Abstract
Zearalenone, a secondary metabolite produced by several plant-pathogenic fungi of the genus Fusarium, has high estrogenic activity in vertebrates. We developed a Saccharomyces cerevisiae bioassay strain that we used to identify plant genes encoding UDP-glucosyltransferases that can convert zearalenone into zearalenone-4-O-glucoside (ZON-4-O-Glc). Attachment of the glucose moiety to zearalenone prevented the interaction of the mycotoxin with the human estrogen receptor. We found that two of six clustered, similar UGT73C genes of Arabidopsis thaliana encode glucosyltransferases that can inactivate zearalenone in the yeast bioassay. The formation of glucose conjugates seems to be an important plant mechanism for coping with zearalenone but may result in significant amounts of “masked” zearalenone in Fusarium-infected plant products. Due to the unavailability of an analytical standard, the ZON-4-O-Glc is not measured in routine analytical procedures, even though it can be converted back to active zearalenone in the digestive tracts of animals. Zearalenone added to yeast transformed with UGT73C6 was converted rapidly and efficiently to ZON-4-O-Glc, suggesting that the cloned UDP-glucosyltransferase could be used to produce reference glucosides of zearalenone and its derivatives.
Zearalenone (ZON) is a secondary metabolite with estrogenic activity synthesized by several species of Fusarium/Gibberella (for reviews, see references 8 and 20) during infection of small-grain cereals and maize. ZON-contaminated feed has been implicated in numerous cases of fertility disturbances in farm animals, especially pigs (20). In a recent risk assessment, the toxicological hazard of ZON was reevaluated (33, 40) by the Scientific Committee on Food of the European Commission and the WHO (Joint FAO/WHO Expert Committee on Food Additives), and the occurrence of ZON and dietary intake were assessed (11). ZON in general cooccurs with other Fusarium toxins, such as deoxynivalenol, although large annual variations occur (26). Legislation establishing maximum tolerable levels for ZON (and other mycotoxins) has recently been enacted in Europe (Commission regulation no. 856/2005 [12]). Products exceeding the maximum levels cannot be used as food ingredients and cannot be blended with products with lower toxin content to reduce the overall level of toxin in the composite product. For example, the maximum level of ZON allowed in unprocessed cereals other than maize is 100 μg/kg, and the maximum level allowed in processed cereal-based foods for infants and young children is 20 μg/kg.
ZON exerts its hormonal effect in animals by binding to estrogen receptors ER-α and ER-β (19, 20) and thereby activating transcription via a hormone-dependent transcription activation domain (38). ZON and its derivatives are cleared from the body predominantly as glucuronides (2, 24). Relatively little is known about the role of ZON in plant-pathogen interactions or the fate of ZON in plants. Maize suspension cultures treated with ZON formed the reduction products α- and β-zearalenol (ZOL) and glucose conjugates of the respective compounds (9, 10, 41), in particular zearalenone-4-β-d-glucopyranoside (ZON-4-O-Glc).
This conjugate can occur in wheat samples naturally infected with Fusarium at concentrations ranging from 17 to 104 μg/kg (32). The glucose conjugate of ZON is not detected by conventional analytical techniques but can be easily hydrolyzed in the digestive tracts of animals (14) and could therefore be an important, but hitherto unaccounted for, source of the mycotoxin in the diet. The lack of a commercially available standard prevents the routine analysis of ZON-4-O-Glc.
The formation of glycoconjugates of small molecules is catalyzed by UDP-glycosyltransferases (UGTs), which use a variety of activated UDP-sugars as donors, in particular, UDP-glucose for glucoconjugates. Members of the UDP-glycosyltransferase superfamily are highly abundant in plant genomes. The genome of Arabidopsis thaliana, for example, contains more than 100 UGT genes with mostly unknown substrate specificities (6, 21, 22). We previously identified an Arabidopsis UGT that could detoxify the Fusarium toxin deoxynivalenol (29). We suggested that, in addition to the presumed role in homeostasis of endogenous plant metabolites, plant UGTs may participate in plant-pathogen interactions in which microbial toxins play a role.
The objective of this work was to identify plant genes encoding glucosyltransferases capable of inactivating ZON. We hypothesized that heterologous expression of plant cDNAs in Saccharomyces cerevisiae and utilization of estrogen-responsive marker genes would provide appropriate means for identification of such cDNAs. We report here the construction of a suitable host strain and the identification of UGTs from Arabidopsis thaliana capable of forming high levels of ZON-4-O-Glc. The availability of ZON-4-O-Glc as a reference material should enable the evaluation of masked ZON in agricultural products.
MATERIALS AND METHODS
Yeast strains and plasmid constructions.
The strains used in this study are derivatives of YPH499 (35). Strain YYM4 (Table 1), a snq2 disruption strain (23), was derived from the pdr5Δ::TRP1 mutant YKK-A7 (3).
TABLE 1.
Genotypes of S. cerevisiae strains used in this study
| Strain | Parental strain | Plasmid (relevant change) | Genotype (episomal plasmid)a |
|---|---|---|---|
| YYM4 | YKK-A7 | 23 | MATaleu2-Δ1 ura3-52 trp1-Δ1 his3-Δ200 lys2-801 ade2-101 pdr5Δ::TRP1 snq2Δ::hisG |
| YZHB796 | YYM4 | pHB107/EcoRI | MATaleu2-Δ1 ura3-52 trp1-Δ1 his3-Δ200 lys2-801 ade2-101 pdr5Δ::trp1::P3xERE-URA3-loxP-KANr-loxP snq2Δ::hisG |
| YZHB824 | YZHB796 | YEp90-HEGO | MATaleu2-Δ1 ura3-52 trp1-Δ1 his3-Δ200 lys2-801 ade2-101 pdr5Δ::trp1::P3xERE-URA3-loxP-KANr-loxP snq2Δ::hisG (2μm HIS3 PPGK1-hER) |
| YZCP908 | YZHB824 | pCP303/SalI+EcoRI | MATaleu2-Δ1 ura3-52 trp1-Δ1 his3-Δ200 lys2-801 P3xERE-ADE2 pdr5Δ::trp1::P3xERE-URA3-loxP-KANr-loxP snq2Δ::hisG (2μm HS3 PPGK1-hER) |
| YZBP67 | YZCP908 | pBP868 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C5) |
| YZBP69 | YZCP908 | pBP874 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C1) |
| YZBP71 | YZCP908 | pBP1378 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C2) |
| YZBP73 | YZCP908 | pBP901 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C3) |
| YZBP75 | YZCP908 | pBP1374 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C4) |
| YZBP77 | YZCP908 | pBP918 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-UGT73C6) |
| YZBP79 | YZCP908 | pBP910 | Same as YZCP908 + (2μm LEU2 PADH1-c-MYC-no insert) |
ERE, estrogen-responsive element; hER, human estrogen receptor.
The trp1::P3xERE-URA3 cassette was amplified with primers TRP1-ATG and TPR1-END (Table 2 shows primer sequences), which bind in the flanking trp1 gene remnants present in the genomic DNA of strain YYM8 (23). The PCR product was cloned by using the pCR2.1-TOPO kit (Invitrogen, Karlsruhe, Germany), yielding plasmid pRM1783. The EcoRI insert of pRM1783 was cloned into the EcoRI site of pUC7 (37) in order to avoid flanking restriction sites in the resulting plasmid, pHB75. A Klenow-treated NotI fragment of the plasmid pUG6 (17) containing the loxP-KANr-loxP cassette was ligated to XbaI-digested, Klenow-treated pHB75. The resulting plasmid, pHB107, was cut with EcoRI to release the trp1::P3xERE-URA3-loxP-KANr-loxP fragment for yeast transformation. Geneticin (G418)-resistant transformants of YMM4 were screened for loss of the Trp+ phenotype, yielding strain YZHB796, in which the P3xERE-URA3 cassette was inserted at pdr5 (Table 1). This strain was transformed with YEp90-HEG0, a 2μm-HIS3 plasmid, resulting in the expression of a human estrogen receptor cDNA under the control of the constitutive yeast PGK1 promoter (25).
TABLE 2.
Primer pairs and their uses (resulting plasmids)
| Primer name | Oligonucleotide sequence | PCR product | Plasmid |
|---|---|---|---|
| TRP1-ATG | 5′-CGTGAGTATACGTGATTAAGCA-3′ | trp1::P3xERE-URA3 cassette | pRM1783 |
| TPR1-END | 5′-ATGTTTAGATCTTTTATGCTTGC-3′ | ||
| ADE2-ATG-FW | 5′-ACCTGCAGGAAACGAAGATAAATCATGGATTCTAGAACAGTTGG-3′a | PURA3-ADE2 cassette with PstI near ATG | pHB137 |
| ADE-DW | 5′-TTGAATTCTATGTATGAAGTC-3′b | ||
| ADEPROM-FOR | 5′-GGGCGTCGACAATCTGCTATGC-3′c | ADE2 upstream region fragment | pCP303 |
| ADEPROM-REV | 5′-GATGATATACTGGTAGTGCGC-3′ | ||
| UGT73C1-FW | 5′-CTAAGCTTGGAATCATGGCATCGGAATTTCG-3′ | N-terminally c-MYC-tagged UGT73C1 | pBP874 |
| UGT73C1-RV | 5′-TAGCGGCCGCATTCATTTCTTGGGTTGTTC-3′ | ||
| UGT73C2-FW | 5′-CTAAGCTTGGAATCATGGCTTTCGAGAAGACC-3′ | N-terminally c-MYC-tagged UGT73C2 | pBP1378 |
| UGT73C2-RV | 5′-TAGCGGCCGCATTCAACTCTTGGATTCTAC-3′ | ||
| UGT73C3-FW | 5′-CTAAGCTTGGAATCATGGCTACGGAAAAAACC-3′ | N-terminally c-MYC-tagged UGT73C3 | pBP901 |
| UGT73C3-RV | 5′-TAGCGGCCGCATTCAATTCTTGAATTGTGC-3′ | ||
| UGT73C4-FW | 5′-CTAAGCTTGGAATCATGGCTTCCGAAAAATC-3′ | N-terminally c-MYC-tagged UGT73C4 | pBP1374 |
| UGT73C4-RV | 5′-TAGCGGCCGCATTCAGTTCTTGGATTTCA-3′ | ||
| UGT73C6-FW | 5′-CTAAGCTTGGAATCATGGCTTTCGAAAAAAACA-3′ | N-terminally c-MYC-tagged UGT73C6 | pBP918 |
| UGT73C6-RV | 5′-TAGCGGCCGCATTCAATTATTGGACTGTGC-3′ |
A PstI site is underlined, while the following 20 bases (italics) of the primer correspond to the first 20 bases of the ADE2 gene.
An introduced EcoRI site is underlined (see Materials and Methods).
A SalI site is underlined.
A P3xERE-ADE2 construct was generated in two steps. First, the HindIII-PstI fragment of pRM1783, which contains the estrogen-responsive elements, was inserted into the HindIII-PstI-digested yeast shuttle vector YCplac111 (15), yielding plasmid pHB83. A PCR product of the yeast ADE2 gene was amplified by using primers ADE2-ATG-FW and ADE-DW (Table 2). The first part of the ADE2-ATG-FW sequence corresponds to the bases −24 to −1 upstream of the URA3 start codon and contains a PstI site (Table 2), while the following 20 bases of the primer correspond to the first 20 bases of the ADE2 gene. Primer ADE-DW binds about 100 bp downstream of the stop codon of ADE2 and creates an EcoRI site. The PCR product was cut with PstI-EcoRI and ligated into the PstI-EcoRI-cut vector pHB83, yielding pHB137. This plasmid was used to transform a yeast ade2 mutant expressing the estrogen receptor for functional testing.
The estrogen-responsive ADE2 gene was then integrated into the chromosomal locus. First, an EcoRI-HindIII trp1-P3xERE-ADE2 cassette from plasmid pHB137 was cloned into pBlueskript SK+, yielding pCP302. Then, a 450-bp fragment of the intergenic region between ADE2 and the upstream gene YOR129c was amplified with primers ADEPROM-FOR (which contains a SalI site [Table 2]) and ADEPROM-REV. The PCR product was cloned into pCR4-TOPO (Invitrogen), and the insert was released as an EcoRI fragment and treated with Klenow enzyme. The blunt-ended ADE2 upstream fragment was cloned into the SalI site of pCP302, which was also treated with Klenow enzyme. A clone containing the upstream fragment in the correct orientation (pCP303) was selected. The integration cassette was released from this plasmid as a SalI-EcoRI fragment, which was used to transform the ade2-101 mutant strain YZHB824. Ade+ colonies were selected on SC-Ade-His medium supplemented with 10 μg/liter (ppb) ZON. The desired transformants had repaired the ade2-101 mutation and replaced the promoter of the chromosomal ADE2 with the estrogen-responsive promoter. These transformants were identified by adenine auxotrophy and the formation of a red pigment on SC-His medium (34) without added estrogen and limiting adenine (5 mg/liter adenine sulfate, final concentration [1/4-Ade]). Strain YZCP908 (the full genotype is given in Table 1) was chosen as the host strain for further work.
Constitutive expression of Arabidopsis UDP-glycosyltransferases in yeast and immunodetection.
The construction of a c-MYC epitope-tagged UGT73C5 yeast expression vector (pBP868) has been described previously (29). To generate yeast cells that expressed the closest homologues of UGT73C5 (At2g36800) in the cluster (UGT73C1, At2g36750; UGT73C2, At2g36760; UGT73C3, At2g36780; UGT73C4, At2g36770; and UGT73C6, At2g36790; http://mips.gsf.de/proj/thal/db/index.html or http://www.arabidopsis.org/), the respective open reading frames of these genes were PCR amplified (Triple Master PCR System; Eppendorf, Wesseling-Berzdorf, Germany) from genomic DNA with gene-specific primers (Table 2) containing flanking HindIII and NotI restriction sites at the 5′ and 3′ ends, respectively.
The PCR products were cloned downstream of a c-MYC epitope tag in frame in the yeast expression vector pYAK7 (2μm LEU2 PADH1-c-MYC-PDR5), by using the HindIII and NotI restriction sites and replacing the PDR5 gene (29). The resulting expression vectors (with the respective UGT genes in parentheses) were pBP874 (UGT73C1), pBP1378 (UGT73C2), pBP901 (UGT73C3), pBP1374 (UGT73C4), and pBP918 (UGT73C6). The empty vector control plasmid pBP910 was produced by deletion of the PDR5 insert in pYAK7 by digestion with HindIII-NotI and religation. Transformants were selected on SC-Leu medium.
The c-MYC-tagged UGT constructs were verified by sequencing and used to transform yeast strain YZCP908 (Table 1). Exponentially growing cultures were diluted to an optical density at 600 nm (OD600) of 0.05 and spotted onto SC-Leu-His plus 1/4-Ade medium containing increasing concentrations of ZON. All mycotoxins used in this study were purchased from Sigma (Vienna, Austria) or Biopure (Tulln, Austria), dissolved in 70% ethanol, and stored at −20°C. For immunodetection, the extraction of proteins from yeast cells was performed as previously described (29). Western blot analysis was conducted with a primary mouse anti c-MYC antibody (clone 9E10).
Synthesis of zearalenone-4-O-β-d-glucopyranoside.
An analytical standard of ZON-4-O-β-d-glucopyranoside was synthesized by using acetobromoglucose (31) according to a protocol modified from that of Grabley et al. (16). The identity and purity of the synthesis product were characterized by 1H-nuclear magnetic resonance and liquid chromatography-tandem mass spectrometry (LC-MS/MS) (data not shown).
HPLC-MS/MS conditions.
LC-MS/MS analysis was performed on a QTrap LC-MS/MS system (Applied Biosystems, Foster City, California) equipped with an electrospray interface and an 1100 series high-performance LC (HPLC) system (Agilent, Waldbronn, Germany). The electrospray interface was used in the negative ion mode at 400°C with the following settings: curtain gas, 20 lb/in2; nebulizer gas, 30 lb/in2; auxiliary gas, 75 lb/in2; ion spray voltage, −4,200 V; collision-activated dissociation gas setting, 6. Chromatographic separation was achieved on a 100- by 4.6-mm (inside diameter), 3-μm Aquasil RP-18 column (Keystone, Waltham, Mass.) at 25°C using methanol-water (70/30 [vol/vol]). The flow rate was set to 0.5 ml/min. For multiple-reaction-monitoring (MRM) MS/MS experiments, a dwell time of 50 ms was used. Deprotonated ZON ([ZON-H]−, corresponding to m/z 317.1; declustering potenial [DP], −51 V) was fragmented to m/z 130.9 (collision energy [CE], −38 eV). For the detection of α- and β-ZOL, the MRM transitions from m/z 319.1 to m/z 129.9 (DP, −71 V; CE, −48 eV) were used.
Isolation and analysis of ZON metabolites in vivo.
The chemical structures of ZON metabolites resulting from enzymatic transformation of the mycotoxin were determined after incubation with the bioassay strain YZCP908 expressing UGT73C6. The yeast cells were grown in selection medium (SC-His-Leu) to an OD600 of 1.0, harvested, and resuspended in medium to yield an OD600 of 7.0. To 5 ml of such suspensions, ZON, α-ZOL, or β-ZOL stock solutions (5 mg/ml in ethanol) were added stepwise, and the mixtures were incubated on a shaker at 30°C. After the initial addition of 8 μl stock solution, another 8 μl was added 3 h from the start, 4 μl more at 5 h, and another 4 μl at 7 h (total additions, approximately 24 mg/liter). The cells were collected by centrifugation 24 h after the first addition, and the culture supernatants were removed. The cells were resuspended in 500 μl of water. Then, 2 ml of methanol was added and the suspensions were sonicated (four times for 15 s and three times for 30 s; output control setting, 45% on a Vibra Cell 50 W [Sonics and Materials, Inc., Danbury, Conn.]). After centrifugation, the supernatants were filtered through a GF-A filter (Whatman International Ltd., Maidstone, United Kingdom) and a glass microfiber filter (1822 025; Whatman) and then through a 0.22-μm syringe filter (Millex GV; Millipore, Billerica, Mass.). The culture supernatants were also filtered as described above. Finally, 10-μl aliquots of the filtered supernatants (medium) and the methanol extracts of the cells were injected into the HPLC-MS/MS system without further cleanup.
Estrogen receptor assay.
The abilities of ZON and ZON-4-O-Glc to bind to the human estrogen receptor were assayed in vitro with the HitHunter EFC Estrogen Chemiluminescence Assay Kit (DiscoveRx Corp., Fremont, Calif.) and the human estrogen receptor α (Sigma, Vienna, Austria). The assay is based on complementation of β-galactosidase activity by two separate fragments, an enzyme acceptor and an enzyme donor peptide. The donor peptide is conjugated to a steroid hormone. Enzyme complementation is prevented if the estrogen receptor binds to the conjugated steroid. The addition of a competitor that binds to the estrogen receptor allows enzyme complementation, which can be measured by the hydrolysis of a luminescent β-galactosidase substrate. The assay was performed according to the manufacturer's instructions. The reaction mixtures were incubated with either ZON or ZON-4-O-Glc (0.5, 1, 5, 10, 50, 100, and 500 nM and 1 μM, dissolved in dimethyl sulfoxide), and hydrolysis of the substrate was measured with a luminometer (Victor 2; Wallac/Perkin-Elmer, Monza, Italy).
RESULTS
Generation of a novel yeast indicator strain.
Inactivation of PDR5 and SNQ2, two S. cerevisiae genes encoding ABC transporter proteins that act as efflux carriers, increases the net uptake of zearalenone in yeast (25). We started strain construction with YYM4 (Table 1), in which the efflux carriers PDR5 and SNQ2 were already disrupted. Our aim was to construct a strain that allowed positive selection of ZON degradation, utilizing the previously described trp1::P3xERE-URA3 construct present in strain PL3 (25, 28), and that allowed visual detection of the loss of estrogenic activity of ZON (and other estrogenic compounds) using ADE2. In the presence of an estrogenic substance, the expressed human estrogen receptor binds to the estrogen-responsive elements engineered into the promoter of the URA3 gene. Such a strain is therefore Ura+ and sensitive to 5-fluoroorotic acid (4) in the presence of zearalenone or other estrogenic substances (data not shown).
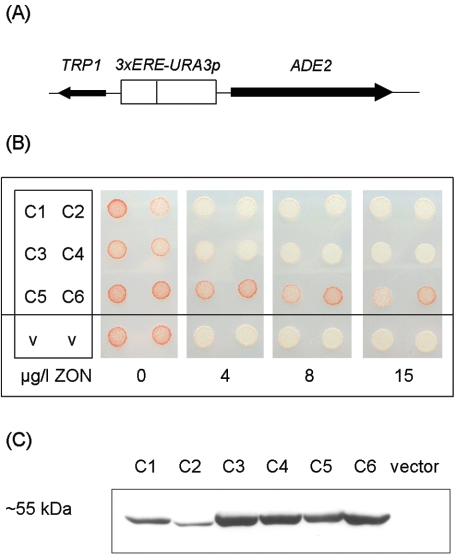
As a secondary reporter for screening, we utilized ade2 mutants, which accumulate a red pigment (7). An estrogen-responsive ADE2 gene (Fig. 1A) was constructed and transferred to the chromosomal ade2 locus of yeast, yielding strain YZCP908. On media with limiting adenine and in the absence of an estrogenic substance, this strain accumulates a red pigment. In the presence of as little as 2 μg/liter zearalenone, the transcription of the engineered ADE2 gene was activated, leading to a color change from red to white (Fig. 1B).
FIG. 1.
Screening of Arabidopsis UGTs of subfamily UGT73C for inactivation of ZON in a yeast bioassay. (A) Schematic illustration of the developed yeast bioassay. The expression of ADE2 is under the control of the engineered 3xERE-URA3 promoter (three estrogen-responsive elements in front of a minimal URA3 promoter). (B) Yeast strains transformed with the indicated members of the UGT73C gene family were spotted on SC-Leu plus 1/4-Ade plates containing the indicated amounts of ZON (ppb, or μg/liter). The strains were bioassay strain YCP908 transformed with the c-MYC-tagged UGTs of subfamily UGT73C, UGT73C1 (C1) to UGT73C6 (C6), and YCP908 transformed with an empty vector as a control (v). (C) Western blot analysis of the yeast strains used in panel B. The N-terminal c-MYC epitope tag introduced into the respective 73C UGT family members was detected. A transformant containing the empty expression vector was used as a control.
Identification of plant UDP-glucosyltransferases inactivating ZON.
We used the bioassay strain we developed, YZCP908, to test individual members of the large UGT family of Arabidopsis (22) for the ability to reduce the estrogenic activity of ZON. We cloned all six genes from a cluster of tandemly repeated UGT genes with very high sequence similarity from chromosome II of Arabidopsis (members of the UGT73C family) and expressed them as fusion proteins with an N-terminal c-MYC epitope tag in strain YZCP908 (Fig. 1C). As little as 4 μg/liter of ZON induced a color change from red to white in yeast transformed with the empty vector pBP910 (Fig. 1B). Yeast transformants (Table 1) expressing UGT73C1, UGT73C2, UGT73C3, and UGT73C4 responded to ZON in a manner similar to that of the control, whereas those expressing UGT73C5 and UGT73C6 remained red even at higher ZON concentrations (Fig. 1B). We interpreted these results to mean that yeast cells expressing UGT73C6 and UGT73C5 tolerated higher concentrations of ZON in the media before ADE2 transcription was activated and that the estrogenic properties of ZON in yeast were altered.
UGT73C6 catalyzes the transfer of glucose to the C-4 positions of ZON, α-ZOL, and β-ZOL.
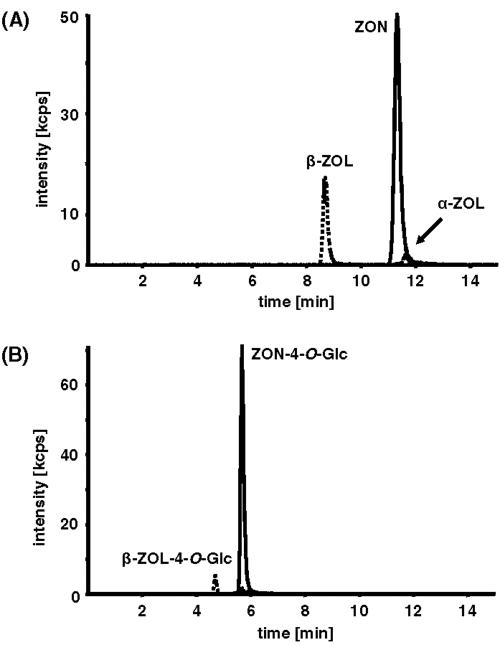
We tested for the presence of ZON-4-O-Glc by LC-MS/MS. This compound elutes at 5.66 min; the fragmentation (MS/MS) of the [M-H]− ions of m/z 479.1 resulted mainly in [ZON-H]− ions with m/z 317.1 due to the loss of glucose (−162 atomic mass units). The same transition was used for ZON-4-glucoside in succeeding MRM experiments (DP, −31 V; CE, −22 eV), while for α- and β-ZOL-glucosides, the MRM transition from m/z 481.1 (corresponding to the deprotonated molecular ions) to m/z 319.1 (deprotonated α- or β-ZOL) was used at a DP of −31 V and a CE of −22 eV.
Yeasts can convert ZON to both α-ZOL and β-ZOL by reduction of the C-6′ keto group to a hydroxy group (5). Both ZON derivatives were therefore included in the analysis. When yeast expressing an empty vector as a control was incubated with ZON, both β-ZOL (the ZON derivative formed predominantly by S. cerevisiae) and (to a lesser extent) α-ZOL were detected in the supernatants (Fig. 2A). No glucosides were detected in this control culture. In the samples from yeast cultures expressing UGT73C5 (data not shown) and UGT73C6 (Fig. 2B), ZON-4-O-Glc was the dominant compound. In addition, minor fractions of both ZOL glucosides were also detected. On a molar basis, 90% of the added ZON was converted to ZON-4-O-Glc and 9% to β-ZOL-4-O-Glc. Feeding experiments with α-ZOL or β-ZOL also showed that the respective glucosides were formed at high yield. Little or none of the parental compound, i.e., ZON, α-ZOL, or β-ZOL, was detected in the UGT73C6 samples, indicating that nearly all of the mycotoxin had been glucosylated.
FIG. 2.
MS/MS analysis of ZON metabolites formed in yeast cells expressing UGT73C6. Yeasts expressing an empty vector as a control (A) and expressing UGT73C6 (B) were incubated with ZON, and the products formed were analyzed by LC-MS/MS (cps, counts per second).
Glucosylation of ZON prevents binding to the estrogen receptor in vitro.
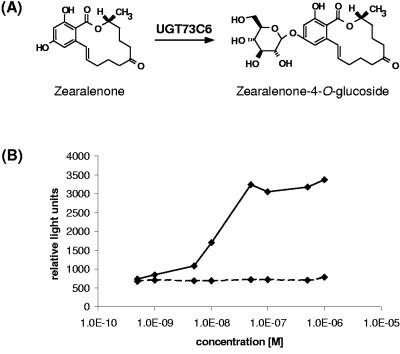
The abilities of ZON and ZON-4-O-Glc to bind to the human estrogen receptor were tested with a commercially available in vitro binding assay. We found that ZON could displace the bound estrogen receptor, leading to a dose-dependent increase in β-galactosidase activity (Fig. 3). However, ZON-4-O-Glc was inactive in the competitive binding assay, which is consistent with the hypothesis that attachment of the glucose moiety to ZON prevents estrogen receptor binding.
FIG. 3.
Glucosylation of ZON reduces its estrogenic activity in vitro. (A) Schematic illustration of the reaction proposed to be catalyzed by UGT73C6. (B) ZON and ZON-4-O-Glc were tested in a competitive estrogen receptor binding assay (HitHunter EFC Estrogen Chemiluminescence Assay Kit). ZON, continuous line; ZON-4-O-Glc, dotted line.
DISCUSSION
We constructed a yeast bioindicator strain that enabled the phenotypic detection of inactivation of zearalenone by a detoxification gene expressed in yeast. In principle, this strain and experimental setup can be used to study the detoxification of other estrogenic substances, e.g., for cloning plant genes involved in the biodegradation of estrogenic pollutants, such as bisphenol A (27). In the constructed strain, both URA3 and ADE2 expression are induced by estrogenic substances. In a typical setup, candidate clones would be selected on 5-fluoroorotic acid plates (selecting for lack of URA3 activation due to reduced estrogenicity of the metabolite formed). The colony color phenotype (lack of ADE2 expression) would then be used in a secondary screen. The constructed strain is compatible with cDNA expression libraries that have either LEU2 or TRP1 as the selectable marker in yeast.
A possible limitation of the system is the stringent requirement for phenotypic change. Highly estrogenic compounds result in estrogen receptor-mediated activation of URA3 and ADE2 expression at very low concentrations. An enzyme metabolizing the estrogenic compound might have little or no activity at that level of the estrogenic compound. The Arabidopsis UGT73C5 and UGT73C6 gene products have very high affinity for ZON and can outcompete the human estrogen receptor for the compound. We also have tested (C. Peterbauer, unpublished data) a lactonohydrolase from the fungus Clonostachys rosea that can inactivate ZON by opening the lactone ring (36). This enzyme is inactive in our assay, probably due to its high Km (>10 μM, or >3 mg/liter).
Comparison of chemically synthesized ZON-4-O-Glc and the yeast product by HPLC-MS/MS indicated that ZON was converted to this glucoside by the Arabidopsis UGT73C5 (data not shown) and UGT73C6 proteins (Fig. 2). At least at the low concentrations used in the yeast in vivo assay, the other four highly similar proteins encoded by the UGT73C cluster do not make this conversion. These results are not consistent with the hypothesis that sequence similarity indicates similar substrate specificities for UGTs. Indeed, such a hypothesis may be a serious oversimplification that can be highly misleading, especially in automated annotations of expressed sequence tags and new genomic sequences. We currently are using the developed yeast system to test hybrids between members of the UGT73C gene family to identify substrate specificity determinants. In addition to ZON and its derivatives, the recombinant UGT73C5 expressed in yeast can catalyze glucosylation of compounds with very different structures, e.g., the trichothecene deoxynivalenol (29) and brassinosteroid plant hormones (30).
The role of ZON in plant-pathogen interactions and the relevance of the detoxification of ZON to ZON-4-O-Glc remain subjects for further studies. The results of microarray experiments showed that ZON produces interesting transcriptional responses in Arabidopsis (39; http://affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid=71), pointing to a possible role for ZON as a suppressor of some plant defense responses. On the other hand, two groups reported recently that disruption of genes required for ZON synthesis did not alter virulence phenotypes (13, 18). As an alternative to concluding that ZON has no role in planta, one could assume that ZON is inactivated efficiently by the tested plants and adds little to the observed phenotypes. Formation of the ZON glucoside seems to be an important mechanism for plants to cope with ZON. The transcriptional response in Arabidopsis is transient, with the exogenous ZON metabolized rapidly to ZON-4-O-Glc and other ZON conjugates by Arabidopsis (1) and wheat (F. Berthiller, unpublished data).
The phenotype in yeast and the in vitro data obtained with the competitive binding assay (Fig. 3) indicate that the ZON-glucoside cannot bind to the human estrogen receptor. However, it can be reactivated in the digestive tracts of animals by glucosidases (14). ZON-4-O-Glc, which may be present in ZON-contaminated wheat (32), is not measured in routine analytical procedures and may be responsible for cases in which higher estrogenic activity is observed in feed than would be expected from ZON measurements. One possibility would be to incorporate a glucosidase treatment of samples into existing analytical procedures. Unfortunately, the commercially available almond β-glucosidase does not efficiently hydrolyze all of the glucosides synthesized by wheat, and the extent of the hydrolysis could alter the accuracy and reliability of the results obtained. A better way to address this problem would be to develop commercially available standards for ZON-4-O-Glc, since it and other glucosides can be easily detected by LC-MS/MS techniques. At present, the chemical synthesis and purification of ZON-4-O-Glc is difficult and time-consuming. The yeast strains developed in this study produce the compound at relatively high yields and could serve as a much cheaper source of ZON-4-O-Glc that would make the detection of this compound economically feasible. Such methodology should further increase the safety of commercial foods and feeds, since the levels of both ZON and its “masked” forms could be reliably detected by routine screening protocols.
Most of the ZON added to a culture of yeast transformed with UGT73C6 was converted to ZON-glucoside and excreted into the medium. We hypothesize that in the pdr5 snq2 mutant strain, ZON is taken up efficiently, rapidly converted to the glucose conjugate in UGT73C6 transformants, and actively exported by an unknown mechanism. Since the enzyme is active in the μg/liter range and ZON was fed in the mg/liter range, very little ZON remained in the medium or was available for side reactions. In the control yeast lacking a UGT gene, a significant portion of ZON was converted to ZOLs (Fig. 2A), probably by the yeast alcohol dehydrogenase. Since our strains take up more ZON, we observed much higher levels of ZOLs than were observed in previous studies with wild-type yeast strains (5). The ZON-glucoside is produced at much higher yields than by chemical synthesis (56%) (16, 31). On a molar basis, about 90% of the added ZON was converted into the glucoside by the UGT73C6-transformed strain, and this compound is easily purified using preparative HPLC.
Acknowledgments
This work was supported by grants from the University of Natural Resources and Applied Life Sciences (BOKU 600.032) and the Austrian Federal Ministry for Education, Science and Culture (Austrian Genome Programme GEN-AU) (GZ 200.051/6-VI/1/2002). B.P. received a DOC fellowship from the Austrian Academy of Sciences.
We thank Karl Kuchler (Medical University Vienna, Max F. Perutz Laboratories) for providing strain YYM4 and Paul Kosma (Department of Chemistry, BOKU) for his assistance in the synthesis of ZON-4-O-Glc. We also thank Rebecca C. Painter, Fiona Doohan, Gerlinde Wiesenberger, and David Steele for critically reading the manuscript.
REFERENCES
- 1.Berthiller, F., U. Werner, M. Sulyok, R. Krska, M. T. Hauser, and R. Schuhmacher. LC-MS/MS determination of phase II metabolites of the mycotoxin zearalenone in the model plant Arabidopsis thaliana. Food Addit. Contam., in press. [DOI] [PMC free article] [PubMed]
- 2.Biehl, M. L., D. B. Prelusky, G. D. Koritz, K. Hartin, W. B. Buck, and H. L. Trenholm. 1993. Biliary excretion and enterohepatic cycling of zearalenone in immature pigs. Toxicol. Appl. Pharmacol. 121:152-159. [DOI] [PubMed] [Google Scholar]
- 3.Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 269:4180-4186. [PubMed] [Google Scholar]
- 4.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 5.Böswald, C., G. Engelhardt, H. Vogel, and P. R. Wallnöfer. 1995. Metabolism of the Fusarium mycotoxins zearalenone and deoxynivalenol by yeast strains of technological relevance. Nat. Toxins 3:138-144. [DOI] [PubMed] [Google Scholar]
- 6.Bowles, D., J. Isayenkova, E. K. Lim, and B. Poppenberger. 2005. Glycosyltransferases: managers of small molecules. Curr. Opin. Plant Biol. 8:254-263. [DOI] [PubMed] [Google Scholar]
- 7.De la Cruz, J., M. C. Daugeron, and P. Linder. 1998. ‘Smart’ genetic screens, p. 267-295. In A. J. P. Brown and M. F. Tuite (ed.), Yeast gene analysis. Methods in microbiology, vol. 26. Academic Press, San Diego, Calif.
- 8.Desjardins, A. E. 2003. Gibberella from a(venaceae) to z(eae). Annu. Rev. Phytopathol. 41:177-198. [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt, G., M. Ruhland, and P. R. Wallnöfer. 1999. Metabolism of mycotoxins in plants. Adv. Food Sci. 21:71-78. [Google Scholar]
- 10.Engelhardt, G., G. Zill, B. Wohner, and P. R. Wallnöfer. 1988. Transformation of the Fusarium mycotoxin zeralenone in maize cell suspension cultures. Naturwissenschaften 75:309-310. [DOI] [PubMed] [Google Scholar]
- 11.European Commission. 2003. SCOOP, task 3.2.10. Collection of occurrence data of Fusarium toxins in food and assessment of dietary intake by the population of EU Member States. European Commission, Directorate-General Health and Consumer Protection, reports on tasks for scientific co-operation, April 2003. [Online.] http://europa.eu.int/comm/food/fs/scoop/task3210.pdf.
- 12.European Commission. 2005. Commission regulation no. 856/2005 of 6 June 2005 amending regulation (EC) no. 466/2001 as regards Fusarium toxins. Official J. Eur. Union L 143:3-8. [Google Scholar]
- 13.Gaffoor, I., D. W. Brown, R. Plattner, R. H. Proctor, W. Qi, and F. Trail. 2005. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 4:1926-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gareis, M., J. Bauer, J. Thiem, G. Plank, S. Grabley, and B. Gedek. 1990. Cleavage of zearalenone-glycoside, a “masked” mycotoxin, during digestion in swine. J. Vet. Med. B 37:236-240. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 16.Grabley, S., M. Gareis, W. Böckers, and J. Thiem. 1992. Glycosylation of mycotoxins. Synthesis 11:1078-1080. [Google Scholar]
- 17.Gueldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y. T., Y. R. Lee, J. Jin, K. H. Han, H. Kim, J. C. Kim, T. Lee, S. H. Yun, and Y. W. Lee. 2005. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 58:1102-1113. [DOI] [PubMed] [Google Scholar]
- 19.Kuiper, G. G., J. G. Lemmen, B. Carlsson, J. C. Corton, S. H. Safe, P. T. van der Saag, B. van der Burg, and J. A. Gustafsson. 1998. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology 139:4252-4263. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper-Goodman, T., P. M. Scott, and H. Watanabe. 1987. Risk assessment of the mycotoxin zearalenone. Regul. Toxicol. Pharmacol. 7:253-306. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y., S. Baldauf, E. K. Lim, and D. J. Bowles. 2001. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276:4338-4343. [DOI] [PubMed] [Google Scholar]
- 22.Lim, E. K., S. Baldauf, Y. Li, L. Elias, D. Worrall, S. P. Spencer, R. G. Jackson, G. Taguchi, J. Ross, and D. Bowles. 2003. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13:139-145. [DOI] [PubMed] [Google Scholar]
- 23.Mahe, Y., Y. Lemoine, and K. Kuchler. 1996. The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J. Biol. Chem. 271:25167-25172. [DOI] [PubMed] [Google Scholar]
- 24.Mirocha, C. J., S. V. Pathre, and T. S. Robison. 1981. Comparative metabolism of zearalenone and transmission into bovine milk. Food Cosmet. Toxicol. 19:25-30. [DOI] [PubMed] [Google Scholar]
- 25.Mitterbauer, R., H. Weindorfer, N. Safaie, R. Krska, M. Lemmens, P. Ruckenbauer, K. Kuchler, and G. Adam. 2003. A sensitive and inexpensive yeast bioassay for the mycotoxin zearalenone and other compounds with estrogenic activity. Appl. Environ. Microbiol. 69:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller, H. M., J. Reimann, U. Schumacher, and K. Schwadorf. 1997. Fusarium toxins in wheat harvested during six years in an area of Southwest Germany. Nat. Toxins 5:24-30. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima, N., Y. Ohshima, S. Serizawa, T. Kouda, J. S. Edmonds, F. Shiraishi, M. Aono, A. Kubo, M. Tamaoki, H. Saji, and M. Morita. 2002. Processing of bisphenol A by plant tissues: glucosylation by cultured BY-2 cells and glucosylation/translocation by plants of Nicotiana tabacum. Plant Cell Physiol. 43:1036-1042. [DOI] [PubMed] [Google Scholar]
- 28.Pierrat, B., D. M. Heery, Y. Lemoine, and R. Losson. 1992. Functional analysis of the human estrogen receptor using a phenotypic transactivation assay in yeast. Gene 119:237-245. [DOI] [PubMed] [Google Scholar]
- 29.Poppenberger, B., F. Berthiller, D. Lucyshyn, T. Sieberer, R. Schuhmacher, R. Krska, K. Kuchler, J. Glossl, C. Luschnig, and G. Adam. 2003. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 278:47905-47914. [DOI] [PubMed] [Google Scholar]
- 30.Poppenberger, B., S. Fujioka, K. Soeno, G. L. George, F. E. Vaistij, S. Hiranuma, H. Seto, S. Takatsuto, G. Adam, S. Yoshida, and D. Bowles. 2005. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102:15253-15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharnhorst, K. 2003. Nachweis von Zearalenon und dem Metaboliten Zearalenon-4-O-β-Glukosid in Mais und Maisprodukten. Diploma thesis. Universität für Bodenkultur Wien und Technische Universität Wien, Vienna, Austria.
- 32.Schneweis, I., K. Meyer, G. Engelhardt, and J. Bauer. 2002. Occurrence of zearalenone-4-β-d-glucopyranoside in wheat. J. Agric. Food Chem. 50:1736-1738. [DOI] [PubMed] [Google Scholar]
- 33.Scientific Committee on Food. 2000. Opinion on Fusarium toxins, part 2: zearalenone. European Commission SCF/CS/CNTM/MYC/22 rev. 3 final. [Online.] http://europa.eu.int/comm/food/fs/sc/scf/out65_en.pdf.
- 34.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi-Ando, N., S. Ohsato, T. Shibata, H. Hamamoto, I. Yamaguchi, and M. Kimura. 2004. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea. Appl. Environ. Microbiol. 70:3239-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 38.Webster, N. J., S. Green, J. R. Jin, and P. Chambon. 1988. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54:199-207. [DOI] [PubMed] [Google Scholar]
- 39.Werner, U. 2005. Characterisation of the effect of the Fusarium mycotoxin zearalenone in Arabidopsis thaliana. Ph.D. thesis. BOKU—University of Natural Resources and Applied Life Sciences, Vienna, Austria.
- 40.World Health Organization. 2000. Zearalenone. Safety evaluation of certain food additives and contaminants, p. 393-482. Prepared by the fifty-third meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). WHO Food Additives Series 44. World Health Organization, Geneva, Switzerland.
- 41.Zill, G., G. Engelhardt, B. Wohner, and P. R. Wallnhöfer. 1990. The fate of the Fusarium mycotoxin zearalenone in maize suspension cultures. Mycotoxin Res. 6:31-48. [DOI] [PubMed] [Google Scholar]