Abstract
A gene cluster for the biosynthesis of a new small cyclic peptide, dubbed trichamide, was discovered in the genome of the global, bloom-forming marine cyanobacterium Trichodesmium erythraeum ISM101 because of striking similarities to the previously characterized patellamide biosynthesis cluster. The tri cluster consists of a precursor peptide gene containing the amino acid sequence for mature trichamide, a putative heterocyclization gene, an oxidase, two proteases, and hypothetical genes. Based upon detailed sequence analysis, a structure was predicted for trichamide and confirmed by Fourier transform mass spectrometry. Trichamide consists of 11 amino acids, including two cysteine-derived thiazole groups, and is cyclized by an N—C terminal amide bond. As the first natural product reported from T. erythraeum, trichamide shows the power of genome mining in the prediction and discovery of new natural products.
Trichodesmium is a genus of marine diazotrophic, nonheterocysteous cyanobacteria. It occurs throughout the open waters of oligotrophic tropical and subtropical oceans and forms filaments (trichomes) of 20 to 200 cells that can further aggregate into colonies several millimeters across. Trichodesmium can form blooms in excess of 100,000 km2 (12), which are most commonly composed of Trichodesmium erythraeum and Trichodesmium thiebautii. Trichodesmium spp. have been the subject of intense research, mainly for two reasons. First, they contribute a significant portion (40% or more) (12) to global oceanic nitrogen fixation, thereby directly affecting the biogeochemical carbon flux in tropical oceans, with implications for the world's climate (1). Second, massive coastal Trichodesmium blooms have been reported to have toxic effects both directly on invertebrates (8, 10) and humans (Trichodesmium or Tamandare fever) (21) as well as indirectly by inducing blooms of other organisms (3, 14) that can potentially be harmful. While cyanobacteria are a prolific source of diverse natural products and toxins (2, 6, 18), to our knowledge, a toxic compound (or any natural product) has not been isolated from a Trichodesmium species, despite some efforts (9).
While most small peptides found in cyanobacteria are biosynthesized by nonribosomal peptide synthetases (17), we recently reported a microcin-like pathway for the biosynthesis of a family of cyclic peptides, the patellamides, from Prochloron didemni, a cyanobacterial symbiont of tropical ascidians (GenBank accession number AY986476) (23). The patellamides are moderately cytotoxic and are composed of a pseudosymmetrical, cyclic dimer, with each substructure having the sequence thiazole-nonpolar amino acid-oxazoline-nonpolar amino acid. Despite these unusual features, patellamide biosynthesis is ribosomal. The pat gene cluster consists of a precursor peptide gene, which codes for the patellamide amino acid sequence, and a number of genes with protease- and other peptide-modifying homologies (23). BLAST searches of GenBank with the pat genes revealed homologs in T. erythraeum IMS101. This led us to investigate the presence of a potential patellamide-like biosynthesis cluster as well as its product, a small cyclic peptide dubbed trichamide, in T. erythraeum.
MATERIALS AND METHODS
Bioinformatics.
Most of the T. erythraeum IMS101 genome was shotgun sequenced by the Joint Genome Institute and is available in the GenBank database (http://www.ncbi.nlm.nih.gov). The contig with accession number NZ_AABK04000003 contains the pat homologs listed previously (23). Nucleotides 785,500 to 803,500 of this contig were downloaded and manually annotated in Artemis (Sanger Institute). Predicted open reading frames (ORFs) were compared to the Joint Genome Institute autoannotation, and BLASTP in GenBank was used to assign putative functions.
Culturing.
T. erythraeum IMS101 (20) was obtained from John Waterbury, Woods Hole Oceanographic Institution, via Katherine Barbeau at the Scripps Institution of Oceanography (SIO). The culture is nonaxenic; i.e., it does contain other heterotrophic bacteria (K. Barbeau, personal communication). Cultures were grown in R medium (modified from John Waterbury's recipe) (25% double-distilled water and 75% natural seawater from the Scripps pier are mixed and amended with 8 μM KH2PO4, 2.5 μM EDTA, 0.1 μM ferric citrate, 0.1 μM MnCl2, 10 nM Na2MoO4, 10 nM ZnSO4, 0.1 nM CoCl2, 0.1 nM NiCl2, and 0.1 nM Na2SeO4) at 25°C under a 12-h light-dark photocycle with slow stirring as well as daily inversion of the culture flasks. All components were filter (0.2 μm) sterilized. T. erythraeum requires a 10% inoculum to start cultures; accordingly, 800 ml of culture was used in 8 liters of R medium. After 12 to 14 days, the culture was vacuum filtered through a 5-μm polycarbonate filter to retain T. erythraeum colonies and remove free-living bacteria. The cell material was rinsed off the filters into a 50-ml Falcon tube with 25% double-distilled water, immediately frozen at −80°C, and later lyophilized. The average yield was ∼10 mg dried cells per liter culture volume.
Extraction and purification.
Lyophilized cyanobacterial pellets were extracted three times with a ∼100-fold excess of methanol. The methanolic extract was dried, yielding a crude extract that was used for initial electron spray ionization (ESI) mass spectrometry (MS). For Fourier transform (FT) MS, the crude extract was purified with a C18 ZipTip (Millipore). A peak at m/z 1,099 corresponded to a new compound, trichamide.
A portion of the crude methanolic extract (23 mg) was further purified by partitioning between ethyl acetate and water. The aqueous part was fractionated over a HP20SS column with 25, 50, 75, and 100% acetone. As determined by ESI-MS, the 25 and 50% acetone (aqueous) fractions contained the peak at m/z 1,099 and were combined. This combined fraction was run on a high-performance liquid chromatography (HPLC) Phenomenex C18 analytical column with the following protocol (all solvents contained 0.01% trifluoroacetic acid): 5 min of water, a 5- to 35-min gradient from 0 to 100% acetonitrile, and 10 min of 100% acetonitrile. Fractions were collected at 1-min intervals. Only fractions eluting at 16 to 17 and 17 to 18 min contained a peak at m/z 1,099 as determined by ESI-MS. These fractions did not contain a single compound, since additional peaks beside the peak at m/z 1,099 were present in the MS analysis. The amount of material in the two HPLC fractions was too low to measure.
In an improved procedure, a methanolic extract (57 mg) was partially purified by a step gradient on a column containing 7 g C18 using solvents containing 0.01% trifluoroacetic acid. Fractions were eluted with water followed by 25%, 50%, and 100% acetonitrile (aqueous). The 100% elution fraction was further purified on a Phenomenex C18 column as described above. A single peak with the correct diode array profile cleanly eluted at 16.6 min. By ESI-MS analysis, this HPLC peak contained the m/z 1,099 ion. The concentration of trichamide was below a measurable limit and was thus estimated by comparison of the diode array absorbance at 240 nm with those for standards of ulithiacyclamide at various concentrations. This intensity should depend mainly upon the concentration of thiazole, since both ulithiacyclamide and trichamide have no other chromophores at this wavelength. By this method, the total amount of trichamide isolated was estimated to be 25 to 50 μg.
Mass spectrometry.
Crude extracts and partially purified fractions were monitored by ESI-MS and by FT-MS on a ThermoFinnigan LTQ-FT at a 100,000 resolution (i.e., mass of 400). FT tandem MS (MS/MS) experiments were run with collision-induced dissociation (CID) and infrared multiphoton dissociation (IRMPD) techniques. Predicted masses were calculated using the following values: C, 12; H, 1.007825; N, 14.003074; O, 15.994914; S, 31.97207.
RESULTS AND DISCUSSION
Biosynthetic genes.
Using genomic data available from GenBank, we have annotated a 12.5-kb gene cluster proposed to be responsible for the biosynthesis of trichamide (hence named the tri cluster). The GC content is 40%, which is higher than the average GC content of T. erythraeum at 34%. It is bordered by tRNA synthetase genes on both sides, potentially implicating horizontal gene transfer. The T. erythraeum genome is not closed, currently residing in 52 contigs at GenBank. The contig containing the tri genes (GenBank accession number NZ_AABK04000003) is 842 kb long and also contains a number of ribosomal proteins. A BLAST analysis of the ribosomal proteins found similarities with other cyanobacteria, so it is assumed that this contig is indeed from T. erythraeum and not from a possible contamination by heterotrophic bacteria.
The tri cluster contains 11 ORFs designated triA-K (Fig. 1 and Table 1). Four of these ORFs (triBCEF) are short and have sequence identity only to conserved hypothetical proteins, while triI is only hypothetical, with no significant sequence identities. Some of these ORFs may not be actively transcribed. This paper will focus on the remaining six genes, for which function may be inferred.
FIG. 1.
The tri gene cluster. Arrows denote ORFs and their direction. Black ORFs are tRNA synthetases, white ORFs are conserved hypothetical genes without a homolog in the pat cluster, green ORFs are pat homologs, and the gene encoding the precursor peptide is orange.
TABLE 1.
The tri cluster proteins and their homologs
| Protein | Length (aa)a | Homolog (GenBank accession no.) | % Identity/ % similarity | Predicted function | GenBank accession no. |
|---|---|---|---|---|---|
| TriA | 769 | PatD (AAY21153) | 57/70 | Adenylation/heterocyclization | ZP_00672901 |
| TriB | 112 | Conserved hypothetical protein (NP_942321) | 53/70 | Unknown | ZP_00672900 |
| TriC | 124 | Conserved hypothetical protein (BAB73591) | 60/78 | Unknown | ZP_00672899 |
| TriD | 475 | PatG, N terminal (AAY21156) | 45/59 | Oxidase | ZP_00672897 |
| TriE | 106 | Transposase-like protein (ZP_00345329) | 79/85 | Unknown | ZP_00672896 |
| TriF | 112 | Conserved hypothetical protein (ZP_00675293) | 78/91 | Unknown | ZP_00672895 |
| TriG | 67 | None | Precursor protein | NZ_AABK04000003b | |
| TriH | 666 | PatA (AAY21150) | 60/72 | Subtilisin-like protease | ZP_00672894 |
| TriI | 72 | None | Unknown | ZP_00672893 | |
| TriJ | 71 | PatB (AAY21151) | 52/70 | Unknown | ZP_00672892 |
| TriK | 702 | PatG, C terminal (AAY21156) | 48/64 | Subtilisin-like protease | ZP_00672891 |
aa, amino acids.
Accession number refers to a cohtig of which the sequence corresponding to positions 794178 to 794381 was translated to obtain TriG.
The product of triG is the putative precursor protein. It was identified by two 5-amino-acid motifs (GPGPS…SYDGD) that closely resemble the proposed cyclization signal found before and after the patellamide A and C sequences in the precursor protein of patellamide biosynthesis, PatE (Fig. 2) (23). Analogous to patellamide biosynthesis, these motifs would define the borders of the 11-amino-acid peptide GDGLHPRLCSC. TriG also contains a leader sequence of 43 amino acids without similarities in GenBank except that five of the first six amino acids are identical to those of PatE.
FIG. 2.
Alignment of the precursor peptides PatE and TriG. The sequences encoding patellamide C, patellamide A, and trichamide (top to bottom) are underlined, and the proposed cyclization signals are in boldface type.
TriA has high similarity to patD, which is proposed to be involved in the heterocyclization of cysteine and/or threonine/serine into thiazoline and oxazoline rings. The putative function was assigned on the basis of low sequence identity to previously characterized proteins: the adenylating enzyme MccB from microcin biosynthesis (7) for the N-terminal part and a possible hydrolase, SagD, from Streptomyces iniae for the C-terminal part (5).
TriD has high similarity to the N-terminal part of PatG and to oxidases. Previously, we predicted that this part of PatG would oxidize the intermediate thiazoline rings into thiazoles (23).
The BLASTP analysis of TriH and TriK gives homology to subtilisin-like proteases. They have high similarity to PatA and the C-terminal part of PatG (23). We predicted that these proteases would be involved in the maturation of PatE by cleaving the product from leader and trailer sequences and assumed the same function in trichamide biosynthesis. It is interesting that TriH and TriK have 48% identity to each other.
TriJ has 50% similarity and 72% identity to PatB. There is no other homolog of either of the two proteins in GenBank. PatB is not required for biosynthesis but seems to improve patellamide yield in heterologous expression experiments with the pat cluster (E. W. Schmidt, unpublished results). The high identity between TriJ and PatB over their entire length and their presence in both clusters suggest that they serve a role in peptide biosynthesis.
There are a few differences between the pat and tri clusters: PatG has two domains, one for oxidation and one for proteolytic cleavage. In T. erythraeum, these functionalities are separated into two proteins, TriD and TriH, respectively. The only pat gene without a homolog in the tri cluster (excluding very short putative ORFs) is patF, which has no significant homologies in GenBank. Overall, the pat and tri clusters have striking similarities. The biosynthetic genes have between 45 and 60% identity, and both gene clusters consist of a heterocyclization enzyme, an oxidase, two proteases, and patB/triJ, a gene of unknown functionality. Also, while there is variability in the length of the precursor protein, in terms of both the leader sequence and product sequence (8 amino acids for patellamide and 11 amino acids for trichamide), the 5-amino-acid cyclization signals before and after the peptide are highly conserved.
Based upon these similarities in biosynthesis genes, we predicted the presence of a patellamide-like compound, trichamide, a cyclic, thiazole-containing peptide, in T. erythraeum. Depending on the pattern of cyclization of the peptide and/or the heterocyclization of serine and cysteine moieties, the possible molecular weight of the compound would be between 1,079 and 1,157.
Mass spectrometry.
Initial screening of a crude extract of T. erythraeum with ESI-MS revealed the presence of a major peak at m/z 1,099. A molecule with this mass can be constructed from the precursor peptide sequence GDGLHPRLCSC by heterocyclization and oxidation of two of the three possible amino acids, cysteine, serine, and cysteine, to thiazoles or oxazoles and cyclization of the entire peptide. Alternatively, this mass is also consistent with heterocyclization of the remaining amino acid to a thiazoline or oxazoline moiety in a linear peptide.
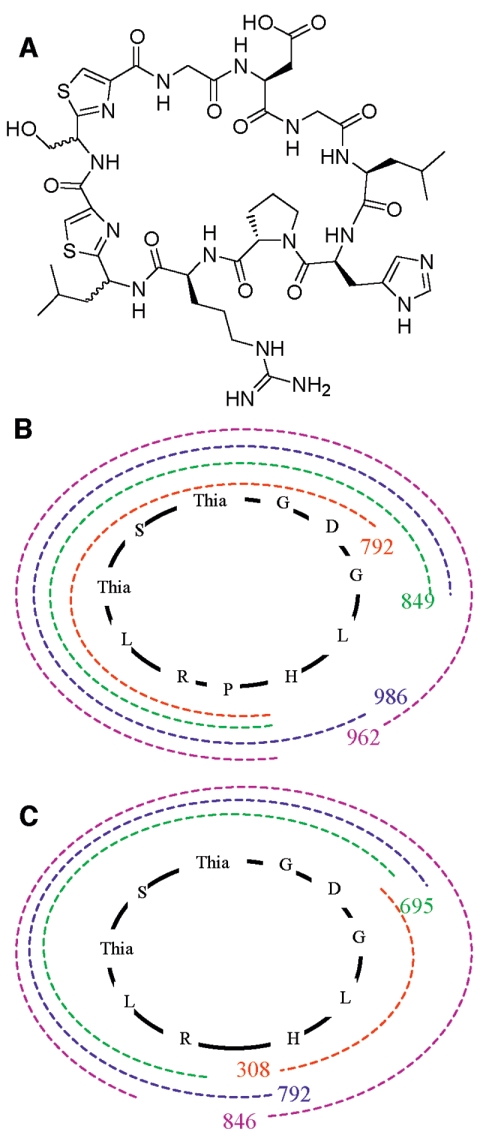
A high-resolution experiment using a Fourier transform MS/MS system gave a molecular ion at (MH2)2+ 550.23166, only 0.022 ppm different from the theoretical value of (MH2)2+ 550.231648 for the predicted structure, validating the presence of a molecule containing the trichamide molecular formula C46H66N16O12S2 (see the supplemental material).
Further MS/MS fragmentation experiments with a mass of 550.2 using CID and IRMPD techniques revealed fragmentation patterns in congruence only with a cyclic peptide (Table 2; see the supplemental material). With the exception of ion A, all masses are within ∼3 ppm of their predicted values. This leaves three possible heterocyclization patterns that have identical masses: (i) thiazole-serine-thiazole, (ii) thiazole-oxazole-cysteine, and (iii) cysteine-oxazole-thiazole. The data are consistent with the first pattern on the basis of three arguments. First, heterocyclization of adjacent amino acids has no precedent in the literature on patellamide structure; in fact, when two cysteine residues are adjacent in the patellamide family as in the ulithiacyclamides (4), only one is cyclized. Second, the patellamide class of compounds does not contain oxazoles but contains only oxazolines. Third, it is highly unlikely that an enzyme would specifically modify one cysteine but not the other.
TABLE 2.
Mass spectrometrya
| Method | Ion | Proposed structure | Mass (m/z)
|
Difference (ppm) | |
|---|---|---|---|---|---|
| Observed | Theoretical | ||||
| FT-MS | I | (MH2)2+, GDGLHPRL-Thia-S-Thia | 550.23166 | 550.231648 | 0.022 |
| II | (MH2)2+, 34S | 551.22845 | 551.22955 | 2.0 | |
| III | (MH)+, 13C2 | 551.23627 | 551.2350025 | 2.3 | |
| CID-MS/MS of 550.2 | A | (MH2)2+ of ion F | 481.20212 | 481.19883 | 6.8 |
| B | (MH2)2+ of parent ion minus C=O | 536.23520 | 536.23520 | 0.0 | |
| C | (MH)+, PRL-Thia-S-Thia-GD | 792.29387 | 792.29213 | 2.2 | |
| D | (MH)+, PRL-Thia-S-Thia-GDG | 849.31559 | 849.31359 | 2.4 | |
| E | (MH)+, PRL-Thia-S-Thia-GDGL | 962.39975 | 962.39766 | 2.2 | |
| F | (MH)+, HPRL-Thia-S-Thia-GDG | 986.37429 | 986.37250 | 1.8 | |
| IRMPD-MS/MS of 550.2 | G | (MH4)4+ of parent ion | 275.11674 | 275.115824 | 3.3 |
| H | (MH)+, GLH | 308.17152 | 308.17220 | 2.2 | |
| I | (MH2)2+ of parent ion | 550.23035 | 550.231648 | 2.4 | |
| J | (MH)+, RL-Thia-S-Thia-GD | 695.23870 | 695.239367 | 1.0 | |
| K | As ion C | 792.29121 | 792.29213 | 1.2 | |
| L | (MH)+, L-Thia-S-Thia-GDGLH | 846.30172 | 846.302694 | 1.2 | |
| M | As ion E | 962.39676 | 962.39766 | 0.9 | |
Proposed peptide structures are in one-letter amino acid code. Thia, cysteine modified to thiazole. Artifacts intrinsic to the machine and visible in other spectra of unrelated molecules constitute the other major peaks in the spectrum and are not tabulated here.
Because of the ribosomal mode of synthesis and in accordance with the patellamides, all of the amino acids in this molecule should adopt the l configuration. Exceptions to this rule may be serine and leucine 2, which are adjacent to thiazole. These stereocenters readily undergo epimerization, and they are often found in either the d or l form in patellamides. We are currently examining the patellamide biosynthetic proteins to determine whether or not this epimerization is enzymatic. The proposed structure of trichamide is shown in Fig. 3.
FIG. 3.
(A) Structure of trichamide. Stereochemistry is inferred and not determined experimentally, as described in the text. (B) Assignment of CID-MS fragments from Table 2 to the trichamide structure. (C) Assignment of IRMPD-MS fragments.
Biosynthetic pathway.
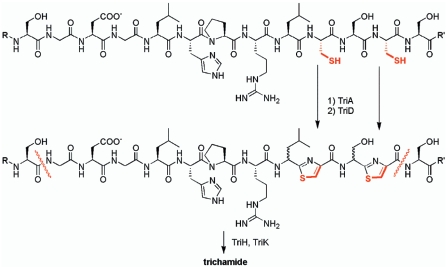
Closely paralleling patellamide biosynthesis, we predict the following pathway for trichamide biosynthesis (Fig. 4). TriG is the precursor protein and forms the substrate for posttranslational modification by TriA, TriD, TriH, and TriK. First, TriA modifies the cysteine moieties of TriG to form thiazoline groups. This could be an ATP-consuming process as in microcin heterocyclization (16), needing the ATP-hydrolyzing functionality of the N-terminal part of TriA, while the reaction itself would be catalyzed by the uncharacterized C-terminal part. Next, TriD oxidizes thiazolines to thiazoles. TriA and TriK cleave the propeptide guided by the conserved motifs GXXXS and XYDG. We propose that one protease cleaves the peptide bond after the header sequence, leading to a free amine group. The other protease would cut the back end and catalyze a transpeptidation reaction between the two ends of the peptide, leading to the mature cyclic form in a mechanism similar to the well-characterized peptidoglycan cyclization by a serine protease, penicillin binding protein (reviewed in reference 22). It is possible that the significant similarities between the two proteases allow them to form a dimer, which catalyzes both the hydrolysis of two peptide bonds and the cyclization in concert. It is interesting that the biosynthetic cluster of the linear peptide goadsporin (19) does not contain the two subtilisin-like proteases found in the tri and pat clusters, in agreement with an involvement of TriHK in cyclization. Recently, Milne et al. published a computational study in which preorganization of patellamides was predicted to lead to cyclization, and an enzyme would thus not be required (15). The differences in size and sequence and the maintenance of dedicated proteases in patellamides and trichamide argue against this possibility. Finally, the absence of a PatF homolog in T. erythraeum and the requirement of PatF in patellamide biosynthesis (E.W. Schmidt, unpublished) implicate PatF in oxazoline formation, which is not part of the trichamide pathway.
FIG. 4.
Proposed biosynthetic pathway to trichamide.
The biosynthetic scheme presented here is in accordance with all available data. Due to the ribosomal mode of biosynthesis, i.e., the coding of the trichamide amino acids in triG, the link between the tri genes and trichamide is direct. Further evidence awaits heterologous expression or knockout experiments with the tri genes.
Patellamide and trichamide biosynthesis may be examples of a more common pathway to small peptides. Besides the above-mentioned goadsporin from Streptomyces sp. strain TP-A0584, at this time, clustered ORFs with 35 to 40% identity to TriA and TriD are present in the genomes of phylogenetically distant bacteria: plut_0880 and plut_0878 in Pelodictyon luteolum, chlorobia (GenBank accession number CP000096), swolDRAFT_1502 and swolDRAFT_1501 in Syntrophomonas wolfei, clostridia (GenBank accession number NZ_AAJG01000002), and blr4538 and blr4539 in Bradyrhizobium japonicum (Rhizobiales) (11) (GenBank accession number BA000040).
Trichamide function.
Trichamide is hydrophilic, partitioning to the aqueous fraction relative to ethyl acetate. In addition, it is found only in the cells and is not excreted in significant quantities in the growth medium (data not shown). These properties suggest an antipredation defense function rather than anticompetitor or communication functions. To test biological activities,T. erythraeum crude methanolic extracts were tested for general cytotoxicity (HCT-116 at 10 μg/ml and CEM-TART at 5 and 50 μg/ml) and anti-human immunodeficiency virus (1 and 10 μg/ml), antifungal (Candida albicans at 10 μg/ml), or antimicrobial (Staphylococcus aureus and Enterococcus faecium at 10 μg/ml) effects. No significant activity was found in these assays (data not shown). A number of algal blooms have neurotoxic effects (2), and neurotoxicity of environmental Trichodesmium spp. in mice has previously been reported (9). The crude methanolic extract of T. erythraeum IMS101 also exhibited neurotoxicity in a mouse assay, but purified trichamide was not the active component (data not shown). Guo and Tester previously found that healthy Trichodesmium sp. cells do not affect the copepod Acartia tonsa, while aged or lysed Trichodesmium cells are toxic (8). This result is consistent with the properties of trichamide, which suggest that the compound is maintained inside healthy cells but would be released from lysed cells into seawater. Testing of the effect of trichamide on health and feeding behavior of copepods and other grazers of Trichodesmium might reveal the ecological function of trichamide. The presence of trichamide should be examined in Trichodesmium bloom waters. It would also be interesting to determine if T. thiebautii, the other major Trichodesmium species, which was proven to be more toxic than T. erythraeum in one report (10), contains trichamide (or a related compound) and the necessary biosynthetic capabilities.
Conclusion.
The ongoing exponential growth of DNA sequence data will lead to the discovery of many natural-product biosynthesis pathways for which no actual product has been characterized. A careful study of these pathways can lead to the discovery of novel products. Lautru et al. previously identified a gene cluster encoding nonribosomal peptide synthetase in the genome of a Streptomyces strain and discovered a novel compound, coelichelin (13). Nonribosomal peptide synthetase pathways are well-known routes to natural products, and the genus Streptomyces is a prolific source. The genus Trichodesmium was not previously known to produce natural products, and trichamide is only the second example, after the pat cluster, of a cyclic peptide biosynthesized in this way. Depending on the type of pathway, genomic mining should encompass careful curation. While the autoannotation of the T. erythraeum genome identified most of the tri genes as ORFs of unknown function, the most essential part of the cluster, the precursor peptide gene, was discovered manually.
This study is also an example of the power of general and timely access to genomic data. Even though the T. erythraeum IMS101 genome has not been completely sequenced or published at the time of this writing, the public availability of the draft sequence data allowed an assignment of function to the tri gene cluster and the discovery of a novel cyclic peptide.
Supplementary Material
Acknowledgments
This work was funded by the National Science Foundation (OCE0327070 to M.G.H.), by UU Startup funds to E.W.S., and by a U.S.-Egypt Joint Science and Technology Fund award (BIO8-002-003) to D.T.A.Y. and E.W.S.
We are grateful to Kelly Roe and Kathy Barbeau (SIO) for the Trichodesmium starter culture as well as helpful advice for maintaining it, to Chad Nelson (UU) for FT-MS analysis, and to Louis Barrows (UU), Baldomero Olivera (UU), William Gerwick (SIO), and William Fenical (SIO) for bioassays.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergmann, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 2.Carmichael, W. W. 1992. Cyanobacteria secondary metabolites—the cyanotoxins. J. Appl. Bacteriol. 72:445-459. [DOI] [PubMed] [Google Scholar]
- 3.Devassy, V. P., P. M. Bhattathiri, and S. Z. Qasim. 1979. Succession of organisms following Trichodesmium phenomenon. Indian J. Mar. Sci. 8:88-93. [Google Scholar]
- 4.Fu, X., T. Do, F. J. Schmitz, V. Andrusevich, and M. H. Engel. 1998. New cyclic peptides from the ascidian Lissoclinum patella. J. Nat. Prod. 61:1547-1551. [DOI] [PubMed] [Google Scholar]
- 5.Fuller, J. D., A. C. Camus, C. L. Duncan, V. Nizet, D. J. Bast, R. L. Thune, D. E. Low, and J. C. De Azavedo. 2002. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect. Immun. 70:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerwick, W. H., L. T. Tan, and N. Sitachitta. 2001. Nitrogen-containing metabolites from marine cyanobacteria. Alkaloids Chem. Biol. 57:75-184. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Pastor, J. E., J. L. San Millan, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, C., and P. A. Tester. 1994. Toxic effect of the bloom-forming Trichodesmium sp. (cyanophyta) to the copepod Acartia tonsa. Nat. Toxins 2:222-227. [DOI] [PubMed] [Google Scholar]
- 9.Hawser, S. P., E. J. Carpenter, G. A. Codd, and D. G. Capone. 1991. A neurotoxic factor associated with the bloom-forming cyanobacterium Trichodesmium. Toxicon 29:277-278. [DOI] [PubMed] [Google Scholar]
- 10.Hawser, S. P., J. M. O'Neil, M. R. Roman, and G. A. Codd. 1992. Toxicity of blooms of the cyanobacterium Trichodesmium to zooplankton. J. Appl. Phycol. 4:79-86. [Google Scholar]
- 11.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 12.Karl, D., A. Michaels, B. Bergman, D. Capone, E. Carpenter, R. Letelier, F. Lipschultz, H. Paerl, D. Sigman, and L. Stal. 2002. Dinitrogen fixation in the world's oceans. Biogeochemistry 57-58:47-98. [Google Scholar]
- 13.Lautru, S., R. J. Deeth, L. M. Bailey, and G. L. Challis. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat. Chem. Biol. 1:265-269. [DOI] [PubMed] [Google Scholar]
- 14.Lenes, J. M., B. P. Darrow, C. Cattrall, C. A. Heil, M. Callahan, G. A. Vargo, R. H. Byrne, J. M. Prospero, D. E. Bates, K. A. Fanning, and J. J. Walsh. 2001. Iron fertilization and the Trichodesmium response on the West Florida shelf. Limnol. Oceanogr. 46:1261-1277. [Google Scholar]
- 15.Milne, B. F., P. F. Long, A. Starcevic, D. Hranueli, and M. Jaspars. 2006. Spontaneity in the patellamide biosynthetic pathway. Org. Biomol. Chem. 4:631-638. [DOI] [PubMed] [Google Scholar]
- 16.Milne, J. C., A. C. Eliot, N. L. Kelleher, and C. T. Walsh. 1998. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry 37:13250-13261. [DOI] [PubMed] [Google Scholar]
- 17.Moore, B. S. 2005. Biosynthesis of marine natural products: microorganisms (part A). Nat. Prod. Rep. 22:580-593. [DOI] [PubMed] [Google Scholar]
- 18.Namikoshi, M., and K. L. Rinehart. 1996. Bioactive compounds produced by cyanobacteria. J. Ind. Microbiol. 17:373-384. [Google Scholar]
- 19.Onaka, H., M. Nakaho, K. Hayashi, Y. Igarashi, and T. Furumai. 2005. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology 151:3923-3933. [DOI] [PubMed] [Google Scholar]
- 20.Prufert-Bebout, L., H. W. Paerl, and C. Lassen. 1993. Growth, nitrogen fixation, and spectral attenuation in cultivated Trichodesmium species. Appl. Environ. Microbiol. 59:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato, S., M. N. Paranangua, and E. Eskinazi. 1963. On the mechanism of red tide of Trichodesmium in Recife north eastern Brazil, with some considerations of the relation to human disease Tamandare fever. Trab. Inst. Oceanogr. Univ. Fed. Pernambuco Recife 5/6:7-50. [Google Scholar]
- 22.Scheffer, D., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69:585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt, E. W., J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.