Abstract
Established methods for quantifying experimental Cryptosporidium infection are highly variable and subjective. We describe a new technique using quantitative real-time PCR (qPCR) that can be used to measure in vitro and in vivo laboratory infections with Cryptosporidium. We show for the first time that qPCR permits absolute quantification of the parasite while simultaneously controlling for the amount of host tissue and correlates significantly with established methods of quantification in in vitro and in vivo laboratory models of infection.
Cryptosporidium spp. are waterborne protozoan parasites that cause a range of diseases in humans, including self-limited diarrhea in immunocompetent hosts, persistent diarrhea in children living in developing countries, and severe infections in individuals with compromised immune systems. A massive waterborne outbreak in 1993 in Milwaukee, Wisconsin, resulted in over 400,000 human cases of cryptosporidiosis, and the parasite continues to cause sporadic water- and food-borne outbreaks in the United States (17, 31). The environmental form of the parasite, the oocyst, is resistant to chlorination, and effective therapies for human infection are limited. The Centers for Disease Control and the National Institutes of Health, acknowledging the potential risk of intentional dissemination, included the parasite on their priority lists of bioterrorism agents (24).
Research on the pathogenesis of C. parvum has been hampered by several factors, including inadequate animal models, inability to propagate the parasite in vitro, and imprecise methods for quantification of infection. Laboratory models of C. parvum infection have a critical role in advancing our understanding of many different aspects of the basic biology of the parasite and are also essential for the testing of potential interventions. Accurate, objective, and reproducible methods of quantifying C. parvum infection are necessary for these studies. Established methods of quantification of experimental C. parvum infection, such as counting intracellular forms of the parasite on hematoxylin-eosin-stained histopathologic specimens (7, 8), counting shed oocysts in stool (13), and enumerating intracellular stages by immunofluorescence assays (IFA) in in vitro samples, are imprecise, time-consuming, and subjective (25).
The use of molecular techniques, PCR in particular, has significantly advanced our understanding of C. parvum epidemiology, taxonomy, and biology (18, 20, 26, 27) and may have applications in the clinical setting (21). Semiquantitative PCR and quantitative real-time PCR (qPCR) have been used to detect C. parvum in environmental (9, 10) as well as fecal (30) samples. Either DNA or RNA that has been reverse transcribed into cDNA can be quantified using this technique. Recently, this technique has also been applied to measurement of in vitro laboratory infections (3, 6, 11, 14, 15). Using qPCR analysis, we developed an objective and reproducible technique that can be used to quantify experimental C. parvum infections in in vitro and in vivo models. We demonstrate for the first time that qPCR analysis positively correlates with established techniques for the measurement of in vivo and in vitro experimental C. parvum infections.
qPCR correlates significantly with the IFA for the measurement of experimental Cryptosporidium infection in vitro.
We used an established technique, IFA, for quantifying in vitro infection (12) and compared the results with those provided by qPCR. The statistical methods used to compare these and all subsequent techniques were as follows: after normal distribution was demonstrated by use of the Kolgorov-Smirnov test, linear regression was then used to correlate qPCR results with the dose of parasite used (in vitro) as well as with results obtained with a “gold standard” method of quantification, such as IFA or the enumeration of the parasite in histological sections (in vitro and in vivo). All in vitro analyses were performed in duplicate or in triplicate. The relationships were analyzed by plotting least-squares linear regression lines and computing coefficients of determination (r2) and corresponding P values. When data were not normally distributed, the Spearman rank correlation was used, and a two-tailed test of significance was used to determine P values. All statistical analyses were performed with SPSS 10.0 (Chicago, IL).
Two intestinal epithelial cell lines which support C. parvum infection in vitro, one of murine (16) and the other of human (12) origin, were used. The murine m-ICcl2 cell line was derived from the small intestine of a transgenic L-PK-tag 1 mouse carrying the simian virus 40 large T antigen under the control of the 5′ regulatory sequence of the L-type pyruvate kinase promoter (2). The human intestinal-like Caco2A cell line was originally obtained by the GRASP digestive disease center from H. Buller (Academic Medical Center, Amsterdam, The Netherlands) (28). Collagen-coated glass chamber slides (BD Falcon, Bedford, MA) were seeded with 4 × 104 cells/well, and 24-well plates (Costar; Corning, NY) were seeded with 2 × 105 cells/well, which were grown to confluence under previously defined conditions (2, 28). Confluent cells grown on glass slides or cultured plates were then infected with various doses of C. parvum oocysts (IOWA isolate; Bunch Grass Farms, Deary, ID) for 24 h as described previously (12). All experiments were performed in triplicate except where stated otherwise.
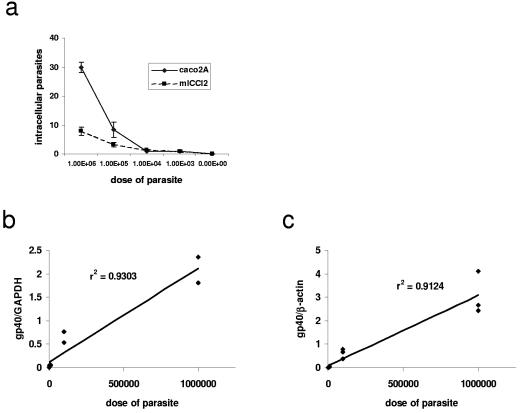
IFA was performed as described previously (12). Twenty-four hours after infection, the cells were washed with complete medium and then fixed with methanol for 30 min at room temperature. After nonspecific binding was blocked with 5% normal goat serum in phosphate-buffered saline (PBS), intracellular stages of the parasite were detected with the monoclonal antibody 4E9, which is specific for a carbohydrate epitope on the sporozoite antigens gp40 and gp900 (4). After being washed with PBS, cells were incubated with Alexafluor 488-conjugated goat anti-rabbit immunoglobulin M and counterstained with 4′,6′-di-amidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Slides were examined by fluorescence microscopy, and parasitic stages were counted in a blinded manner in 20 high-powered fields and expressed in terms of the number of parasites per high-powered field. Using IFA, we found that murine m-ICcl2 cells contained fewer parasites than human Caco2A cells after infection with equivalent doses of C. parvum, which is in agreement with a previous report (see Fig. 2) (16).
FIG. 2.
(a) IFA used to quantify C. parvum infection in Caco2A and m-ICcl2 cells. (b and c) Linear regression analysis comparing qPCR result (y axis) and parasite inoculum (x axis) after a 24-hour infection in m-ICcl2 (b) and Caco2A (c) cells. The parasite inoculum ranged from 103 to 106/ml, and these data are representative of two independent experiments performed in triplicate (for each experiment, n = 15).
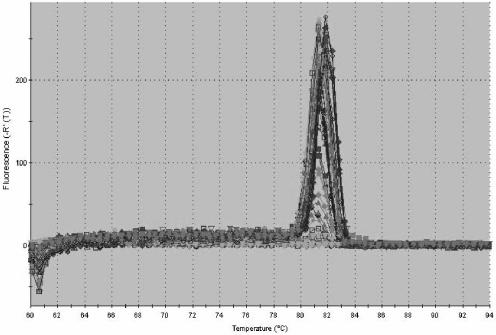
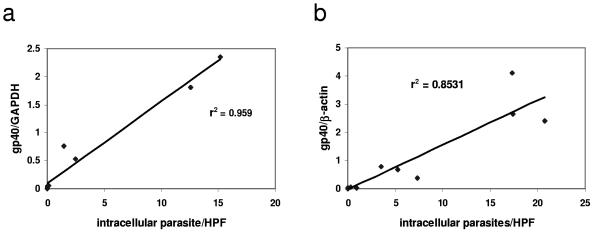
We then used qPCR to measure C. parvum infection from cells infected in parallel to those in which infection was quantified by IFA. Total RNA was isolated with an RNeasy kit (QIAGEN, Valencia, CA). After DNase (Ambion, Austin, TX) treatment, a reverse transcription reaction was performed using Stratascript (Stratagene, La Jolla, CA). Total DNA was extracted from intestinal tissue by using a GNOME DNA kit, (Qbiogene, Irvine, CA). qPCR was performed by using primers designed with Primer Express (Applied Biosystems, Foster City, CA) to amplify a conserved region of the gp15 portion of the Cpgp40/15 gene (4, 27). The sequences of these primers were as follows: forward, 5′-TCA TTT GTA ATG TGG TTC GGA GAA-3′, and reverse, 5′-AGG GTA AAG GCA AAC AAA TCG A-3′. Primers designed to amplify the murine housekeeping genes encoding nidogen (22) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (23) and the human gene encoding β-actin (1) were used as the internal standard as previously described. qPCR was performed using an ABI Prism 7700 thermocycler (Applied Biosystems) with the following conditions: 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 30 s. The Cpgp40/15 primers produced a single PCR product when used to measure C. parvum in in vitro and in vivo samples, thus demonstrating specificity for the target gene (Fig. 1). For each sample, infection was quantified by dividing the number of copies of Cpgp40/15 by the number of copies of the respective endogenous control as described previously (22). We found that in comparison with qPCR performed using cDNA, qPCR performed using DNA could not reliably distinguish between viable and heat-inactivated parasites in vitro (data not shown). Therefore, qPCR using cDNA was used for all subsequent in vitro experiments. Linear regression was then used to correlate the results of qPCR with the dose of parasite used to infect the intestinal epithelial cells. We found that the values obtained by qPCR positively correlated with the dose of parasite used to infect both the m-ICcl2 (r2 = 0.930; P < 0.0001) and the Caco2A (r2 = 0.912; P < 0.0001) cells over a fourfold log range (Fig. 2). Again, we used linear regression to correlate the results of qPCR and IFA. We found that qPCR positively correlated with the established technique of IFA for measurement of infection in both m-ICcl2 (r2 = 0.959; P < 0.0001) and Caco2A (r2 = 0.853; P < 0.0001) cells (Fig. 3). Therefore, qPCR correlates well with IFA when used to measure C. parvum in vitro infection over a wide range of parasite doses.
FIG.1.
Melting-curve analysis of gp40 primers for qPCR performed using SYBR green.
FIG. 3.
Linear regression analysis comparing qPCR result (y axis) and IFA (x axis) to quantify parasites after a 24-hour infection in m-ICcl2 (a) and Caco2A (b) cells. The parasite inoculum ranged from 103 to 106/ml, and these data are representative of two independent experiments performed in triplicate (panel a; n = 15) or in duplicate (panel b; n = 10). HPF, high-powered field.
qPCR correlates significantly with histological measurement of experimental Cryptosporidium infection in vivo.
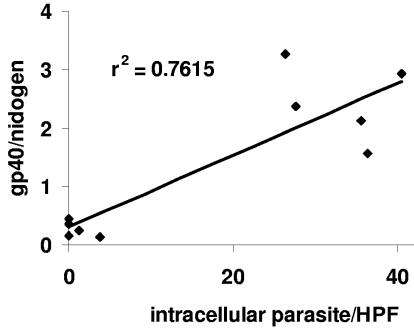
C. parvum-infected intestinal samples from a previously published study were used to compare techniques for quantifying infection (19). For histological analysis, portions of the terminal ilea of 10 C. parvum-infected mice were removed and flushed with PBS prior to either fixation in 10% buffered formalin or freezing in liquid nitrogen. After paraffin embedding, sections of the intestine were stained with hematoxylin-eosin. The parasite burden for each animal was determined by counting the mean number of intracellular forms of the parasite within a 10-by-10 grid counted in five separate high-powered fields within each sample (7). C. parvum infection was also quantified in parallel by qPCR performed using DNA and extracted from a segment of the terminal ilea adjacent to that used for histological analysis. qPCR was performed as described above for the in vitro experiments, and the parasite burden was expressed as the number of copies of Cpgp40/15 divided by the number of copies of the endogenous control, the gene encoding nidogen. Comparison of qPCR with histological analysis using linear regression demonstrated a statistically significant positive correlation (r2 = 0.761 and P = 0.001) (Fig. 4). qPCR was performed in parallel on cDNA obtained after reverse transcription of RNA extracted from adjacent segments of terminal ileum. We found that the results obtained by use of cDNA correlated poorly with histological analyses and therefore did not use them for further analysis (data not shown).
FIG. 4.
Linear regression analysis comparing qPCR result (y axis) and histological measurement (x axis) to quantify parasites after a 24-hour infection in mice. The data are representative of two independent experiments (n = 10). HPF, high-powered field.
In summary, we found that qPCR compared favorably with other established methods of quantification used in experimental infections with C. parvum. Unlike other microbial organisms that can readily be propagated in cell-free medium in vitro and are thus more easily quantified, the parasite C. parvum has traditionally required direct visualization for its enumeration. However, techniques such as IFA and histological measurements are labor-intensive and prone to interobserver variability. The development of objective and less time-consuming assays to measure infection is therefore desirable. Enzyme-linked immunosorbent assay-based assays are an objective and reliable alternative and have been used successfully for in vitro (29) and in vivo (5) infections. However, these assays have several drawbacks, including an inability to absolutely quantify the parasite and a reliance on oocyst shedding, which may be temporally dissociated from the actual infection in in vivo experiments. While there have been several other recent reports demonstrating that qPCR can be used to quantify in vitro C. parvum infection (3, 6, 14, 15), qPCR was not validated by comparison with gold standard techniques. Additionally, only one of these studies used host genes to correct for the amount of sample when quantifying C. parvum in vitro (3). Importantly, we demonstrate that qPCR can be used to measure parasite burden in an in vivo model of C. parvum infection and have correlated this technique with histological quantification. To our knowledge there has been only one previous study in which qPCR was used to detect, but not quantify, C. parvum in a tissue sample (11).
In this report we show that qPCR can be used to quantify parasites in in vitro and in vivo laboratory models of C. parvum infection. Objective and reproducible assays for the measurement of experimental infection in animals are particularly relevant for studies of the host immune responses to C. parvum. In these circumstances, targeted gene deletions or neutralization of target proteins affecting the immune response of the host may result in slight but significant differences in parasite burden which might not be detectable by use of more-subjective methods of quantification. Our technique permits absolute quantification of parasites while simultaneously controlling for the amount of host tissue and thus may be useful in such future studies.
Acknowledgments
We thank Carla Ciugini for initial assistance with qPCR and Robin Ruthazer for assistance with statistical methods.
This work was supported by NIAID grant AI51518A, the Center for Gastroenterology Research on Absorptive and Secretory Processes (GRASP), NIDDK P30 DK-34928-19, and the Center for AIDS Research (CFAR) P30 AI42853-06 (B.A.L.) and AI52786 and AI068535 (H.D.W.).
REFERENCES
- 1.Behera, A. K., C. M. Thorpe, J. M. Kidder, W. Smith, E. Hildebrand, and L. T. Hu. 2004. Borrelia burgdorferi-induced expression of matrix metalloproteinases from human chondrocytes requires mitogen-activated protein kinase and Janus kinase/signal transducer and activator of transcription signaling pathways. Infect. Immun. 72:2864-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bens, M., A. Bogdanova, F. Cluzeaud, L. Miquerol, S. Kerneis, J. P. Kraehenbuhl, A. Kahn, E. Pringault, and A. Vandewalle. 1996. Transimmortalized mouse intestinal cells (m-ICc12) that maintain a crypt phenotype. Am. J. Physiol. 270:C1666-C1674. [DOI] [PubMed] [Google Scholar]
- 3.Cai, X., K. M. Woods, S. J. Upton, and G. Zhu. 2005. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob. Agents Chemother. 49:4437-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cevallos, A. M., X. Zhang, M. K. Waldor, S. Jaison, X. Zhou, S. Tzipori, M. R. Neutra, and H. D. Ward. 2000. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 68:4108-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosyns, M., S. Tsirkin, M. Jones, R. Flavell, H. Kikutani, and A. R. Hayward. 1998. Requirement of CD40-CD40 ligand interaction for elimination of Cryptosporidium parvum from mice. Infect. Immun. 66:603-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Giovanni, G. D., and M. W. LeChevallier. 2005. Quantitative-PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enriquez, F. J., and C. R. Sterling. 1991. Cryptosporidium infections in inbred strains of mice. J. Protozool. 38:100S-102S. [PubMed] [Google Scholar]
- 8.Fayer, R., A. Guidry, and B. L. Blagburn. 1990. Immunotherapeutic efficacy of bovine colostral immunoglobulins from a hyperimmunized cow against cryptosporidiosis in neonatal mice. Infect. Immun. 58:2962-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontaine, M., and E. Guillot. 2003. An immunomagnetic separation-real-time PCR method for quantification of Cryptosporidium parvum in water samples. J. Microbiol. Methods 54:29-36. [DOI] [PubMed] [Google Scholar]
- 10.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, J. A., R. Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323-337. [DOI] [PubMed] [Google Scholar]
- 12.Joe, A., R. Verdon, S. Tzipori, G. T. Keusch, and H. D. Ward. 1998. Attachment of Cryptosporidium parvum sporozoites to human intestinal epithelial cells. Infect. Immun. 66:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, T. C., and B. L. Ungar. 1994. Comparison of sequential, random, and hemacytometer methods for counting Cryptosporidium oocysts. J. Parasitol. 80:816-819. [PubMed] [Google Scholar]
- 14.Keegan, A. R., S. Fanok, P. T. Monis, and C. P. Saint. 2003. Cell culture-Taqman PCR assay for evaluation of Cryptosporidium parvum disinfection. Appl. Environ. Microbiol. 69:2505-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King, B. J., A. R. Keegan, P. T. Monis, and C. P. Saint. 2005. Environmental temperature controls Cryptosporidium oocyst metabolic rate and associated retention of infectivity. Appl. Environ. Microbiol. 71:3848-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacroix-Lamande, S., R. Mancassola, M. Naciri, and F. Laurent. 2002. Role of gamma interferon in chemokine expression in the ileum of mice and in a murine intestinal epithelial cell line after Cryptosporidium parvum infection. Infect. Immun. 70:2090-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leav, B. A., M. Mackay, and H. D. Ward. 2003. Cryptosporidium species: new insights and old challenges. Clin. Infect. Dis. 36:903-908. [DOI] [PubMed] [Google Scholar]
- 18.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leav, B. A., M. Yoshida, K. Rogers, S. Cohen, N. Godiwala, R. S. Blumberg, and H. Ward. 2005. An early intestinal mucosal source of gamma interferon is associated with resistance to and control of Cryptosporidium parvum infection in mice. Infect. Immun. 73:8425-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao, S. Y., T. N. Liao, B. L. Chiang, M. S. Huang, C. C. Chen, C. C. Chou, and K. H. Hsieh. 1996. Decreased production of IFN gamma and increased production of IL-6 by cord blood mononuclear cells of newborns with a high risk of allergy. Clin. Exp. Allergy 26:397-405. [PubMed] [Google Scholar]
- 21.Morgan, U. M., L. Pallant, B. W. Dwyer, D. A. Forbes, G. Rich, and R. C. Thompson. 1998. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J. Clin. Microbiol. 36:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison, T. B., Y. Ma, J. H. Weis, and J. J. Weis. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J. Clin. Microbiol. 37:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbergh, L., D. Valckx, M. Waer, and C. Mathieu. 1999. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 11:305-312. [DOI] [PubMed] [Google Scholar]
- 24.Rochelle, P. A., M. M. Marshall, J. R. Mead, A. M. Johnson, D. G. Korich, J. S. Rosen, and R. De Leon. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 68:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slifko, T. R., D. Friedman, J. B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63:3669-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Beers, E. H., R. H. Al, E. H. Rings, A. W. Einerhand, J. Dekker, and H. A. Buller. 1995. Lactase and sucrase-isomaltase gene expression during Caco-2 cell differentiation. Biochem. J. 308:769-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdon, R., G. T. Keusch, S. Tzipori, S. A. Grubman, D. M. Jefferson, and H. D. Ward. 1997. An in vitro model of infection of human biliary epithelial cells by Cryptosporidium parvum. J. Infect. Dis. 175:1268-1272. [DOI] [PubMed] [Google Scholar]
- 30.Verweij, J. J., R. A. Blange, K. Templeton, J. Schinkel, E. A. Brienen, M. A. van Rooyen, L. van Lieshout, and A. M. Polderman. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 42:1220-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoder, J. S., B. G. Blackburn, G. F. Craun, V. Hill, D. A. Levy, N. Chen, S. H. Lee, R. L. Calderon, and M. J. Beach. 2004. Surveillance for waterborne-disease outbreaks associated with recreational water—United States, 2001-2002. Morb. Mortal. Wkly. Rep. 53(SS-8):1-22. [PubMed] [Google Scholar]