaction curves (40).
Soil is believed to be one of the most complex environments for microbial life (6, 7, 11, 12, 14), containing (per gram) about 109 bacterial cells and an estimated ∼104 distinct species. Novel isolation strategies, based on expanding the number of cultivation variables, are contributing to our increasing knowledge of cultivated bacteria from soil (6, 8, 23-25, 37, 43, 47). In particular, different laboratories have shown that when exhaustive plating conditions and long incubation times are employed, 4 to 7% of the total microbial community can be recovered (8, 24, 37, 43).
We have been interested in the biotechnological potential of the uncultivated portion of bacteria, especially strains belonging to the class Actinobacteria, because we assume that truly novel strains could yield structurally novel compounds. In particular, we are using an approach in which morphologically unusual strains are evaluated for phylogenetic novelty by 16S rRNA gene analysis (10). This approach has led to recent descriptions of novel Actinobacteria genera (1, 4). During this program, we identified filamentous bacterial isolates that were initially mistakenly identified as actinomycetes due to morphological similarities. Here, we describe these novel isolates, which appear to belong to a new bacterial phylum.
MATERIALS AND METHODS
Strain isolation.
Soil samples were obtained from an internal collection of samples from natural sources, where they are maintained desiccated in sealed plastic containers at room temperature. Before use, soil aliquots were dried in a vacuum oven for 7 days at 30°C and finely ground with a mortar. Using a sterile foam tip, a few milligrams of soil was directly deposited on HSA5 plates, as described by Busti et al. (1). After 8 weeks of incubation at 28°C, colonies were individually transferred to ISP3 agar (42), acidified to pH 5.5 to 6.0 with HCl, and incubated for 3 to 4 weeks at 28°C. The strains described in this paper are listed in Table 1. Cells from ISP3 agar were maintained in nutrient broth containing glycerol (3 g/liter beef extract, 5 g/liter peptone in 20% glycerol) at −80°C.
TABLE 1.
Bacterial strains and 16S rRNA gene sequence identities
| Strain | Sourcea | Accession no.b | Described species (accession no., % identity)c | 16S rRNA clone accession no. (% identity)c |
|---|---|---|---|---|
| SOSP1-0 | 11943 | AM180153 | R. castenholzii (AB041226, 77.6) | AY425790 (87.2) |
| SOSP1-1 | 10170 | AM180154 | C. aerophila (AB067647, 78.7) | AY425790 (88.1) |
| SOSP1-9 | 10170 | AM180155 | C. aerophila (AB067647, 78.6) | AY425790 (89.4) |
| SOSP1-21 | 11943 | AM180156 | C. aerophila (AB067647, 79.3) | AY425790 (88.6) |
| SOSP1-30 | 11943 | AM180157 | R. castenholzii (AB041226, 77.3) | AY425790 (87.3) |
| SOSP1-52 | 5646 | AM180158 | R. castenholzii (AB041226, 77.5) | AY425790 (87.1) |
| SOSP1-63 | 9572 | AM180159 | S. thermophilus (AJ420142, 78.9) | AY425785 (87.5) |
| SOSP1-79 | 11671 | AM180160 | C. aerophila (AB067647, 78.8) | AY425786 (90.1) |
| SOSP1-85 | 11943 | AM180161 | R. castenholzii (AB041226, 77.3) | AJ536876 (87.1) |
| SOSP1-142 | 11943 | AM180162 | C. aerophila (AB067647, 79.3) | AY425790 (88.6) |
| SOSP1-165 | 12095 | AM180163 | C. aerophila (AB067647, 78.5) | AY425790 (87.1) |
The numbers indicate the following soils: 5646, soil collected from a pine woods in an unknown location in Spain; 9572, soil collected from a solfatara in Pozzuoli, Italy; 10170, soil collected from an ant house in an unknown location in Honduras; 11671, soil collected from a cereal field in an unknown location in Egypt; 11943, soil collected from a black locust woods in Gerenzano, Italy; and 12095, soil collected from an uncultivated field in Bardello, Italy. The pHs, as measured after desiccation, were 6.5, 4.5, 6.2, 6.1, 4.2, and 5.6 for the 5646, 9572, 10170, 11671, 11943, and 12095 soils, respectively.
Accession numbers for the 16S rRNA gene sequences of the strains.
Closest relatives of the 16S rRNA sequences (described bacterial species and environmental clones). Abbreviations: C., Caldilinea; R., Roseiflexus; S., Sphaerobacter.
Cultural and physiological characterization.
Submerged growth was obtained in 500- and 50-ml baffled Erlenmeyer flasks containing 100 and 20 ml, respectively, of ATSB medium (1) by incubation for 5 to 8 days at 28°C on a rotary shaker at 200 rpm. Cells were collected, washed three times with sterile water, and used as inocula for cultural and physiological characterization or were freeze-dried for chemotaxonomic analyses.
The pH range for SOSP1-21T growth was determined on ISP2 medium (42); the pH was adjusted to the desired values with HCl or NaOH after autoclaving. The temperature range for growth was determined using acidified (pH 5.5 to 6.0) ISP2 medium. NaCl tolerance and lysozyme tolerance were also determined with the same medium by adding appropriate amounts of filter-sterilized stock solutions. The ability to reduce nitrates to nitrites was evaluated after 3 and 7 days of incubation in ATSB medium supplemented with 2 g liter−1 KNO3, using Bacto Nitrite test strips (Difco) and the procedures recommended by the manufacturer. Catalase production was evaluated qualitatively by determining the appearance of bubbling after a few drops of freshly prepared 3% hydrogen peroxide were added to an ISP3 medium culture. Gelatin liquefaction was evaluated as described by Gottlieb (15). Hydrolysis of keratin, casein (skim milk powder), chitin, starch, xylan, or cellulose was evaluated on antibiotic medium 1 (AM1) agar (Penassay seed agar; Difco) adjusted to pH 5.5 to 6.0 with HCl and supplemented with each substrate at a concentration of 0.5 to 1% (wt/vol); the presence of a transparent halo around the growth indicated that there was hydrolysis. Resistance to selected antibiotics was evaluated on AM1 agar. H2S formation was detected 2 days after sterile lead acetate filter paper strips (Fluka) were inserted into the necks of culture tubes containing acidified ISP6 medium (42). Aerobiosis-related properties were analyzed on acidified ISP2 medium under an aerobic, microaerophilic (gas generating kit for Campylobacter; catalog no. BR056A; Oxoid), or anaerobic (GasPak Pouch; catalog no. 260651; BBL) atmosphere. Unless indicated otherwise, physiological characteristics were evaluated after 3 weeks of incubation at 28°C.
Microscopy.
The morphology of an aerial mass was examined directly on HSA5 plates by using a light microscope equipped with a ×40 long-working-distance objective (model ULWD-CDPlan; Olympus) and with a 3CCD camera (Sony). Spore motility was examined as previously described (1). For field emission scanning electron microscopy (FESEM), strains grown on agar plates were fixed with a solution containing 5% formaldehyde and 2% glutaraldehyde in cacodylate buffer (0.1 M cacodylate, 0.01 M CaCl2, 0.01 M MgCl2, 0.09 M sucrose; pH 6.9) for 3 h on ice and washed with cacodylate buffer. Samples were then dehydrated with a graded acetone series (10, 30, 50, 70, 90, and 100% acetone) on ice for 30 min for each step. The samples in 100% acetone were allowed to reach room temperature before another change of 100% acetone. Samples were then subjected to critical-point drying with liquid CO2 (CPD030; Balzers, Liechtenstein). The dried samples were covered with an approximately 10-nm-thick gold film by sputter coating (SCD040; Balzers Union, Liechtenstein) before examination with a Zeiss DSM 982 Gemini field emission scanning electron microscope using an Everhart Thornley SE detector and the in-lens detector at a 50:50 ratio at an acceleration voltage of 5 kV. Data were stored digitally on MO disks, and contrast and brightness were adjusted using Adobe Photoshop 7.0. The morphological description of strain SOSP1-21T below is based on colonies examined after 3 to 4 weeks of incubation at 28°C.
Chemical analyses.
Gram staining and acid-fast staining were performed with cells from 3-week-old ISP3 acid agar cultures, using standard protocols. For analysis of the amino acid composition and determination of the peptidoglycan structure we used a previously described method (39), as modified by Willems et al. (44). The molar ratio of peptidoglycan amino acids was determined by gas chromatography and gas chromatography/mass spectrometry as described by MacKenzie (29). The N-terminal amino acid of the peptidoglycan interpeptide bridge was determined by dinitrophenylation as described by Schleifer (38). Cellular fatty acid methyl esters were obtained as described previously (34). Identification and quantification of the fatty acid methyl esters were performed using the standard MIS library generation software (Microbial ID Inc.). Isoprenoid quinones and polar lipids were analyzed as described by Groth et al. (16). The DNA base composition was determined by reversed-phase high-performance liquid chromatography of nucleosides performed as described by Mesbah et al. (33).
16S rRNA gene sequencing and phylogenetic analysis.
Sequencing and comparative analysis of the nearly complete 16S rRNA gene were performed as described previously (35). Phylogenetic analyses were performed with programs of the PHYLIP package (13). For maximum likelihood analyses, a model assuming a gamma distribution of rates of evolution across sites was used. The coefficient of variation of the substitution rate and the transition/transversion ratio were empirically estimated by performing iterative analyses with the same data sets in order to find the values that maximized the likelihood. These values were also used in bootstrap analyses.
PCR analysis of secondary metabolism genes.
PCRs for amplification of gene segments encoding type I and II polyketide synthases were performed as described previously (2). The primers described by Carnio et al. (3) were used for amplification of nonribosomal peptide synthetase (NRPS) gene segments. Cloning and sequencing of PCR products were performed as described previously (2).
Nucleotide accession numbers.
The nucleotide sequences of the 16S rRNA genes and the NRPS gene segments have been deposited in the GenBank database under accession numbers AM180153 through AM180163 and AM180164 through AM180170, respectively.
RESULTS
Strain morphology.
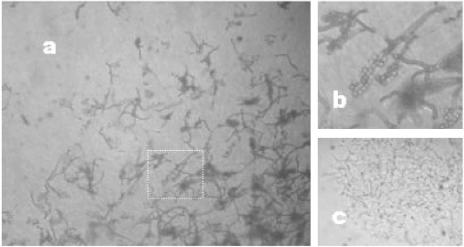
Strain SOSP1-21T was isolated from a soil sample collected in Gerenzano, Italy. It was initially mistakenly identified as an actinomycete because of its resemblance to Thermomonospora spp. (31). On solid medium, strain SOSP1-21T produces both vegetative and aerial mycelia (Fig. 1A). Moreover, the aerial hyphae, some of which have particularly large diameters, bear spherical elements, presumed to be spores (Fig. 1B). Filamentous growth also occurred in submerged cultures, which contained the branched mycelia typical of actinomycetes (Fig. 1C).
FIG. 1.
Light micrographs of strain SOSP1-21T. (a) Aerial morphology on an HSA5 plate after incubation for 4 weeks. Magnification, ×400. (b) Magnified (ca. ×2.5) image of the area in panel a indicated by the box, showing aerial hyphae and spherical spores. (c) Mycelial growth in a submerged culture after 3 days.
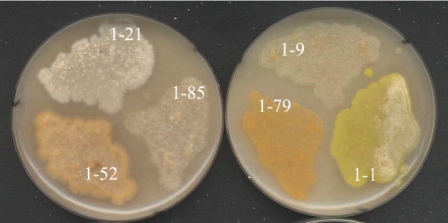
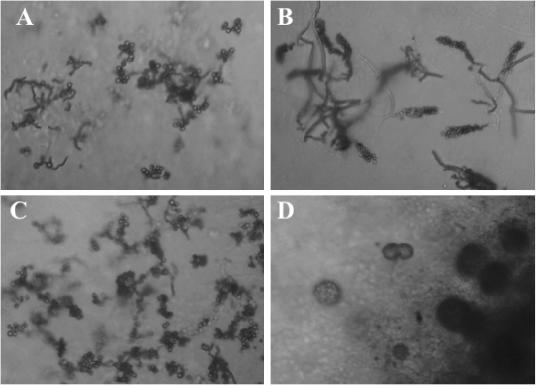
In subsequent isolation campaigns, several strains that resembled SOSP1-21T and were subsequently shown to be phylogenetically related to it (see below) were recovered from soil samples obtained from different locations (Table 1). These newly isolated strains had cream to yellow or orange pigmentation on acidic ISP3 agar (Fig. 2). Figure 3 shows details of the aerial structures. All strains produced rounded spores, although they were arranged differently on the aerial hyphae. Strain SOSP1-52 and SOSP1-85 spores formed clusters (Fig. 3A and B); SOSP1-1 spores looked like maize ears (Fig. 3C); and strain SOSP1-9 spores were enclosed in globular vesicles (possibly sporangia) (Fig. 3D).
FIG. 2.
Appearance of strains on acidic ISP3 agar after 3 weeks. 1-21, strain SOSP1-21T; 1-85, strain SOSP1-85; 1-52, strain SOSP1-52; 1-9, strain SOSP1-9; 1-79, strain SOSP1-79; 1-1, strain SOSP1-1.
FIG. 3.
Light micrographs (obtained with a ULWD objective) of strains SOSP1-52 (A), SOSP1-1 (B), SOSP1-85 (C), and SOSP1-9 (D) on HSA5 plates after incubation for 4 weeks. Magnification, ×400.
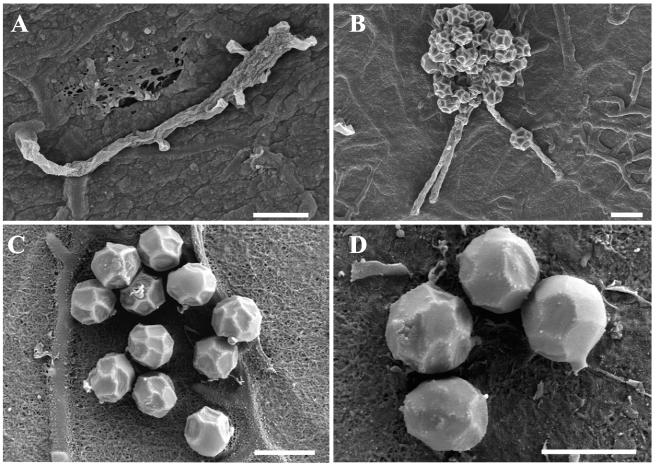
FESEM of strain SOSP1-21T (Fig. 4) confirmed the presence of vegetative and aerial hyphae. Consistent with the light microscopy observations (Fig. 1B), peduncles emerged from the terminal part of an aerial hypha (Fig. 4A), which could be interpreted as an initial stage of spore production or as residual sporophores after spore detachment. The diameters of hyphae were 0.5 to 0.7 μm (Fig. 4B), similar to the diameters of actinomycete hyphae. The interlacing spores originating from three or more sporophore hyphae formed a cluster, giving strain SOSP1-21T a grape-like appearance (Fig. 1B). Details of spores on an agar surface are shown in Fig. 4C and 4D. The spores were spherical and had a diameter of 1.6 to 1.8 μm. They appeared to have an internal scaffold, reminiscent of a football, covered by a sheet that we inferred was tight in young spores, giving them a smooth surface (Fig. 4D), but collapsed on the scaffold (Fig. 4C) as dehydration proceeded, giving the final corrugated appearance shown in Fig. 4B. It should be noted that the overall appearance of the SOSP1-21T spores was very similar to that of Thermoactinomyces vulgaris endospores (32). However, T. vulgaris does not produce the spore clusters observed with SOSP1-21T. In addition, T. vulgaris is a thermophilic, fast-growing bacterium (28), in contrast to SOSP1-21T (see below). Recently, the genus Thermoactinomyces, together with the newly described genera Laceyella, Seinonella, and Thermoflavimicrobium, was assigned to the family Bacillaceae (46).
FIG. 4.
FESEM of strain SOSP1-21T. Bars = 2 μm. See the text for details.
Phylogenetic analysis.
Nearly complete 16S rRNA gene sequences of the new isolates were determined. The highest levels of binary similarity with GenBank entries (87.1 to 90.1% identity) were the levels of similarity with environmental clones, while the levels of identity with cultured strains were less than 80% (Table 1). In particular, strain SOSP1-21T exhibited the highest level of identity (88.6%) with environmental clone 1959-6, which was recovered from a volcanic deposit in Hawaii (GenBank accession number AY425790). Among cultured strains, the closest relative (79.3%) was Caldilinea aerophila, a recently described member of Chloroflexi subphylum 1 (41). This phylum has subsequently been amended by inclusion of the class Thermomicrobia (21). For the other 10 strains, the levels of binary similarity ranged from 87.1 to 90.1% for environmental clones and from 77.3 to 79.3% for cultured strains (Table 1).
The sequences of the 11 strains in Table 1 were aligned with the sequences of representatives of the major bacterial lineages, and phylogenetic trees were constructed by using different methods. In all analyses (Fig. 5), the 11 sequences appeared to form two separate lineages, one consisting of only strains in Table 1 (seven strains, including SOSP1-21T) and the other consisting of the other four strains in Table 1 and clones from a Hawaiian volcanic deposit.
FIG. 5.
Maximum likelihood tree based on 1,202 aligned positions of the 16S rRNA gene. The tree was rooted using the 16S rRNA gene sequence from Methanococcus jannaschii (accession no. M59126) as the outgroup. The numbers at nodes are bootstrap values based on 100 replicated data sets; only values greater than 65 are shown. Scale bar = 10 inferred substitutions per 100 nucleotides.
As Fig. 5 shows, the newly isolated strains appear to cluster with the Chloroflexi, but the bootstrap values are too low to support a monophyletic origin. Since the highest levels of binary similarity were the levels of similarity with representatives of the Chloroflexi genera Caldilinea, Roseiflexus, and Sphaerobacter (Table 1), the 16S rRNA sequences of the strains were also aligned with representatives of all the subdivisions of the Chloroflexi (including both cultured and uncultured strains), and phylogenetic trees were constructed (Fig. 6). The 11 strains, together with sequences of environmental clones from the Hawaiian volcanic deposits, formed a cluster supported by a bootstrap value of 100%. The five described subdivisions of the Chloroflexi formed coherent clusters, which were clearly distinguished from the cluster comprising our strains. Similar results were obtained when treeing methods based on parsimony and on matrices of genetic distances were applied. The results show that the strains in Table 1 appear to be distinct lineages, which are referred to as clades GER1 through GER3. Clade GER1 consists of five strains (including SOSP1-21T) exhibiting >97.9% 16S rRNA gene identity, and clade GER2 comprises SOSP1-165 and SOSP1-1, which exhibit 98.3% 16S rRNA gene identity. The levels of pairwise identity between members of clade GER1 and members of clade GER2 are around 92%, suggesting that these two clades represent distinct families (or higher-rank taxa) with a monophyletic origin supported by a bootstrap value of 88%. The remaining four strains exhibit 86.6 to 90% identity to strains in either clade GER1 or clade GER2. Strains SOSP1-9 and SOSP1-79, which exhibit 96.5% 16S rRNA gene sequence identity, are clearly associated (clade GER3), but relatively low bootstrap values weakly support common ancestry with the other two isolates (SOSP1-63 and SOSP1-142) and with a group of environmental clones, including three sequences with >96.2% identity (clade HAW1).
FIG. 6.
Maximum likelihood tree based on 1,168 aligned positions of the 16S rRNA gene. The tree was rooted using the Aquifex pyrophilus 16S rRNA sequence (accession no. M83548) as the outgroup. The numbers at nodes are bootstrap values based on 100 replicated data sets; only values greater than 65 are shown. Scale bar = 10 inferred substitutions per 100 nucleotides. The numbers in parentheses indicate the subphylum-level groups proposed by Hugenholtz and Stackebrandt (21), as follows: 1, “Anaerolineae”; 2, “Dehalococcoidetes”; 3, Chloroflexi; 5, Thermomicrobia. There are no cultivated representatives for lineage 4. The clades of highly related sequences are clades GER1 to GER3 for isolates and clade HAW1 for environmental clones (see the text for details).
Overall, the levels of identity between our strains and members (either cultivated strains or environmental clones) of the five known Chloroflexi subdivisions range from 74.3 to 79.3%. These values are comparable to the values obtained for members of different Chloroflexi subdivisions and also to the values typically found in interdivision comparisons. In fact, Hugenholtz et al. (22) suggested a cutoff of 85% identity for 16S rRNA sequences for proposing a candidate phylum. Thus, phylogenetic data alone are not sufficient to decide whether the novel isolates represent a novel subdivision of the Chloroflexi or if they belong to a novel phylum.
Chemotaxonomic characteristics.
Strains SOSP1-1, SOSP1-9, SOSP1-21T, SOSP1-52, and SOSP1-85 were gram positive and acid fast negative. The G+C content of SOSP1-21T genomic DNA was 53.9 mol%. SOSP1-21T cells contained MK-9(H2), which was the only menaquinone. The polar lipids consisted of phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, and an unknown glycolipid. The peptidoglycan contained ornithine, alanine, glutamic acid, serine, and glycine at a molar ratio of approximately 0.7:1.8:1.0:0.8:1.9. Serine represented the N terminus of the interpeptide bridge. Although a detailed peptidoglycan structure could not be determined from these data, the presence of the characteristic peptide l-Ala-d-Glu in the partial peptidoglycan hydrolysates suggested the occurrence of the A-type of cross-linkage (39). The cellular fatty acid analysis revealed an unusual abundance of C16:1 2OH, which was the major component (30%) together with i-C17:0 (25%). All of the fatty acids detected are shown in Table 2.
TABLE 2.
Cellular fatty acids of strain SOSP1-21T
| Fatty acid | % |
|---|---|
| C16:1 2OH | 29.65 |
| i-C17:0 | 25.00 |
| i-C16:0 | 11.54 |
| ai-C17:0 | 9.61 |
| C16:010Me | 7.79 |
| C16:0 | 6.66 |
| i-C15:0 | 2.37 |
| C18:0 | 2.01 |
| C18:1 w9c | 1.21 |
| i-C16:1 | 1.00 |
| C15:0 3OH | 0.76 |
| i-C18:0 | 0.67 |
| C16:1 w7c | 0.54 |
| C17:010Me | 0.37 |
| ai-C15:0 | 0.29 |
| i-C18:1 | 0.27 |
| C15:0 | 0.26 |
Cultivation characteristics.
Strain SOSP1-21T formed solid colonies that were up to 3 to 4 mm in diameter on some agar media. It was able to grow well at 28 to 30°C on acidified IPS2, ISP3, and AM1 agar media, and the pigmentation ranged from cream to pinkish orange on all media; no diffusible pigments were evident in any medium, except that ISP3 medium occasionally turned pinkish. Occasionally, a thin, white aerial mycelium was produced on ISP3 medium. A characteristic of ISP3 medium-grown cultures was the production of a volatile compound(s) with a characteristic sweetish smell. As shown in Fig. 2, the other isolates produced pigmentation ranging from cream (SOSP1-9 and SOSP1-85) to orange (SOSP1-52 and SOSP1-79) to yellow with production of a whitish aerial mycelium (SOSP1-1). Strains SOSP1-63 and SOSP1-142 lacked aerial mycelium, and the substrate mycelium was cream to orange (not shown).
Strain SOSP1-21T was able to grow well at pH values between 4.8 and 6.8, and the apparent optimum pH was around 6. After growth, the final pH was always around 6. Scant growth was observed at pH 4.2 and 7.2, and there was no growth at pH 3.9 and 7.5. Good growth was observed at 22, 28, 33, and 37°C, and the optimal temperature was 28 to 33°C. Strain SOSP1-21T grew poorly at 17 and 40°C and not at all at 14 and 45°C. NaCl added at a concentration of 10 g per liter did not inhibit growth, but higher concentrations retarded (20 g/liter) or inhibited (30 g/liter) growth. Lysozyme (at a concentration of 100 μg/ml) had no effect on growth. All isolates grew well in the pH range from 5.5 to 6.5 and at temperatures between 28 and 33°C. For some strains, optimal growth also occurred at 37 and 42°C.
Strain SOSP1-21T was able to grow aerobically in different solid media. Growth did not occur on acidified ISP3 medium under anaerobic conditions, while a microaerophilic atmosphere allowed growth comparable to the growth under aerobic conditions. In liquid media, the best growth was obtained with agitation using baffled flasks, and reduced growth was observed under static incubations.
Strain SOSP1-21T could hydrolyze starch, casein, gelatin, and (to a lesser extent) keratin. No hydrolysis was observed with cellulose, xylan, or chitin. Strain SOSP1-21T was catalase positive and produced H2S but could not reduce nitrates. The behavior of strains SOSP1-1, SOSP1-9, SOSP1-52, and SOSP1-85 was identical to the behavior of SOSP1-21T except for a lack of keratin hydrolysis by all strains except SOSP1-1 and except for a lack of starch hydrolysis by SOSP1-1.
The antibiotic resistance profiles indicated that the five strains tested (SOSP1-9, SOSP1-21T, SOSP1-52, SOSP1-79, and SOSP1-85) were all sensitive to 5 μg/ml novobiocin or ramoplanin and to 20 mg/ml apramycin and the glycopeptide A40926. The strains in clade GER1 (SOSP1-21T, SOSP1-52, and SOSP1-85) were resistant to rifampin and thiostrepton (5 μg/ml each) and sensitive to nalidixic acid (5 μg/ml), apramycin, and kanamycin (20 mg/ml), while the other two strains (belonging to clade GER3) showed the opposite behavior. All strains belonging to clade GER1 and strain SOSP1-79 were resistant to 20 μg/ml rifampin, while SOSP1-9 was sensitive.
None of the strains in Table 1 was able to produce antimicrobial activities under the cultivation conditions employed (data not shown). However, all strains analyzed yielded a group of bands ranging from 0.5 to 0.8 kbp when they were analyzed with NRPS gene-specific primers. The PCR products of three strains were analyzed (seven clones each). In database searches 13 clones showed the highest matches with NRPS genes, and five clones resembled long-chain fatty acyl-coenzyme A ligases (both sequences belong to the acyl adenylate-forming enzyme superfamily [30]). The remaining three clones matched unrelated sequences. Overall, two distinct NRPS gene segments were recovered from strain SOSP1-21T; one NRPS gene segment was recovered from SOSP1-142, and four NRPS gene segments were recovered from SOSP1-30 (data not shown). Thus, it appears that these strains have the genetic potential for nonribosomal synthesis of peptides.
No band was observed with primers specific for type I or type II polyketide synthases. However, the primers were biased for high-G+C-content DNA.
DISCUSSION
General characteristics.
Strain SOSP1-21T is an aerobic, moderately acidophilic bacterium that is also able to grow under microaerophilic conditions but not anaerobically. On the basis of preliminary analyses, the other strains in Table 1 have similar physiological characteristics and are capable of transforming organic compounds, with a preference for protein substrates over polysaccharides. In addition, they were retrieved from different soils having pH values below 6.5 (Table 1). The Hawaiian volcanic deposit, from which SOSP1-21T-related sequences were retrieved, might also have an acidic pH (27). The method used for isolation of SOSP1-21T and related strains did not allow estimation of the abundance in soil. However, the relative ease of retrieval suggests that, as in many other instances (8, 24, 25, 37, 43), the combination of a slightly acidic nutrient-poor medium with long incubation times was critical for isolation of these organisms. At the moment, the role of SOSP1-21T and related strains in the soil is not known. They do not appear to produce antibiotic substances under the conditions that we tested; the NRPS gene-like sequences in their genomes may encode compounds for metal uptake, which are usually expressed under metal-limiting conditions (5). Perhaps the ability of these organisms to grow well under microaerophilic conditions may provide a competitive advantage.
Strain SOSP1-21T and the related strains have a unique mixture of features observed in other bacterial lineages. The mycelial growth is reminiscent of the growth of Actinomycetales, while the G+C content and the overall shapes of the spores are consistent with properties of the Bacillaceae. The strains analyzed are gram positive and, like gram-positive bacteria, are sensitive to high-molecular-weight antibiotics, such as ramoplanin. The high level of 2-hydroxy C16:1, the major component of cellular fatty acids, and the menaquinone profile are uncommon, while the amino acid composition of SOSP1-21T peptidoglycan occurs in other gram-positive strains.
Comparison of strain SOSP1-21T with Chloroflexi.
Are SOSP1-21T and the related strains phylogenetically related to Chloroflexi? The highest levels of binary similarity of the 16S rRNA sequences were the levels of similarity with representatives of this phylum (Table 1), but the association with Chloroflexi was not supported by high bootstrap values in phylogenetic trees. In addition, it was difficult to find similarities with characterized members of this phylum. The major morphological peculiarity of strain SOSP1-21T is the production of a branched mycelium. In contrast, species belonging to the genera Roseiflexus (18), Anaerolinea and Caldilinea (41), Oscillochloris (26), Chloroflexus (17), and Herpetosiphon (20), all described as filamentous, produce unbranched, multicellular filaments and not true mycelia. Moreover, none of them is described as a spore-forming organism. Other characteristics found in most representatives of the Chloroflexi are the temperature ranges for growth, as all organisms except Oscillochloris are thermophilic (optimal temperature, more than 50°C), and the pH ranges, as all organisms except Herpetosiphon grow at pHs higher than 6. All the strains that we describe here are gram positive, while most Chloroflexi strains are gram negative; the only exception is Sphaerobacter thermophilus, which has only recently been reclassified as a member of this phylum (21). The structure of the cell wall, however, clearly differentiates this organism from our strains, as S. thermophilus has the peptidoglycan type A3β (l-Orn←β-Ala) and contains the completely unsaturated menaquinone MK-8 (9).
In conclusion, morphological, physiological, and chemotaxonomic data suggest that the strains described here are not associated with the Chloroflexi. In addition, the fact that, despite a relatively low level of 16S rRNA gene similarity, strains in clades GER1 and GER3 have similar morphological features that clearly differentiate them from known Chloroflexi support the hypothesis that the strains in Table 1, together with the phylogenetically related environmental clones, constitute a new bacterial division of filamentous, spore-forming, gram-positive bacteria. However, we cannot exclude the possibility that the Chloroflexi is a highly heterogeneous phylum with respect to phylogenetic, morphological, and physiological characteristics.
The 11 strains represent at least five families (or higher-rank taxa), as deduced from 16S rRNA gene similarities. In addition, they are phylogenetically distant enough from other cultivated Chloroflexi to justify a proposal for a new lineage comprising SOSP1-21T and the other strains in Table 1. At this time, we do not propose a new bacterial division to accommodate our strains. Strain SOSP1-21T is phylogenetically distinct from previously characterized bacterial strains and represents a new genus and species, for which we propose the name Ktedobacter racemifer.
Description of Ktedobacteria classis nov.
Ktedobacteria (Kte.do.bac.te′ri.a. N.L. masc. n. Ktedobacter, type genus of the class; -ia, suffix denoting a class; N.L. fem. pl. n. Ktedobacteria, the Ktedobacter class).
On the basis of 16S rRNA gene sequence analyses, this group may represent one of the primary lineages in the phylum Chloroflexi. Although we are aware that further analyses might indicate that Ktedobacteria is not part of the phylum Chloroflexi, we do not propose a new phylum at this time. The class Ktedobacteria currently comprises only the order Ktedobacterales.
Description of Ktedobacterales ord. nov.
Ktedobacterales (Kte.do.bac.ter.a′les. N.L. masc. n. Ktedobacter, type genus of the order; -ales, suffix denoting an order; N.L. fem. pl. n. Ktedobacterales, the Ktedobacter order).
The description is the same as that for the genus Ktedobacter. The order contains the family Ktedobacteraceae.
Description of Ktedobacteraceae fam. nov.
Ktedobacteraceae (Kte.do.bac.ter.a′ce.ae. N.L. masc. n. Ktedobacter, type genus of the family; -aceae, suffix denoting a family; N.L. fem. pl. n. Ktedobacteraceae, the Ktedobacter family).
The description is the same as that for the genus Ktedobacter. The family contains the type genus Ktedobacter.
Description of Ktedobacter gen. nov.
Ktedobacter (Kte.do.bac′ter. Gr. n. ktedon, fiber; N.L. masc. n. bacter, rod, bacterium, prokaryote; N.L. masc. n. Ktedobacter, filamentous bacterium). The filamentous, spore-forming bacteria are gram positive. Strains grow as mesophilic aerobic heterotrophs and can also grow under microaerophilic conditions. They contain ornithine, alanine, glutamic acid, serine, and glycine as the peptidoglycan amino acids. C16:1 2OH is the major component of the cellular fatty acids, and MK-9(H2) is the major menaquinone. The G+C content of the genomic DNA of strain SOSP1-21, the type strain of the type species, Ktedobacter racemifer, is 53.9%.
Description of Ktedobacter racemifer sp. nov.
Ktedobacter racemifer (ra.ce′mi.pher. L. adj. masc. racemifer, carrying clusters of grapes). In addition to the properties given in the genus description, this species has the following characteristics.
Gram-positive, non-acid-fast, aerobic, heterotrophic organism that produces branched vegetative mycelium in solid and liquid cultures. It also produces aerial hyphae that can bear spherical spores that are 1.6 to 1.8 μm in diameter. Spores emerge singly on short sporophores, but the dense spores and the interlacing of different spore-forming hyphae result in clusters of spores. Spores are not motile. Colonies are solid with cream to pink or orange pigmentation. Grows at pH 4.2 to 7.2; the optimal pH is around 6, and there is no growth at pH 3.9 and 7.5. The temperatures tolerated range from 17 to 40°C, and the optimum temperatures range from 28 to 33°C; no growth occurs at 14°C and 45°C. Lysozyme at a concentration of 100 μg/ml and 10 g/liter NaCl do not inhibit growth, while 30 g/liter NaCl does inhibit growth. Gelatin, casein, keratin, and starch are hydrolyzed, while chitin, xylan, and cellulose are not hydrolyzed. Catalase positive. Grows well in a microaerophilic atmosphere. Produces H2S but does not reduce nitrates. The type strain is SOSP1-21 (= DSM 44963).
REFERENCES
- 1.Busti, E., L. Cavaletti, P. Monciardini, P. Schumann, M. Rohde, M. Sosio, and S. Donadio. Catenulispora acidiphila gen. nov., sp. nov., a novel mycelium-forming actinomycete and proposal of Catenulisporaceae fam. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 2.Busti, E., P. Monciardini, L. Cavaletti, R. Bamonte, A. Lazzarini, M. Sosio, and S. Donadio. 2006. Antibiotic producing ability by previously uncultured groups of actinomycetes. Microbiology 152:675-683. [DOI] [PubMed] [Google Scholar]
- 3.Carnio, M. C., T. Stachelhaus, K. P. Francis, and S. Scherer. 2001. Pyridinyl polythiazole class peptide antibiotic micrococcin P1, secreted by foodborne Staphylococcus equorum WS2733, is biosynthesized nonribosomally. Eur. J. Biochem. 268:6390-6400. [DOI] [PubMed] [Google Scholar]
- 4.Cavaletti, L., P. Monciardini, P. Schumann, M. Rohde, R. Bamonte, E. Busti, M. Sosio, and S. Donadio. Actinospica acidiphila gen. nov., sp. nov., and Actinospica robiniae gen. nov., sp. nov.; proposal for Actinospicaceae fam. nov. and Catenulisporinae subordo nov. in the order Actinomycetales. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 5.Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochemistry 6:601-611. [DOI] [PubMed] [Google Scholar]
- 6.Chin, K.-J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, K. E. R., S. J. Jospeh, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demharter, W., R. Heusel, J. Smida, and E. Stackebrandt. 1989. Sphaerobacter thermophilus gen. nov., sp. nov. A deeply rooting member of the actinomycetes subdivision isolated from thermophilically treated sewage sludge. Syst. Appl. Microbiol. 11:261-266. [Google Scholar]
- 10.Donadio, S., E. Busti, P. Monciardini, R. Bamonte, P. Mazza, M. Sosio, and L. Cavaletti. 2005. Sources of polyketides and nonribosomal peptides, p. 19-41. In W. Wohlleben, T. Spelling and B. Müller-Tiemann (ed.), Biocombinatorial approaches for drug finding, vol. 51. The Ernst Schering Research Foundation, Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 11.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, R. J., P. Morgan, A. J. Weightman, and J. C. Fry. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 69:3223-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package), version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- 14.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb, D. 1961. An evaluation of criteria and procedures used in the description and characterization of the streptomycetes. Appl. Microbiol. 9:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth, I, P. Schumann, F. A. Rajney, K. Martin, B. Schuetze, and K. Augsten. 1997. Bogoriella caseilytica gen. nov., sp. nov., a new alkaliphilic actinomycete from a soda lake in Africa. Int. J. Syst. Bacteriol. 47:788-794. [DOI] [PubMed] [Google Scholar]
- 17.Hanada, S., A. Hiraishi, K. Shimada, and K. Matsuura. 1995. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int. J. Syst. Bacteriol. 45:676-681. [DOI] [PubMed] [Google Scholar]
- 18.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52:187-193. [DOI] [PubMed] [Google Scholar]
- 19.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt, J. G., and R. A. Lewin. 1968. Herpetosiphon aurantiacus gen. et sp. n., a new filamentous gliding organism. J. Bacteriol. 95:2407-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugenholtz, P., and E. Stackebrandt. 2004. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description). Int. J. Syst. Evol. Microbiol. 54:2049-2051. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen, P. H., A. Schuhmann, E. Mörschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keppen, O. I., T. P. Tourova, B. B. Kuznetsov, R. N. Ivanovsky, and V. M. Gorlenko. 2000. Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria, and emended description of Oscillochloris and Oscillochloris trichoides in comparison with further new isolates. Int. J. Syst. Evol. Microbiol. 50:1529-1537. [DOI] [PubMed] [Google Scholar]
- 27.King, G. M. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69:4067-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacey, J. 1989. Thermoactinomycetes, p. 2573-2585. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 29.MacKenzie, S. L. 1984. Amino acids and peptides, p. 157-204. In G. Odham, L. Larsson, and P. Mardh (ed.), Gas chromatography/mass spectrometry applications in microbiology. Plenum Publishing Corporation, New York, N.Y.
- 30.Marahiel, M. 1997. Protein templates for the biosynthesis of peptide antibiotics. Chem. Biol. 4:561-567. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy, A. 1989. Thermomonospora and related genera, p. 2552-2561. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 32.McVittie, A., H. Wildermuth, and D. A. Hopwood. 1972. Fine structure and surface topography of endospores of Thermoactinomyces vulgaris. J. Gen. Microbiol. 71:367-381. [Google Scholar]
- 33.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 34.Miller, L. T. 1982. Single derivatization method for routine analysis of bacterial whole-cell, fatty acids methyl esters, including hydroxy acids. J. Clin. Microbiol. 16:584-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monciardini, P., M. Sosio, L. Cavaletti, C. Chiocchini, and S. Donadio. 2002. New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 42:419-429. [DOI] [PubMed] [Google Scholar]
- 36.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 37.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 38.Schleifer, K. H. 1985. Analysis of the chemical composition and primary structure of murein. Methods Microbiol. 18:123-156. [Google Scholar]
- 39.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekiguchi, Y., T. Yamada, S. Hanada, A. Ohashi, H. Harada, and Y. Kamagata. 2003. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53:1843-1851. [DOI] [PubMed] [Google Scholar]
- 42.Shirling, E. B., and D. Gottlieb. 1966. Methods of characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16:313-340. [Google Scholar]
- 43.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willems, A., W. E. Moore, N. Weiss, and M. D. Collins. 1997. Phenotypic and phylogenetic characterization of some eubacterium-like isolates containing a novel type B wall murein from human feces: description of Holdemania filiformis gen. nov., sp. nov. Int. J. Syst. Bacteriol. 47:1201-1204. [DOI] [PubMed] [Google Scholar]
- 45.Woese, C. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon, J. H., I. G. Kim, Y. K. Shin, and Y. H. Park. 2005. Proposal of the genus Thermoactinomyces sensu stricto and three new genera, Laceyella, Thermoflavimicrobium and Seinonella, on the basis of phenotypic, phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 55:395-400. [DOI] [PubMed] [Google Scholar]
- 47.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]