Abstract
By generating a calcineurin mutant of the Candida albicans wild-type strain SC5314 with the help of a new recyclable dominant selection marker, we confirmed that calcineurin mediates tolerance to a variety of stress conditions but is not required for the ability of C. albicans to switch to filamentous growth in response to hypha-inducing environmental signals. While calcineurin was essential for virulence of C. albicans in a mouse model of disseminated candidiasis, deletion of CMP1 did not significantly affect virulence during vaginal or pulmonary infection, demonstrating that the requirement for calcineurin for a successful infection depends on the host niche.
Calcineurin is a conserved Ca2+/calmodulin-activated, serine/threonine-specific protein phosphatase that regulates a variety of physiological processes in eukaryotic organisms. We and others had recently investigated the requirement of calcineurin for virulence of Candida albicans, the major fungal pathogen of humans. C. albicans mutants in which the gene encoding the catalytic (CMP1/CNA) or the regulatory (CNB1) subunit of calcineurin was inactivated were hypersensitive to salt, alkaline, and membrane stress and avirulent in a mouse model of disseminated candidiasis (1, 2, 4, 11). All these calcineurin mutants had been constructed using the URA3 gene as a selection marker for targeted gene inactivation in a ura3 auxotrophic host strain. However, it has recently become evident that using the URA3 marker for the generation of C. albicans mutants can cause phenotypes that are unrelated to target gene inactivation (13). An alternative approach avoiding all potential problems related to the use of auxotrophic markers and host strains is inactivation of the target gene in a C. albicans wild-type strain with the help of a recyclable dominant selection marker (10, 17). In the present work we used this strategy to confirm the role of calcineurin in resistance to various types of stresses and to investigate its importance for the virulence of C. albicans in different host niches.
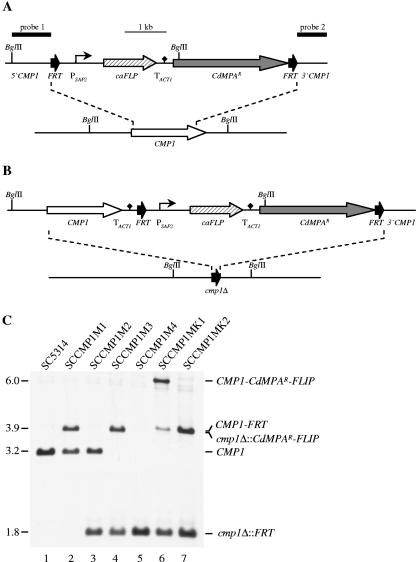
The previously described MPAR marker, which confers resistance to mycophenolic acid (MPA), is a mutated derivative of the C. albicans IMH3 gene encoding IMP dehydrogenase, the target of MPA. When MPAR was used in C. albicans, sometimes a high proportion of MPA-resistant transformants apparently had substituted the MPAR marker for the genomic IMH3 gene but did not contain the mutagenesis cassette inserted into the target gene locus (16). In contrast, when the MPAR marker was used for targeted gene inactivation in the related species Candida dubliniensis, virtually all MPA-resistant transformants had specifically integrated the cassette into the target locus, because the sequence divergence between the two species prevented integration of the C. albicans-derived MPAR marker into the orthologous C. dubliniensis IMP dehydrogenase gene IMD1 (14). These observations suggested that a similar MPAR marker derived from C. dubliniensis should also improve targeted insertion into the C. albicans genome. Therefore, we cloned the IMD1 gene from C. dubliniensis and introduced the same A251T mutation that conferred MPA resistance on IMP dehydrogenase from C. albicans (8). The resulting CdMPAR marker was substituted for the MPAR marker in the MPAR flipper cassette (17) to generate the CdMPAR flipper, which was then inserted between flanking sequences of the CMP1 gene (Fig. 1A). This deletion cassette was used to transform the C. albicans wild-type strain SC5314 to MPA resistance. Southern hybridization analysis demonstrated that 10 out of 17 tested MPA-resistant transformants had specifically inserted the CdMPAR flipper cassette into one of the two CMP1 alleles. One correct transformant, strain SCCMP1M1 (Fig. 1C, lane 2), was used to excise the CdMPAR flipper by FLP-mediated recombination, generating the MPA-sensitive heterozygous cmp1 mutant SCCMP1M2 (Fig. 1C, lane 3). After transformation of this strain with the same deletion cassette, integration was successfully targeted to the remaining wild-type CMP1 allele in the resulting strain SCCMP1M3 (Fig. 1C, lane 4), and subsequent excision of the CdMPAR flipper generated the homozygous cmp1-null mutant SCCMP1M4 (Fig. 1C, lane 5). To exclude the possibility that any phenotype of the cmp1Δ mutant was caused by a nonspecific mutation, an intact copy of the CMP1 gene was reintroduced into its original genomic locus. For this purpose, the 5′ CMP1 flanking region in the deletion construct was replaced by the complete CMP1 open reading frame (ORF) and upstream sequences (Fig. 1B). The complementation cassette was used to transform the cmp1Δ mutant SCCMP1M4, resulting in strain SCCMP1MK1, in which the CMP1 ORF, together with the CdMPAR flipper, was reinserted into one of the inactivated cmp1Δ alleles (Fig. 1C, lane 6). Subsequent excision of the CdMPAR flipper generated the complemented strain SCCMP1MK2 (Fig. 1C, lane 7). This experimental design ensured that the cmp1Δ mutant differed from the wild-type parent and the complemented strain only by the deletion of both CMP1 alleles, but not by the presence or absence of a selection marker.
FIG. 1.
Construction of the C. albicans cmp1 deletion mutant and complemented strain. (A) Structure of the CMP1 deletion cassette (top) and genomic structure of the CMP1 locus in the parent strain SC5314 (bottom). The CMP1 coding region is represented by the white arrow, and the upstream and downstream sequences by the solid lines. The direct repeats of the 34-bp FRT site (black arrows) bordering the CdMPAR flipper cassette are not drawn to scale. The SAP2 promoter (PSAP2) is indicated by the bent arrow, the caFLP gene by the hatched arrow, the transcription termination sequence of the ACT1 gene (TACT1) by the black diamond, and the CdMPAR marker by the gray arrow. The diagnostic BglII sites are shown, and the DNA fragments used for Southern hybridization analysis of the mutants are indicated by thick bars (probe 1 and probe 2). (B) Structure of the DNA cassette (top) which was used for reintegration of an intact CMP1 copy (white arrow) into one of the inactivated cmp1Δ alleles (bottom). (C) Southern hybridization of BglII-digested genomic DNA of the parent strain SC5314 and mutant derivatives with the CMP1-specific probe 1. The sizes of the hybridizing fragments (in kilobases) are given on the left of the blot, and their identities are indicated on the right.
We first tested the sensitivity of the cmp1Δ mutant to various stress conditions using previously described assays (1). Compared with the parental strain SC5314, the cmp1Δ mutant SCCMP1M4 exhibited increased susceptibility to elevated salt concentrations (NaCl, LiCl, MnSO4, or CaCl2), sodium dodecyl sulfate, fluconazole, and alkaline pH. Reintroduction of an intact CMP1 copy into the cmp1Δ mutant restored growth to wild-type levels, demonstrating that all the phenotypes of the cmp1Δ mutant were caused by inactivation of the CMP1 gene and not by nonspecific mutations. As reported previously (1), deletion of CMP1 did not affect the ability of the mutants to switch to filamentous growth in all solid and liquid hypha-inducing media tested (synthetic low-ammonium dextrose medium, Lee's medium, 10% serum).
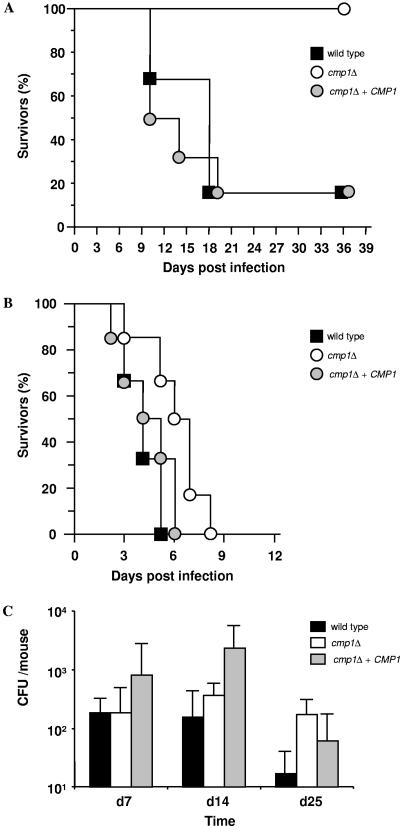
To assess whether the previously reported attenuated virulence of cmp1Δ mutants constructed in the ura3 strain CAI4 was due to a position effect of URA3 or the loss of CMP1 function, we compared the virulence of the cmp1Δ mutant SCCMP1M4, its parental strain SC5314, and the complemented strain SCCMP1MK2 in a mouse model of systemic candidiasis. BALB/c mice (Charles River Breeding Laboratories, Sulzfeld, Germany) were inoculated with 5 × 105 viable cells by intravenous injection and monitored for survival essentially as described previously (1, 12). Kaplan-Meier survival curves were compared using the log rank test (9). A P value of <0.05 was considered significant. Five out of six mice infected with the wild type or the complemented strain died within 18 and 19 days, respectively, whereas all mice infected with the cmp1Δ mutant survived beyond day 70 (P < 0.02) (Fig. 2A). These results confirmed our previous observation and those of others that calcineurin is essential for the ability of C. albicans to cause a systemic infection via the bloodstream (1, 2, 11).
FIG. 2.
Virulence of the C. albicans cmp1Δ mutant in different infection models. (A and B) Survival of mice after intravenous (A) or intranasal (B) infection with the wild-type parental strain SC5314, the cmp1Δ mutant SCCMP1M4, or the reconstituted strain SCCMP1MK2 (cmp1Δ plus CMP1). (C) Vaginal fungal burden in mice infected intravaginally with the cmp1Δ mutant and control strains was measured by determining the CFU in vaginal lavage fluid at the indicated times.
To investigate whether calcineurin is generally required for virulence of C. albicans in a mammalian host, we assessed the capacities of the strains to infect other host niches. Infection of the vaginal canal was initiated by inoculating estradiol-treated mice intravaginally with 5.0 × 104 stationary-phase cells in 20 μl of phosphate-buffered saline (6, 15). Vaginal lavage was performed using 100 μl of phosphate-buffered saline at specified time points following infection. The number of C. albicans CFU was measured by limiting dilution analysis and calculated by the most-probable-number (MPN) method according to the Poisson distribution (3, 7). The number of negative wells was used to calculate the MPN of live C. albicans bacteria in lavage fluids (7). For pulmonary infection, mice treated with Endoxan (200 mg kg of body weight−1 given intraperitoneally; Baxter Oncology, Frankfurt, Germany) were anesthetized with 200 mg of Ketavet kg of body weight−1 (Pharmacia & Upjohn, Erlangen, Germany), and 2.0 × 105 blastoconidia were administered intranasally (5). The immunosuppressed status of mice was maintained for the duration of the experiment. Generation and comparison of survival curves were performed as mentioned above.
In the pulmonary model of C. albicans infection, the cmp1Δ mutant exhibited virulence similar to that of the control strains, although killing of animals infected with the cmp1Δ mutant was slightly delayed compared with the parental strain SC5314 or the complemented strain SCCMP1MK2 (P > 0.1) (Fig. 2B). Similarly, in the mouse model of vaginal candidiasis, no significant differences in the number of CFU grown from the lavage fluids at various times after infection were measured between the cmp1Δ mutant and the control strains (P > 0.5) (Fig. 2C). These results demonstrate that the requirement of calcineurin for the virulence of C. albicans depends on the host niche. Both Sanglard et al. (11) and Blankenship et al. (2) reported that C. albicans calcineurin mutants are unable to survive in serum, a phenotype that had not been investigated in our previous study (1). For a direct comparison with the cnb1 mutants, serum sensitivity tests with our cmp1Δ mutants and complemented strains constructed in the wild-type strain SC5314 and from strain CAI4 were performed in the laboratory of J. Heitman. All mutants were efficiently killed by serum, and reintroduction of a functional CMP1 copy restored serum resistance (J. Reedy and J. Heitman, personal communication). The serum sensitivity may therefore explain why calcineurin mutants were avirulent after infection via the bloodstream but retained virulence during vaginal or pulmonary infections.
Acknowledgments
We thank Jen Reedy and Joe Heitman for performing the serum sensitivity tests with our calcineurin mutants and communicating the results to us. Sequence data for Candida albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida.
This study was supported by a grant from the NRC-HGF Science and Technology Fund (grant 01SF0201/2.2). N.A. and G.K. were supported by NIH grants AI33317 and DE12940, respectively. Sequencing of Candida albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
Editor: A. Casadevall
REFERENCES
- 1.Bader, T., B. Bodendorfer, K. Schröppel, and J. Morschhäuser. 2003. Calcineurin is essential for virulence in Candida albicans. Infect. Immun. 71:5344-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blankenship, J. R., F. L. Wormley, M. K. Boyce, W. A. Schell, S. G. Filler, J. R. Perfect, and J. Heitman. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cochran, W. G. 1950. Estimation of bacterial densities by means of the “most probable number.” Biometrics 6:105-116. [PubMed] [Google Scholar]
- 4.Cruz, M. C., A. L. Goldstein, J. R. Blankenship, M. Del Poeta, D. Davis, M. E. Cardenas, J. R. Perfect, J. H. McCusker, and J. Heitman. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fallon, K., K. Bausch, J. Noonan, E. Huguenel, and P. Tamburini. 1997. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect. Immun. 65:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fidel, P. L., Jr., M. E. Lynch, and J. D. Sobel. 1993. Candida-specific cell-mediated immunity is demonstrable in mice with experimental vaginal candidiasis. Infect. Immun. 61:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garthright, W. E., and R. J. Blodgett. 2003. FDA's preferred MPN methods for standard, large or unusual tests, with a spreadsheet. Food Microbiol. 20:439-445. [Google Scholar]
- 8.Köhler, G. A., X. Gong, S. Bentink, S. Theiss, G. M. Pagani, N. Agabian, and L. Hedstrom. 2005. The functional basis of mycophenolic acid resistance in Candida albicans IMP dehydrogenase. J. Biol. Chem. 280:11295-11302. [DOI] [PubMed] [Google Scholar]
- 9.Mantel, N., and W. Haenszel. 1959. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22:719-748. [PubMed] [Google Scholar]
- 10.Reuß, O., Å. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 11.Sanglard, D., F. Ischer, O. Marchetti, J. Entenza, and J. Bille. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959-976. [DOI] [PubMed] [Google Scholar]
- 12.Schweizer, A., S. Rupp, B. N. Taylor, M. Röllinghoff, and K. Schröppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 13.Staab, J. F., and P. Sundstrom. 2003. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 11:69-73. [DOI] [PubMed] [Google Scholar]
- 14.Staib, P., G. P. Moran, D. J. Sullivan, D. C. Coleman, and J. Morschhäuser. 2001. Isogenic strain construction and gene targeting in Candida dubliniensis. J. Bacteriol. 183:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor, B. N., P. Staib, A. Binder, A. Biesemeier, M. Sehnal, M. Röllinghoff, J. Morschhäuser, and K. Schröppel. 2005. Profile of Candida albicans-secreted aspartic proteinase elicited during vaginal infection. Infect. Immun. 73:1828-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirsching, S., S. Michel, G. Köhler, and J. Morschhäuser. 2000. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirsching, S., S. Michel, and J. Morschhäuser. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856-865. [DOI] [PubMed] [Google Scholar]