Abstract
Bacteroides species are promising indicators for differentiating livestock and human fecal contamination in water because of their high concentration in feces and potential host specificity. In this study, a real-time PCR assay was designed to target Bacteroides species (AllBac) present in human, cattle, and equine feces. Direct PCR amplification (without DNA extraction) using the AllBac assay was tested on feces diluted in water. Fecal concentrations and threshold cycle were linearly correlated, indicating that the AllBac assay can be used to estimate the total amount of fecal contamination in water. Real-time PCR assays were also designed for bovine-associated (BoBac) and human-associated (HuBac) Bacteroides 16S rRNA genes. Assay specificities were tested using human, bovine, swine, canine, and equine fecal samples. The BoBac assay was specific for bovine fecal samples (100% true-positive identification; 0% false-positive identification). The HuBac assay had a 100% true-positive identification, but it also had a 32% false-positive rate with potential for cross-amplification with swine feces. The assays were tested using creek water samples from three different watersheds. Creek water did not inhibit PCR, and results from the AllBac assay were correlated with those from Escherichia coli concentrations (r2 = 0.85). The percentage of feces attributable to bovine and human sources was determined for each sample by comparing the values obtained from the BoBac and HuBac assays with that from the AllBac assay. These results suggest that real-time PCR assays without DNA extraction can be used to quantify fecal concentrations and provide preliminary fecal source identification in watersheds.
The determination of the sources of fecal pollution is a critical issue in complying with the Clean Water Act (Federal Water Pollution Control Act amendments of 1973 and 1977). A particular need is the ability to differentiate fecal microbial contamination of water resulting from animal operations versus that from human sources, such as leaking septic tanks, sewer overflows, or illegal discharges, and wildlife (13, 38). The use of fecal bacteria to determine the host animal source of fecal contamination is based on the assumption that certain strains of fecal bacteria are associated with specific host animals and that strains from different host animals can be differentiated based on phenotypic or genotypic markers (38, 43). Escherichia coli has been used as an indicator microorganism for fecal source tracking because it is easily cultured and is used as the primary regulatory indicator for pathogen contamination in recreational waters (38, 42). Problems associated with using E. coli as a source identifier include a high degree of genetic diversity not attributable to a specific host animal source, the potential for E. coli to replicate outside of the host, and geographic and temporal variabilities (43). Bacteria belonging to the genus Bacteroides have been suggested as alternative fecal indicators to E. coli or fecal coliforms (14, 22) because they make up a significant portion of the fecal bacterial population (25), have little potential for growth in the environment (14, 23), and have a high degree of host specificity that likely reflects differences in host animal digestive systems (11). The approach for using Bacteroides spp. as indicators of the type of host animal serving as the source of fecal pollution differs from the approach used for E. coli in two significant ways. First, no attempt is made to culture individual Bacteroides isolates; the whole Bacteroides population in the fecal sample is examined. Second, Bacteroides-based methodologies are designed to target specific diagnostic sequences within the Bacteroides 16S rRNA gene present in feces from different animals (4-7, 10, 11, 22, 37). The goal of directly targeting genotypes is to design assays that are specific for the host animal regardless of geographic location. PCR primers targeting the Bacteroides 16S rRNA gene have been designed to differentiate human- and ruminant-associated Bacteriodes (4, 22) and, more recently, to identify swine- and equine-associated Bacteroides 16S rRNA genes (11). Real-time PCR with fluorogenic probes is faster than traditional PCR and offers the user the ability to simultaneously identify and quantify specific genes, thus making real-time PCR a diagnostic tool of choice for measuring bacteria in food, water, and fecal and tissue samples (3, 16, 21, 26, 33, 34, 36). Multiple real-time PCR assays targeting different members of a bacterial community can also be used to measure microbial population dynamics because of the large number of samples that can be assayed quickly (2, 24). However, nucleic acid extraction is one step in the use of real-time PCR that slows sample analysis, increases costs, and is a source of variability in real-time PCR (12). In water samples with low concentrations of humic acids or other PCR inhibitors, it may be possible to use direct PCR (15) without DNA extraction, which would improve the speed of sample analysis and minimize variability introduced by DNA extraction.
Bacteria typically comprise approximately one-third of feces by weight (25), and Bacteroides organisms make up approximately 30 to 40% of the amount of total fecal bacteria (18, 20, 26, 31, 35, 44); therefore, Bacteroides may comprise approximately 10% of the fecal mass and thus provide an abundant target for identifying fecal contamination. Thus, quantification of the Bacteroides 16S rRNA genes may provide a reliable and accurate method to estimate fecal concentrations in water samples. In this study, a real-time PCR assay was designed to detect Bacteriodes 16S rRNA genes present in all mammalian fecal samples and determine whether the quantity of Bacteroides 16S rRNA genes present in a water sample was related to the fecal concentration. Other real-time PCR assays were designed to detect Bacteriodes 16S rRNA genes present in bovine or human feces. This study differs from a recently published study that used a real-time PCR assay for the detection of Bacteroides in waste water treatment plants to quantify Bacteroides 16S rRNA genes but did not attempt to differentiate between fecal sources or quantify fecal concentrations (10). The assays developed in the current study were tested against cloned Bacteroides 16S rRNA gene sequences, DNA extracted from fecal samples, and fecal samples without DNA extraction to determine the specificities and sensitivities of the assays. Finally, the three assays were used to estimate the amount of fecal contamination and the percentage of contamination attributable to bovine or human sources in surface water samples from three watersheds.
MATERIALS AND METHODS
Fecal samples and construction of Bacteroides 16S rRNA gene libraries.
Individual fresh fecal samples were collected from apparently healthy human and animal sources. Bovine feces were obtained from pastured animals in Tennessee, Texas, and Pennsylvania. Bovine fecal sources included beef and dairy cattle and cattle of different breeds, including Hereford and Jersey, as well as adults and calves. Canine samples were obtained from local pet owners and represented several different breeds. Equine fecal samples were obtained from local horse owners and the University of Tennessee animal science farm. All swine fecal samples originated from the same farm in Tennessee. For all animal types, feces from individual animals were mixed separately in a volume of sterile distilled water equal to the weight of the feces and frozen at −80°C until processed. For DNA extraction, the fecal samples were thawed on ice and diluted another 10-fold in sterile distilled water for processing with the FastDNA SPIN kit for soil (Qbiogene, Carlsbad, CA). For each extraction, 300 μl of fecal slurry was mixed in lysis matrix E tubes and processed following the manufacturer's protocols. The final product was 50 μl of application-ready DNA.
Bacteroides 16S rRNA genes from fecal DNA extracts were amplified using 20 pmol of the primers Bac32F and Bac708R (4) and 2 μl of DNA extract in a 25-μl total volume with ready-to-go PCR beads (Amersham Pharmacia, Piscataway, NJ). Amplification was performed using a touch-down temperature protocol consisting of 5 min at 94°C, followed by 10 cycles at 94°C for 15 s, 65°C for 45 s (decreasing 1°C per cycle), and 72°C for 60 s, followed by 30 cycles consisting of 94°C for 15 s, 55°C for 45 s, and 72°C for 60 s, ending with a final extension time of 10 min at 72°C. The PCR product was cloned into the pCR4.0 TOPO vector, transformed into chemically competent Escherichia coli one-shot TOP10 cells, and selected on LB plates containing 50 μg/ml kanamycin according to the manufacturer's instructions (TA cloning kit; Invitrogen, Carlsbad, CA). Plasmids were isolated from individual colonies and screened for the presence of inserts using EcoRI restriction digests. Complete plasmid inserts (approximately 675 bp) were initially sequenced in one direction using M13f or M13r primers at the Molecular Biology Resource Center at the University of Tennessee. DNA sequences were compared to DNA sequences at the National Center for Biotechnology Information (NCBI) by using the BLAST program (1) and were aligned in Clustal X (version 1.64b) (41). Phylogenetic trees were displayed using TreeView (30). Selected 16S rRNA genes were resequenced in both directions to verify sequences.
Real-time PCR assays.
Gene targets as well as the probe and primer sequences and amplicon size for the three real-time PCR assays used in this study are summarized in Table 1. The Bacteroides species (AllBac), human-associated (HuBac), and bovine-associated (BoBac) assays were designed from alignments of partial Bacteroides 16S rRNA genes obtained from fecal source libraries and sequences available in GenBank. The DNA sequence regions chosen were conserved in all Bacteroides species or conserved in only Bacteroides species from bovine or human fecal samples. From these DNA sequence regions, primers and probes were selected based on the guidelines provided by Applied Biosystems (Foster City, CA). Oligonucleotide melting temperatures and self-complementarity were determined using the oligonucleotide properties calculator (www.basic.northwestern.edu/biotools/oligocalc.html). Oligonucleotide specificity for all Bacteroides 16S rRNA genes or for human-associated and bovine-associated Bacteroides 16S rRNA genes was verified using the BLAST program at the NCBI (1) and the probe match program of the Ribosomal Database Project (8). Oligonucleotide primers and 6-carboxyfluorescein (FAM)-BHQ probes were obtained from Biosearch Technologies.
TABLE 1.
Real-time PCR assays used to detect Bacteriodes 16S rRNA genes, the primers and probe used for each assay, and the annealing temperature used for each assay
| Assay | Primer/probe name and sequence (5′-3′)a | Size (bp) of product | Annealing temp (°C) |
|---|---|---|---|
| AllBac (all Bacteroides) | AllBac296f, 5′-GAGAGGAAGGTCCCCCAC-3′ | 106 | 60 |
| AllBac412r, 5′-CGCTACTTGGCTGGTTCAG-3′ | |||
| AllBac375Bhqr, 5′-(FAM)CCATTGACCAATATTCCTCACTGCTGCCT(BHQ-1)-3′ | |||
| BoBac (bovine cluster of Bacteroides) | BoBac367f, 5′-GAAG(G/A)CTGAACCAGCCAAGTA-3′ | 100 | 57 |
| BoBac467r, 5′-GCTTATTCATACGGTACATACAAG-3′ | |||
| BoBac402Bhqf, 5′-(FAM)TGAAGGATGAAGGTTCTATGGATTGTAAACTT(BHQ-1)-3′ | |||
| HuBac (human cluster of Bacteroides) | HuBac566f, 5′-GGGTTTAAAGGGAGCGTAGG-3′ | 116 | 60 |
| HuBac692r, 5′-CTACACCACGAATTCCGCCT-3′ | |||
| HuBac594Bhqf, 5′-(FAM)TAAGTCAGTTGTGAAAGTTTGCGGCTC(BHQ-1)-3′ |
Numbers within the primer/probe name indicate the nucleotide position within the Bacteroides 16S rRNA gene.
All real-time PCR assays were performed using QuantiTect PCR mix (QIAGEN, Valencia, CA), with 15 pmol of the primer and 5 pmol of the probe. PCR assays were run with three different sample types. First, plasmid DNA containing 16S rRNA genes from Bacteroides were run as standards using 10-fold dilutions of the plasmid ranging from 2.5 × 107 copies to 25 copies per PCR. Second, 0.1 to 3 ng genomic DNA extracted from fecal samples was added in 2.5-μl volumes. Third, 0.25-ng to 2.5-μg fecal samples without DNA extraction were added in 2.5-μl volumes to the PCRs. PCR amplification protocols consisted of 50°C for 2 min, followed by 95°C for 10 min and up to 50 cycles of 95°C for 30 s and 57°C (BoBac assay) or 60°C (AllBac and HuBac assays) for 45 s. PCR amplification and detection of the fluorescent signal was performed using the DNA Engine Opticon continuous fluorescence detection system (MJ Research, Waltham, MA). The threshold cycle (CT) value for all measurements was determined as the cycle at which fluorescence reached 5 standard deviations above the background, averaged over 5 cycles collected within the first 15 cycles of PCR amplification. For all PCR runs, standards, negative controls (no DNA), and samples were run in triplicate. Gene copies or fecal concentrations were calculated from standard curves based on the log transformation of known concentrations versus the threshold cycle. Linear correlations were determined using SigmaPlot 2002 (version 8.02) (SPSS).
Determination of fecal concentration in water samples without DNA extraction.
A bovine fecal slurry sample was diluted and mixed thoroughly in sterile distilled water to result in a fecal concentration of 10,000 mg of feces/liter of water (mg/liter). The reproducibility of measuring fecal concentration in water samples was determined by performing a series of 1:5 dilutions on a bovine fecal sample with a starting concentration of 3,000 mg/liter. Triplicate 0.5-ml samples were frozen in 1.5-ml tubes at −80°C. Direct PCR using the AllBac assay was performed on thawed samples on three separate dates, and samples were refrozen between assays.
Application of real-time PCR assays to creek water samples.
Single water samples (approximately 250-ml grab samples) were obtained from three creeks with different land use patterns. The Tennessee Department of Environment and Conservation (9) lists portions of all three watersheds on the 303(d) list for not meeting recreational water quality use as determined by E. coli measurements (geometric mean of five samples in 30 days of >126 CFU/100 ml or a single value of >487 CFU/100 ml). Land use in one watershed (NS-1 and NS-3 sites) is a mix of animal grazing and rural and small subdivision housing. Land use around the second site (U2) is urban. The third watershed, containing sites R07 and R20, is a mixture of resort development and undeveloped forest land.
The ColiBlue24 assay (MEL/MF total coliform lab; Hach Company, Ames, IA) was performed to determine the concentrations of E. coli and total coliforms in CFU/100 ml. Samples (100 μl to 1,000 μl) were diluted in 50 ml phosphate-buffered saline and collected by vacuum filtration on a membrane filter (diameter, 47 mm; pore size, 0.45 μm) placed on top of a filter funnel. The sides of the funnel were washed with 25 ml phosphate-buffered saline, and excess liquid was removed by suction. The filter membrane was placed on an absorbent pad in a petri dish soaked with 1 ampoule of m-ColiBlue 24 broth. All assays were performed in triplicate. The petri dishes were incubated at 35°C for 20 h. The colonies on the plates were enumerated, with blue colonies indicating E. coli and the sum of the red colonies plus the blue colonies indicating coliforms.
Direct PCR without DNA extraction (15) was performed on 2.5-μl creek water samples in 25-μl PCRs containing QuantiTect master mix and primers and probes as described above. Sterile Tris buffer (10 mM) was used as a negative control. In addition to the test samples, each assay plate also contained two types of standard curves, a plasmid dilution standard curve and a fecal dilution standard curve. Each dilution was run in triplicate for both standard curves. For the AllBac and HuBac assays, human fecal samples ranging in concentration from 5,000 mg/liter to 0.32 mg/liter were used as the standard for calculating the concentration of total feces and human-associated feces in each sample. For the BoBac assay, a bovine fecal sample ranging in concentration from 10,000 mg/liter to 1.0 mg/liter was used as the standard for calculating the concentration of bovine-associated feces in each sample. For each assay, the fecal concentration was determined using triplicate 2.5-μl creek water samples. The potential for PCR inhibition was measured by adding 2.5 × 105 copies of plasmid DNA to a fourth well containing 2.5 μl of the creek water sample. The amount of PCR inhibition was measured by determining the recovery of the copies in the presence of the creek water sample as calculated from the plasmid DNA standard curve [percent recovery = (measured copies in water sample spiked with 2.5 × 105 plasmid copies − measured copies in unspiked water sample)/(2.5 × 105) × 100]. The percentage of plasmid recovery was measured in each creek water sample using all three real-time PCR assays, and the means and standard deviations were determined.
Nucleotide sequence accession numbers.
Bacteroides 16S rRNA gene sequences from fecal samples were deposited into GenBank and received accession numbers AY597127 through AY597206.
RESULTS
Analysis of Bacteroides 16S rRNA genes from animal fecal samples.
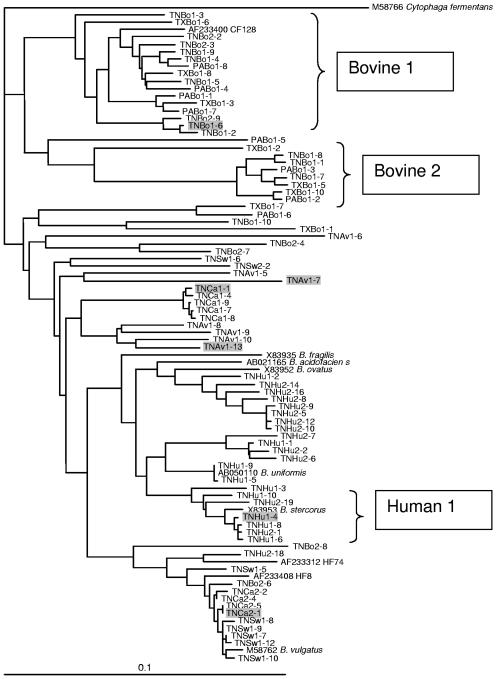
Bacteroides 16S rRNA gene libraries were constructed using DNA extracted from one chicken (avian), two equine, two canine, two human, two swine, and four bovine fecal samples. All of the sequences from the human, avian, and canine libraries and 97% of the sequences from the bovine libraries had greater than 90% similarity to 16S rRNA gene sequences published in GenBank (NCBI). Based on alignment of the clone sequences, the clones were separated into Bacteroides-like and Prevotella-like categories. All of the sequences isolated from equine fecal samples were Prevotella-like, whereas none of the sequences obtained from human samples were Prevotella-like. Prevotella-like sequences from the other fecal sources ranged from 6% in bovines to 40% in swine. Phylogenetic analysis of the Bacteriodes-like 16S rRNA sequences demonstrated that the sequences from bovine fecal samples grouped into two distinct clusters, bovine 1 and bovine 2, with the bovine 1 cluster containing sequences from all four bovine fecal libraries (Fig. 1). Approximately one-third of the 16S rRNA gene sequences obtained from each bovine fecal sample were closely related (>95% similarity) to uncultured Bacteroides sequence AF233400 (C123) from bovine feces in Oregon (5). Bacteroides 16S rRNA gene sequences obtained from swine, canine, and human fecal samples did not form distinct clusters. For instance, five sequences from the TNSw1 (swine) sample were 99% similar to four sequences from the TNCa2 (canine) sample. In addition, these nine sequences were greater than 98% similar to Bacteroides vulgatus (M58762) and 97% similar to the HF8 from a human fecal sample (5). However, one cluster of sequences containing sequences from both human fecal sample libraries (Fig. 1) was found and was used to design the human-associated real-time PCR assays.
FIG. 1.
Phylogenetic dendrogram showing the relationship of cloned Bacteroides 16S rRNA gene sequences from different animal fecal sources. For each clone in this study, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas), the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Av, chicken; Ca, dog; Sw, swine; and Hu, human), and the final number indicates the individual clone within the library. Sequences were aligned, and a bootstrap consensus tree was created using Clustal X (version 1.64b). The root was determined using the 16S rRNA gene sequence from Cytophaga fermentans (M58766) as an outgroup. References for cultured and uncultured Bacteroides or 16S rRNA gene sequences indicated on the tree were M58766 and M58762 (17); X83935, X83952, and X83953 (32); AB050110 (Y. Miyamoto, unpublished data) and AB021165 (27); and AF233400 and AF233408 (5). Plasmids containing the shaded sequences were used to determine the effect on sequence mismatches on real-time PCR assays in Fig. 2.
Design of real-time PCR assays.
Based on DNA sequences obtained from the Bacteroides libraries, the AllBac PCR assay was designed with no mismatches to both the human- and bovine-derived Bacteroides 16S rRNA gene sequences. The primers and probe were later found to have no mismatches to Bacteroides 16S rRNA gene sequences obtained from avian (chicken), canine, and swine fecal samples. The probe check program of the Ribosomal Database Project (8) was used to determine the specificity of the AllBac primer and probe sequences. The forward and reverse primers and probe had perfect homology (no base pair mismatches) to 4,181 (94%), 4,069 (93%), and 4,181 (94%) of the 4,445 classified Bacteroides genus 16S ribosomal genes, respectively. The primers and probes were also evaluated for no mismatches to nontarget 16S ribosomal genes. The forward and reverse primers and probe had no mismatches to 162 (2%), 0, and 803 (10%) of the 8,228 16S rRNA gene sequences present in other classes of bacteria within the Bacteroides phylum and had no mismatches to only 11 (<0.1%), 6 (<0.1%), and 13 (<0.1%) of the 172,026 16S rRNA gene sequences belonging to phyla other than Bacteroides. These combined results indicate that the primers and probes had a high specificity to 16S rRNA gene sequences belonging to Bacteroides genus and very little cross-hybridization to bacteria outside of the Bacteroides class.
The bovine real-time PCR assay (BoBac assay) was designed to target the group of sequences in the bovine 1 cluster in Fig. 1. The BoBac primers and probe had no mismatches to six clones from TN-Bo1, three clones from TN-Bo2, three clones from TX-Bo1, and four clones from PABo-1 libraries. The BoBac primers and probe also had zero mismatches to clone C157 (AF233401) present in GenBank (5). The BoBac primer and probe sequences had at least six mismatches to the Bacteroides 16S rRNA gene sequences obtained from human feces and other nonbovine animal feces.
The human real-time PCR assay (HuBac PCR assay) was designed to match the human 1 cluster of human-associated Bacteroides (Fig. 1). The primer and probe sequences also had no mismatches to the following Bacteroides 16S rRNA gene sequences in GenBank: B. eggerthi (AB050107), B. stercoris (X83953), and B. uniformis (AB050110). At the time of primer and probe design, the HuBac primers and probe had at least one mismatch to Bacteroides 16S rRNA gene sequences obtained from other animal feces.
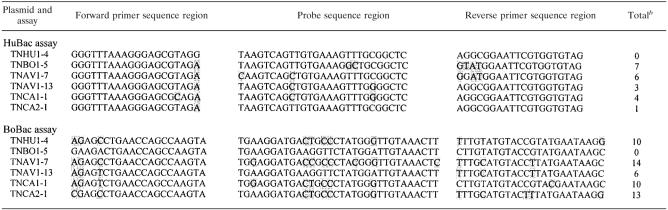
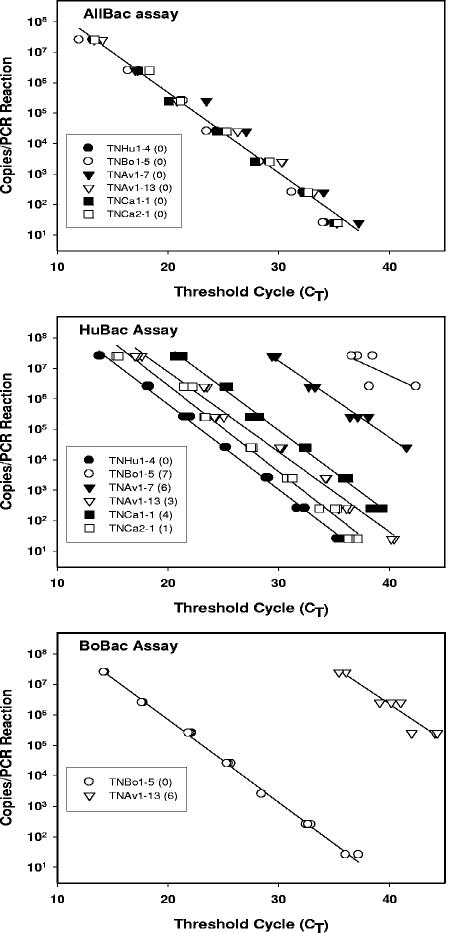
The effect of the total number of mismatches present in the primers and probe on PCR amplification efficiency and quantification was determined for the HuBac and BoBac assays. Six cloned Bacteroides 16S rRNA gene sequences were identified as having a total of 1 to 14 mismatches to the primers and probes designed for either the BoBac or the HuBac assay (Table 2). A series of 10-fold dilutions of the six selected plasmids were made, resulting in 25 to 2.5 × 107 16S rRNA gene targets/PCR. The consistency of the AllBac assay and plasmid dilutions was demonstrated by the linear regression fit of r2 = 0.98 to all six plasmid dilutions, with a slope of −0.26 and a Y intercept of 10.95 (Fig. 2).
TABLE 2.
Location of nucleotide mismatches to primers and probes used for Bacteroides real-time PCR assaysa
Mismatches are indicated by shading.
Total number of mismatches.
FIG. 2.
The effect of sequence mismatches on PCR amplification in real-time PCR assays. Serial dilutions of six different plasmids were performed to generate standard curves from 2.5 × 107 copies to 25 copies. Real-time PCR assays were performed as follows: top, AllBac assay with 0 sequence mismatches to all plasmids; middle, HuBac assay with 0 to 7 sequence mismatches; and bottom, BoBac assay with 0 to 14 sequence mismatches. Base pair mismatches between each plasmid and the primer and probe used in the real-time PCR assay are in parentheses in the legend boxes. Plasmids having more than seven mismatches to the primers and probe did not amplify.
The efficiency of PCR amplification, as indicated by the slope of the line as a function of copies versus CT, did not decrease as the number of mismatches increased from none to six (Fig. 2). However, PCR amplification was less efficient, with seven mismatches (HuBac assay with TNBo1-5 plasmid), and no PCR amplification occurred when more than seven mismatches were present.
Although the PCR amplification efficiency did not change with plasmid and assay combinations having zero to six mismatches, the threshold cycles for each plasmid concentration increased compared to the plasmid assay combination with zero mismatches as the number of mismatches increased. Thus, the number of product copies obtained in each PCR decreased with an increasing number of mismatches in the primers and probes. This was particularly evident for the HuBac assay, which had one to six sequence mismatches to plasmids TNCa2-1 and TNAV1-7, respectively (Fig. 2). The percent product yield relative to the no-mismatch control was calculated for each plasmid with one to seven mismatches to the primers and probe (Table 3). These results indicated that the decrease in product increases with the sum of the mismatches in the primers and probe. Thus, a single mismatch in either the primer or the probe resulted in an approximately 66% reduction in the product relative to that with no mismatches in the primers or probe. The amount of product obtained from plasmids with six or more mismatches to the primers and probe was significantly less than 1% relative to the plasmids having no mismatches to the primers and probe.
TABLE 3.
Percentages of product yield from PCR with plasmids containing zero to seven mismatches to the primers and probe in a real-time PCR assay
| Plasmid | Assay | No. of mismatches | % Product yielda |
|---|---|---|---|
| TNHu1-4 | HuBac | 0 | 100 |
| TNBo1-5 | BoBac | 0 | 100 |
| TNCa2-1 | HuBac | 1 | 33.5 ± 18 |
| ANAv1-13 | HuBac | 3 | 7.6 ± 5.8 |
| TNCa1-1 | HuBac | 4 | 3.2 ± 1.5 |
| TNAv1-7 | HuBac | 6 | 4.2 × 10−3 ± 2.0 × 10−3 |
| TNAv1-13 | BoBac | 6 | 9.7 × 10−3 ± 2.6 × 10−3 |
| TNBo1-5 | HuBac | 7 | 2.1 × 10−5 ± 1.9 × 10−5 |
Means and standard deviations of the percent product yield for each plasmid concentration ranging from 25 to 2.5×107 copies. Percent product yield for each plasmid concentration = (copies calculated from 0 mismatch standard curve/expected copies)×100.
Determination of fecal concentration in water samples without DNA extraction.
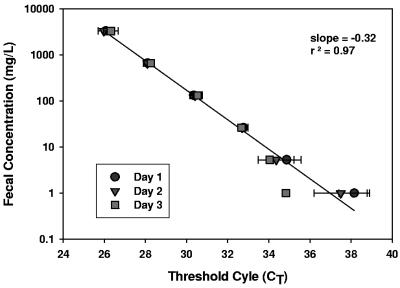
The AllBac assay was tested as a method to calculate the concentration of fecal contamination in a water sample without DNA extraction. Known amounts of bovine feces were added to water samples, resulting in concentrations ranging from 0.3 to 10,000 mg feces/liter of water. The samples were frozen, thawed, and assayed on three separate days (Fig. 3). In this experiment, the concentration of feces and the CT were linear over 3 orders of magnitude, with a detection limit of 1 mg/liter (Fig. 3). Variability was low at concentrations greater than 10 mg/liter but increased markedly below this value, indicating that fecal concentration measurements below 10 mg/liter will be less precise. Assays performed on three separate days were highly reproducible, with a combined r2 of 0.96, suggesting that repeated freezing and thawing did not negatively impact sample integrity.
FIG. 3.
Threshold cycle measurements using the AllBac real time PCR assay in water samples containing bovine feces. Real-time PCR assays were performed on three separate days using triplicate samples for each dilution.
Discriminatory capability of assays with extracted DNA.
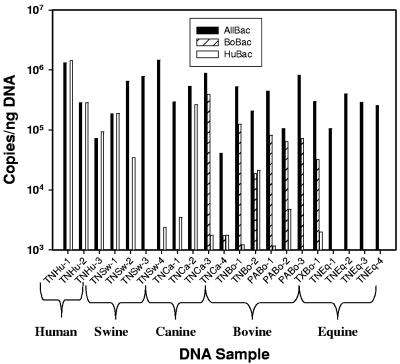
Real-time PCR assays for Bacteroides 16S rRNA genes were used to quantify the relative amount of Bacteroides in DNA extracted from three human, four swine, four canine, four equine and six bovine fecal samples. The Bacteroides 16S rRNA gene concentration determined using the AllBac assay was fairly consistent in DNA extracted from the fecal samples, with a mean of 4.7 (±3.9) × 105 copies per nanogram DNA for all samples (Fig. 4). This suggests that the AllBac assay may be used as a general assay for determining the Bacteroides spp. concentration in feces from a range of mammals.
FIG. 4.
Concentration of ribosomal genes in DNA extracts from animal fecal samples determined by the AllBac, HuBac, and BoBac assays. For each sample, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas) and the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Ca, dog; Sw, swine; and Hu, human).
The HuBac assay measured 5.1 (±6.2) × 105 copies of human-associated Bacteroides 16S rRNA genes per nanogram of DNA in three human fecal samples. These values were similar to the results from the AllBac assay, suggesting that the HuBac assay detected the majority of Bacteroides organisms in human fecal samples. However, the HuBac assay also measured more than 1 × 105 Bacteroides 16S rRNA gene copies per nanogram of DNA in one swine fecal sample (25% of the samples) and one canine fecal sample (25% of the samples), indicating that either the Bacteroides strains in these hosts were similar to Bacteroides strains in humans or that the Bacteroides strains in these hosts had 16S rRNA genes with few mismatches to the HuBac primers and probe. The HuBac assay measured more than 1 × 104 Bacteroides 16S rRNA gene copies per nanogram of DNA in one bovine sample and between 1 × 103 and 1 × 104 Bacteroides 16S rRNA gene copies per nanogram of DNA in other bovine samples, indicating cross-amplification of <1 to 10% with bovine-associated Bacteroides 16S rRNA genes by the HuBac assay.
The BoBac assay measured 6.6 (±3.8) × 104 copies of bovine-associated Bacteroides 16S rRNA genes per nanogram of DNA in six bovine fecal samples (Fig. 4). This represented approximately 20% of the total Bacteroides genes found in cattle, suggesting that other Bacteroides genes also exist in bovine fecal samples. The BoBac assay was more specific than the HuBac assay, with only one canine sample (25% of the canine samples) showing potential cross amplification of more than 1 × 105 copies per nanogram of DNA (Fig. 4).
Discriminatory capability of assays without DNA extraction.
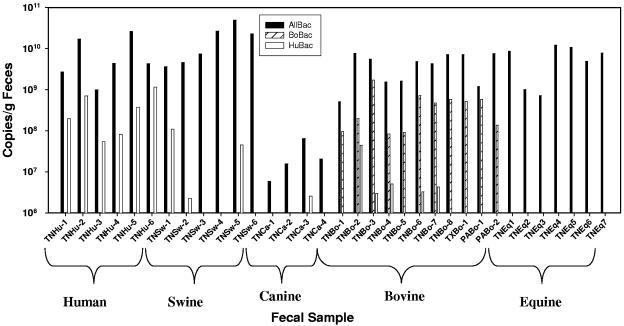
Real-time PCR assays for Bacteroides genes were used to quantify the relative amount of Bacteroides genes in 6 human, 6 swine, 4 canine, 7 equine, and 11 bovine fecal samples (Fig. 5). In these experiments, 600 to 2,000 mg feces/liter water were analyzed. Gene copies were calculated for each PCR using standard curves generated from plasmid DNA and normalized to gram (wet weight) of feces. For the most part, the results of the real-time PCR assays on the fecal samples without DNA extraction were similar to the results of the real-time PCR assays on DNA extracts from fecal samples. However, several differences were noted. First, the AllBac signal was considerably lower in the canine samples (mean of 2.7 × 107 copies/g feces) than were the mean values from fecal samples from all other species (means range from 4.5 × 109 to 1.9 × 1010 copies/g feces) (Fig. 5). Given that concentrations of Bacteroides 16S rRNA genes in the feces of various animal species were similar (Fig. 4), this result suggests that canine fecal samples may have been less efficiently lysed during PCR amplification than were other fecal samples. The HuBac and BoBac assays measured greater than 107 copies rRNA genes/g of feces in the human and bovine fecal samples and represented 2 to 30% of the total Bacteroides rRNA genes as measured by the AllBac assay (Fig. 5).
FIG. 5.
Concentration of ribosomal genes in unextracted animal fecal samples determined by the AllBac, HuBac, and BoBac assays. For each sample, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas) and the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Ca, dog; Sw, swine; and Hu, human).
The results from Fig. 5 were used to determine the percentages of fecal samples that would be correctly and incorrectly identified. Samples were considered to be correctly classified if >107copies/g of feces were detected in the proper animal host (e.g., human and bovine fecal samples for the HuBac and BoBac assays, respectively). Samples were considered to be incorrectly classified if >106 copies/g of feces (minimum detection limit) were detected in the nonhost fecal samples for each assay. Both assays had a 100% correct identification rate toward their target fecal samples, but as seen with the DNA extracts, the HuBac had a higher rate of incorrect classification (32%) than did the BoBac assay (0%). The HuBac assay measured greater than 106 copies/g in three swine fecal samples, one canine fecal sample, and five bovine fecal samples, whereas the BoBac assay did not detect more than 106 copies/g of feces in any nonbovine fecal samples.
Generation and validation of human and bovine fecal standard curves.
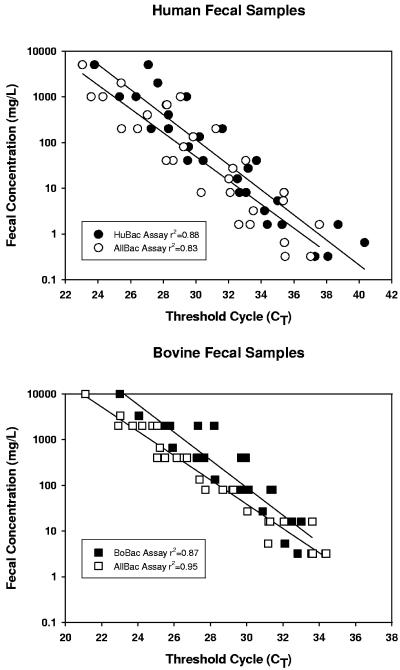
The ability to measure fecal concentrations without calculating gene copies was determined using dilution series of five human fecal slurries from 5,000 mg/liter to 0.32 mg/liter and six bovine fecal slurries from 10,000 mg/liter to 3.2 mg/liter. The human fecal slurries were assayed using the AllBac and HuBac assays. The bovine slurries were analyzed using the BoBac assay and AllBac assays. The amplification efficiencies for all sample types by assay were similar (Fig. 6). However, the detection limit in the human fecal samples was lower (0.3 mg/liter) with both the HuBac and AllBac assays than the detection limit was for the bovine fecal samples (3 mg/liter) with either the BoBac or the AllBac assay. The potential for PCR inhibition by fecal samples was determined by adding 2.5 × 105 copies of the plasmid TNBo1-5 to the PCR wells containing 2.5 μl of a human fecal slurry (ranging in concentration from 5,000 to 0.32 mg/liter) and 2.5 μl of canine fecal slurry (ranging in concentration from 10,000 to 0.64 mg/liter). These fecal samples were chosen because the canine fecal sample and the human fecal sample do not cross-hybridize with the BoBac assay. Real-time PCR was performed using the BoBac assay, and copies in each well were determined based on the addition of approximately 2.5 × 105 copies of the TNBo1-5 plasmid (Table 4). An analysis of variance (ANOVA) was performed on the number of plasmid copies recovered in the samples containing human fecal dilutions and in the control, and a separate ANOVA was performed on the number of plasmid copies recovered in the samples containing the canine fecal dilutions. ANOVA of the human fecal data set indicated that there were no significant differences in the numbers of plasmid copies measured in any samples (P = 0.05). ANOVA of the canine fecal data set indicated that there was a significant difference at a P level of 0.05 but not a P level of 0.01. Further analysis of the canine data set indicated that this difference resulted exclusively from the sample containing 2,000 mg/liter feces. However, the higher-than-expected number of plasmid copies obtained in this sample is not likely due to significant cross-hybridization because there is no increasing trend with fecal concentration. The percent recovery for each sample was calculated by dividing the mean of the plasmid copies measured in samples with feces by the control. For all fecal concentrations, the mean percentages of recovery of plasmid were 96 (±20)% and 91 (±21)% in human and canine fecal dilutions, respectively, suggesting that the feces did not significantly inhibit PCR amplification.
FIG. 6.
Comparison of AllBac and HuBac assays performed on serial dilutions from five individual human fecal samples (top), and the AllBac and BoBac assays run at 57°C on serial dilutions of six individual bovine fecal samples (bottom).
TABLE 4.
Recovery of 2 ×105 plasmid DNA copies spiked into human and canine fecal dilutions
| Fecal source | Sample fecal concn (mg/liter) | Measured no. of recovered DNA copies | % PCR amplification of plasmid relative to controla |
|---|---|---|---|
| Human | 5,000 | 2.6 (±0.02) × 105 | 91 |
| 1,000 | 3.3 (±1.0) × 105 | 116 | |
| 200 | 2.7 (±0.2) × 105 | 96 | |
| 40 | 3.7 (±0.8) × 105 | 124 (±29) | |
| 8 | 2.9 (±0.1) × 105 | 99 (±4.5) | |
| 1.6 | 2.6 (±0.5) × 105 | 89 (±16) | |
| 0.32 | 2.9 (±0.4) × 105 | 99 (±13) | |
| Canine | 10,000 | 2.2 (±0.4) × 105 | 75 (±15) |
| 2,000 | 3.6 (±0.8) × 105 | 123 (±26) | |
| 400 | 2.9 (±0.1) × 105 | 100 (±4.1) | |
| 80 | 2.7 (±0.2) × 105 | 91 (±8.3) | |
| 16 | 2.3 (±0.6) × 105 | 78 (±20) | |
| 3.2 | 2.4 (±0.4) × 105 | 82 (±14) | |
| 0.64 | 2.6 (±0.4) × 105 | 88 (±15) | |
| Control | 0 | 2.8 (±0.1) × 105 | 100 |
Percent PCR amplification = (measured no. of copies in the presence of feces/measured no. of copies in the control) × 100.
Estimating total, bovine-associated, and human-associated fecal concentrations in surface water samples.
E. coli concentrations and fecal concentrations using the AllBac, BoBac, and HuBac assays were measured at two separate locations in a rural watershed (NS-1 and NS-3) at low and high water flows, one location in an urban area (U2), and in two locations in a resort area (R07 and R20). In the rural watershed, the expected primary sources of fecal contamination were cattle from small grazing operations and human fecal contamination from failing or leaking septic tanks. No swine or poultry operations were present in this watershed; however, horses and wildlife were present and may be contributors to fecal contamination. In the urban area, the primary source of fecal contamination was expected to be human via sewer line leaks or overflows. Humans were also expected to be the main source of fecal contamination in the resort area. The R20 site is in a nonsewered area, so fecal contamination may be through straight pipe discharges to the stream or through failing septic tanks. The other site, R07, was in a forested area traversed by sewer lines.
In these water samples, E. coli concentrations ranged from below the state-recommended single sample limit of 487 CFU/100 ml for recreational water use during low-flow conditions to E. coli concentrations 100-fold greater than the single-sample limit after a rainfall event (high-flow conditions) (Table 5). Fecal concentrations, as estimated by the AllBac assay, were also higher in samples collected during the storm event than in those collected during the low-flow conditions and were correlated with E. coli (r = 0.86) when all seven samples were considered. PCR inhibition was not an apparent problem as the percentage of the added spike recovered in these samples ranged from 63 to 112% and was within the measurement variability attributable to real-time PCR (11) (Table 5). The mean percentages of PCR recovery by assay were 68 ± 17% for the AllBac assay, 78 ± 14% for the HuBac assay, and 90 ± 27% for the BoBac assay.
TABLE 5.
E. coli concentrations, fecal concentrations, and percentages of bovine-associated and human-associated fecal concentrations in watersheds with different land use patterns
| Land use type | Site name (flow) | E. coli concna (CFU/100 ml) | Real-time PCR assay result (mg/liter) for:
|
% Spiked plasmid recoveryb | % Recoveryc that was
|
|||
|---|---|---|---|---|---|---|---|---|
| AllBac | BoBac | HuBac | Attributable to bovines | Attributable to humans | ||||
| Pasture/rural housing | NS-1 (low flow) | 267 | 17.4 ± 8.5 | 2.3 ± 1.5 | 0.8 ± 0.7 | 77 ± 18 | 13 | 4 |
| NS-1 (high flow) | 17,800 | 100 ± 7.6 | 15.4 ± 8.9 | 19 ± 8.7 | 97 ± 1 | 15 | 19 | |
| NS-3 (low flow) | 60 | 5.6 ± 3.7 | 2.3 ± 3.1 | 1.2 ± 0.5 | 67 ± 19 | 41 | 21 | |
| NS-3 (high flow) | 39,600 | 452 ± 68 | 144 ± 33 | 37 ± 7.2 | 91 ± 4 | 32 | 8 | |
| Urban | U2 (low flow) | 1,967 | 70.9 ± 21 | 2.1 ± 1.5 | 14 ± 4.0 | 85 ± 42 | 3 | 21 |
| Resort/forest | R07 (low flow) | 600 | 17.7 ± 6.3 | NDd | 0.6 ± 0.3 | 63 ± 41 | ND | 3 |
| R20 (low flow) | 3,160 | 231 ± 1.7 | 1.2 ± 1.8 | 243 ± 62 | 113 ± 35 | 0.5 | 105 | |
Single-sample E. coli concentration.
Percent recovery = (sample spiked with 2.5 × 105 copies − measured copies in unspiked sample)/(2.5 × 105) × 100. The percent recovery was determined for each assay and then averaged.
Percent total = [concentration (mg/liter) of BoBac or HuBac/concentration (mg/liter) of AllBac] × 100.
ND, not detected in this sample.
In the water samples, the BoBac and HuBac assays were used first to estimate a fecal concentration attributable to bovines or humans and then to determine the percentage of the total fecal concentration attributable to bovines or humans (Table 5). Fecal concentrations were estimated by comparison of the sample CT values with CT values obtained from standard curves generated from appropriate fecal dilutions at the same time as the samples. Both human and bovine feces were detected in all four samples (NS-1 and NS-3 and low and high flow) in the rural watershed and were consistent with the expected mixed land use pattern (Table 5). The amount of feces measured in the HuBac assay relative to the amount measured in the AllBac assay in sample R20 suggests that human fecal contamination is a dominant source of fecal contamination at this site (Table 5). This response is not likely to result from swine fecal contamination (the fecal type with the highest potential to cross-hybridize with the HuBac assay) because there are no swine operations in the watershed. In the other two samples with potential human fecal contamination (U2 and R07), the amount of fecal contamination attributable to humans was less than that found in the R20 sample (Table 5). Interestingly, in the R07 sample, both the BoBac and HuBac assays produced very low values, suggesting that other unmeasured Bacteroides spp. were present.
DISCUSSION
Bacteroides spp. have been advocated as both fecal indicator (14) and as fecal source indicator (4-7, 10, 11, 22) bacteria for water quality measurements. Although most previous studies have detected Bacteroides in surface water samples by traditional PCR (4, 11, 23) and reported the results as either present or absent, real-time PCR can be used to rapidly quantify Bacteroides genes (10, 37). In this study, a real-time PCR assay (AllBac) was designed to target the 16S rRNA genes of Bacteroides spp., which are among the most numerically abundant bacteria present in warm-blooded animal feces (18, 20, 26, 35, 44). This assay was shown empirically to be proportional to the concentration of human, bovine, and equine feces in water and thus can be used to estimate fecal concentrations without calculating the number of Bacteroides cells in the sample. When the AllBac assay was applied to water samples from three different watersheds, the log of the measured fecal concentrations was linearly correlated with the log of the E. coli concentrations (r2 = 0.85). Fecal concentrations were measurable by the AllBac assay in a sample with low E. coli concentrations (60 CFU/100 ml) and were still within the linear range of detection in samples with E. coli concentrations greater than 10,000 CFU/100 ml. These results suggest that the AllBac assay provides a rapid direct measurement of fecal contamination in water and may complement E. coli as a fecal indicator.
Bacteroides spp. also have several desirable characteristics for serving as fecal source identifiers, including quantitative assessment, broad geographic stability, and broad distribution in the target host animal (43). The high sequence similarity of the Bacteroides 16S rRNA gene sequences obtained in this study to those in GenBank supports the assertion that similar bovine- and human-associated Bacteroides spp. are present in their respective host animals from different geographic locations. Although the original premise of this study was that Bacteroides spp. reflect host animal specificity, primarily through host animal phylogeny, a recent publication (11) suggests that Bacteroides spp. specificity reflects animal digestive tract physiology and diet rather than host animal phylogeny. For instance, although swine and bovines are in the same order (Artiocactyla), the Bacteroides 16S rRNA gene sequences obtained from swine feces were more closely related to Bacteroides 16S rRNA gene sequences obtained from human feces than to bovine feces (Fig. 1), reflecting the higher similarity between the swine and human digestive tracts than between the swine and bovine digestive tracts (39). In this study, the clustering of the bovine-associated Bacteroides 16S rRNA gene sequences was exploited to design a real-time PCR assay with high specificity towards the Bacteroides spp. present in bovine feces. The BoBac assay showed no incorrect classification results and is expected to be a reliable indicator of bovine fecal contamination.
In contrast to the bovine-associated Bacteroides 16S rRNA gene sequences, the human-associated Bacteroides 16S rRNA gene sequences did not form a cohesive cluster. Some of the human-associated Bacteroides 16S rRNA gene sequences were similar to published Bacteroides 16S rRNA gene sequences from swine. The similarity of Bacteroides spp. 16S rRNA gene sequences from other omnivorous animals with human-associated Bacteroides 16S rRNA genes made the design of a human-associated real-time PCR assay more challenging. The resulting HuBac assay is selective rather than fully specific for human-associated Bacteroides spp. (100% correct classification of human fecal samples and 32% false-positive classification), suggesting that additional improvements in the specificity of a human-associated Bacteroides assay may be warranted. However, the specificity of the current assay appears to be comparable to the reported specificity of E. coli for correct host identification, which ranged from 49% for ribotyping with HindIII to 100% for ribotyping with EcoRI (human/nonhuman classification) (40) and 44% by antibiotic resistance analysis and 69% by ribotyping with HindIII (28). The 32% false-positive rate found in this study is similar to other reported false-positive rates for both culture-dependent and molecular methods (up to 57% for E. coli [29] and 39% for fecal streptococci using antibiotic resistance analysis [19]).
When PCR methods are applied to environmental samples, the potential for PCR inhibition is of concern. In this study, a lack of PCR inhibition was demonstrated by adding 2.5 × 105 copies of an appropriate target, carried in a plasmid, to fecal dilution samples (Table 4) and creek water samples (Table 5). PCR may be inhibited by compounds readily found in environmental samples, including humic acids and metals (45). In water samples, the potential for PCR inhibition is increased when large volumes of water are concentrated in order to detect targets, such as viruses, present at very low concentrations. Although DNA extraction and additional purification steps may remove PCR inhibitors, DNA extraction methods reduce the volume of the sample and may result in an inadvertent concentration of PCR inhibitors which copurify with the DNA (45). An alternative approach to extensive nucleic acid purification is to prevent PCR inhibition by avoiding the concentration of water samples and using small sample volumes. In this study, fecal and water samples were not concentrated and the sample volume was 10% of the total PCR volume. The direct PCR method (15) also reduces the risk of concentrating PCR inhibitors during nucleic acid extraction.
A disadvantage to the direct PCR method is that the minimum amount of target detectable is determined by the small sample volumes (a few microliters) used in the reaction. Since the minimum number of target genes copies detectable per PCR is 1, assuming equal distributions of the targets, a 1-μl water sample must contain at least 1 copy of the target (1 × 106 copies per liter) in order for a positive signal to occur. The advantage of measuring Bacteroides rRNA genes in fecal samples suspended in water is the high gene copy number (>1010 copies) per gram. Thus, 1 g of feces in 1 liter of water contains approximately 1010 Bacteroides 16S rRNA gene copies. Assuming a minimum threshold of 1 × 106 copies per liter (1 copy/μl), the detection limit would be approximately 0.1 mg feces/liter water, which is consistent with the measured detection limits in this study.
In summary, the AllBac assay allows estimation of total fecal contamination, whereas the use of the BoBac assay allows the estimation of the amount and percentage of bovine-associated fecal contamination relative to the total fecal contamination in water samples. The HuBac assay may also provide an estimate of the amount and percentage of human-associated fecal contamination; however, because of the potential for cross-amplification with other omnivores (canine and swine) in the HuBac assay, the use of follow-up PCR assays with other recently described species-specific primers (11) may be warranted. In addition, in some samples, the percentage of feces attributable to humans and bovines was not 100%, indicating that additional assays are needed to fully identify sources of fecal pollution. The simplicity of performing these assays by direct PCR of water samples suggests that these assays may be field deployable and thus would aid data collection in watersheds with inherently high spatial and temporal variabilities.
Acknowledgments
This research was funded in part by the Tennessee Department of Environment and Conservation (TDEC) (grant numbers Z020074100 and Z02008749), the Water Resources Research Institute Program (WRRIP), and the Waste Management Research and Education Institute at the University of Tennessee.
We thank Sherry Wang from the TDEC Division of Water Pollution Control and Jonathon Burr from the TDEC for their support. We also thank Sharyce Banks of Knoxville College for technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Becker, S., P. Böger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belanger, S. D., M. Boissinot, N. Clairoux, F. J. Picard, and M. G. Bergeron. 2003. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 41:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., T. Goyard, M. T. Simonich, and K. G. Field. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 37:909-913. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, A. B., J. A. Fuhrman, R. D. Mrše, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denton, G. M., A. D. Vann, and S. H. Wang. 2000. The status of water quality in Tennessee. Year 2000 305(b) report. Tennessee Department of Environment and Conservation, Division of Water Pollution Control, Nashville, Tenn.
- 10.Dick, L. K., and K. G. Field. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroides bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dionisi, H. M., G. Harms, A. C. Layton, I. R. Gregory, J. Parker, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Power analysis for real-time PCR quantification of genes in activated sludge and analysis of the variability introduced by DNA extraction. Appl. Environ. Microbiol. 69:6597-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field, K. G., A. E. Bernhard, and T. J. Brodeur. 2003. Molecular approaches to microbiological monitoring: fecal source detection. Environ. Monit. Assess. 81:313-326. [PubMed] [Google Scholar]
- 14.Fiksdal, L., J. S. Maki, S. J. LaCroix, and J. T. Staley. 1985. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl. Environ. Microbiol. 49:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fode-Vaughan, K. A. F., C. F. Wimpee, C. C. Remsen, and M. L. Perille Collins. 2001. Detection of bacteria in environmental samples by direct PCR without DNA extraction. BioTechniques 31:598-607. [DOI] [PubMed] [Google Scholar]
- 16.Frahm, E., and U. Obst. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 52:123-131. [DOI] [PubMed] [Google Scholar]
- 17.Gherna, R., and C. R. Woese. 1992. A partial phylogenetic analysis of the “flavobacter-bacteroides” phylum: basis for taxonomic restructuring. Syst. Appl. Microbiol. 15:513-521. [DOI] [PubMed] [Google Scholar]
- 18.Harmsen, H. J. M., G. R. Gibson, P. Elfferich, G. C. Raangs, A. C. M. Wildeboer-Veloo, A. Argaiz, M. B. Roberfroid, and G. W. Welling. 1999. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125-129. [DOI] [PubMed] [Google Scholar]
- 19.Harwood, V. J., B. Wiggins, C. Hagedorn, R. D. Ellender, J. Gooch, J. Kern, M. Samadpour, A. C. Chapman, B. J. Robinson, and B. C. Thompson. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J. Water Health 1:153-166. [PubMed] [Google Scholar]
- 20.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 21.Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 67:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreader, C. A. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl. Environ. Microbiol. 64:4103-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layton, A. C., H. Dionisi, H.-W. Kuo, K. G. Robinson, V. M. Garrett, A. Meyers, and G. S. Sayler. 2005. Emergence of competitive dominant ammonia-oxidizing bacterial populations in a full-scale industrial wastewater treatment plant. Appl. Environ. Microbiol. 71:1105-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madigan, M. M., J. M. Martinko, and J. Parker (ed.). 2003. Brock biology of microorganisms, 10th ed. Prentice Hall, Upper Saddle River, N.J. 07458.
- 26.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto, Y., and K. Itoh. 2000. Bacteroides acidifaciens sp. nov., isolated from the caecum of mice. Int. J. Syst. Evol. Microbiol. 50:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Moore, D. F., V. J. Harwood, D. M. Ferguso, J. Lukasik, P. Hannah, M. Getrich, and M. Brownell. 2005. Evaluation of antibiotic resistance analysis and ribotyping for identification of faecal pollution sources in an urban watershed. J. Appl. Microbiol. 99:618-628. [DOI] [PubMed] [Google Scholar]
- 29.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B. K. Hahm, P. G. Hartel, H. Yampara-Lquise, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 30.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Rigottier-Gois, L., V. Rochet, N. Garrec, A. Suau, and J. Doré. 2003. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridization combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 26:110-118. [DOI] [PubMed] [Google Scholar]
- 32.Ruimy, R., I. Podglajen, J. Breuil, R. Christen, and E. Collatz. 1996. A recent fixation of cfiA genes in a monophyletic cluster of Bacteroides fragilis is correlated with the presence of multiple insertion elements. J. Bacteriol. 178:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. A. Wareing, and D. L. A. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejeuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santo Domingo, J. W., S. C. Siefring, and R. A. Haugland. 2003. Real-time PCR method to detect Enterococcus faecalis in water. Biotechnol. Lett. 25:261-265. [DOI] [PubMed] [Google Scholar]
- 35.Saua, A., R. Bonnent, M. Sutern, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savill, M. G., S. R. Murray, P. Scholes, E. W. Maas, R. E. McCormick, E. B. Moore, and B. J. Giplin. 2001. Application of polymerase chain reaction (PCR) and TaqMan PCR techniques to the detection and identification of Rhodococcus coprophilus in faecal samples. J. Microbiol. Methods. 47:355-368. [DOI] [PubMed] [Google Scholar]
- 37.Seurinck, S. T. Defoirdt, W. Verstraete, and S. D. Siciliano. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249-259. [DOI] [PubMed] [Google Scholar]
- 38.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 39.Stevens, C. E. 1988. Comparative physiology of the vertebrate digestive system. Cambridge University Press, New York, N.Y.
- 40.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggens. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Environmental Protection Agency. 2002. Implementation guidance for ambient water quality criteria for bacteria. EPA-823-B-02-003. [Online.] http://www.epa.gov/ost/standards/bacteria/bacteria.pdf.
- 43.U.S. Environmental Protection Agency. 2005. Microbial source tracking guide document. EPA/600-R-05-064. [Online.] www.epa.gov/ORD/NRMRL/pubs/600r05064/600r05064.pdf.
- 44.Wang, R.-F., W.-W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantification of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]