Abstract
Listeria monocytogenes can be used to deliver protein antigens or DNA and mRNA encoding such antigens directly into the cytosol of host cells because of its intracellular lifestyle. In this study, we compare the in vivo efficiencies of activation of antigen-specific CD8 and CD4 T cells when the antigen is secreted by L. monocytogenes or when antigen-encoding plasmid DNA or mRNA is released by self-destructing strains of L. monocytogenes. Infection of mice with self-destructing L. monocytogenes carriers delivering mRNA that encodes a nonsecreted form of ovalbumin (OVA) resulted in a significant OVA-specific CD8 T-cell response. In contrast, infection with L. monocytogenes delivering OVA-encoding DNA failed to generate specific T cells. Secretion of OVA by the carrier bacteria yielded the strongest immune response involving OVA-specific CD8 and CD4 T cells. In addition, we investigated the antigen delivery capacity of a self-destructing, virulence-attenuated L. monocytogenes aroA/B mutant. In contrast to the wild-type strain, this mutant exhibited only marginal liver toxicity when high doses (5 × 107 CFU per animal administered intravenously) were used, and it was also able to deliver sufficient amounts of secreted OVA into mice. Therefore, the results presented here could lay the groundwork for a rational combination of L. monocytogenes as an attenuated carrier for the delivery of protein and nucleic acid vaccines in novel vaccination strategies.
The use of virulence-attenuated bacteria as carriers for heterologous protein antigens is a versatile vaccination approach (for a review see reference 15). Bacteria directly target vaccine antigens to the appropriate cells of the immune system, such as dendritic cells (DC), for antigen presentation (17), and they act as strong immune potentiators due to their naturally produced bacterial components (pathogen-associated molecular patterns) that stimulate and modulate early innate immune responses, encouraging the generation of robust and long-lasting adaptive immune responses (11).
In the majority of cases so far, virulence-attenuated strains of enteric bacterial pathogens like Salmonella, Shigella, Yersinia, Vibrio, Escherichia coli, or Listeria have been used for vaccine delivery (15). These bacteria, which can be inoculated orally, are able to cross the intestinal mucosa and induce systemic immunity as well as mucosal immunity. Therefore, virulence-attenuated bacteria are optimal candidates as carriers for heterologous protein antigens, antigen-encoding DNA (DNA vaccines), and other molecules for vaccination or other therapeutic purposes (15).
In particular, the bacteria that are able to replicate and release antigen within the cytosol of eukaryotic host cells are attractive candidates for the elicitation of cell-mediated immunity against heterologous antigens. Listeria monocytogenes, a gram-positive bacterium, has been used successfully in several studies performed with different animal models to deliver tumor, viral, or parasite antigens, as well as different cytokines either as secreted protein (13, 23, 34) or as plasmid-encoded DNA (21, 32, 35). A number of biological properties make L. monocytogenes a promising platform for the development of vaccines, particularly vaccines against infectious diseases or tumors. An infection with L. monocytogenes is accompanied by a strong innate immune reaction that leads to pronounced specific cellular immune responses, including both CD8 and CD4 T-cell activation of a type 1-biased Th phenotype (for a review see reference 22). Furthermore, since the genomes of different L. monocytogenes strains have been sequenced (8), many genetically well-defined virulence-attenuated mutants that meet the requirements for promising vaccine carrier candidates have already been constructed (1, 3, 36, 38).
The most commonly used strategy to deliver heterologous antigens by L. monocytogenes is to express them under the control of suitable listerial promoters that are active in vivo. Additionally, antigens have been fused to appropriate secretion signals for export from bacterial cells into the cytosol (33).
An alternative strategy is to deliver plasmid DNA by attenuated carrier strains. The composition of a suitable plasmid DNA corresponds to the composition of common DNA vaccine plasmids except that a L. monocytogenes-specific origin of replication has to be inserted (27). Recently, we were able to demonstrate that infection of host cells with so-called self-destructing L. monocytogenes strains results in disruption of the bacterial carrier cells within the cytosol. The transported plasmid DNA is efficiently released into this compartment, resulting in the expression and presentation of plasmid-encoded model antigens by antigen-presenting cells (APC) (6, 27). These self-destructing L. monocytogenes carriers express an L. monocytogenes-specific endolysin (phage lysin 118 [Ply118]) under the control of a cytosolically activated listerial promoter, which leads to cell wall degradation as soon as a portion of the endolysin is set free, likely after death of some intracellular listerial cells (27).
The main difference between these so-called “protein delivery” and “DNA delivery” strategies is that in the former antigens are delivered in form of protein synthesized by the bacterial carrier, whereas in the latter L. monocytogenes delivers plasmid DNA into the eukaryotic target cell, where it is translated and also posttranslationally modified.
Recently, a novel bacterial delivery approach has been described by our group (31). In this approach L. monocytogenes strains that produce and deliver plasmid-encoded mRNA of a candidate protein under the control of a T7 promoter variant are used. At the 5′ end, the mRNA is genetically fused to an internal ribosomal entry site (IRESEMCV) element, resulting in 5′ cap-independent eukaryotic translation by the infected host cell. To enable mRNA transcription from a T7 promoter, the L. monocytogenes RNA delivery strains carry a second plasmid encoding the T7 RNA polymerase gene under the control of a cytosolically active promoter. This so-called “RNA delivery” strategy was shown to be much faster and more efficient for transferring a candidate protein (enhanced green fluorescent protein) into infected host cells (including human monocyte-derived DC) than L. monocytogenes-based delivery of plasmid DNA is (31). Furthermore, macrophages that were infected in vitro with L. monocytogenes strains delivering ovalbumin (OVA) mRNA were shown to present the H-2Kb restricted epitope OVA 257-264 (SIINFEKL), leading to activation of a specific CD8 T-cell line (31).
Therefore, the aim of the present work was to investigate whether self-destructing L. monocytogenes carrier strains (6, 27, 31) are able to deliver antigen-encoding mRNA into mice and to generate an antigen-specific immune response. Furthermore, since it is not known which L. monocytogenes-based delivery strategy is the most efficient for eliciting immune responses to a candidate antigen, we compared the delivery of a protein antigen synthesized by the carrier with the delivery of an antigen-encoding plasmid DNA or mRNA with respect to the ability to introduce an antigen into antigen presentation pathways and to generate antigen-specific CD8 and CD4 T cells in vivo. Finally, we also tested the efficiency of a recently described virulence-attenuated L. monocytogenes strain with deletions in the basic branch of the aromatic amino acid pathway (27, 36) for delivery of protein antigens or DNA encoding antigens in vivo.
We show here that mRNA encoding the model antigen OVA delivered by L. monocytogenes carriers leads to presentation of OVA in the context of major histocompatibility complex (MHC) class I molecules in vivo and to induction of a specific CD8 T-cell response. Furthermore, we demonstrate that L. monocytogenes-based delivery of prokaryotically expressed (and secreted) OVA leads to more efficient antigen presentation in vivo than bacterial delivery of ovalbumin-encoding DNA or mRNA leads to. We also determined cellular immune responses to OVA following immunization with L. monocytogenes strains containing different delivery systems.
Our findings should be helpful in determining and optimizing the appropriate delivery strategy for various potential applications of this promising live vaccine carrier.
MATERIALS AND METHODS
Animals.
Female C57BL/6 mice were purchased from Harlan Winkelmann GmbH, Germany, or Charles River Laboratories, Germany, and were used when they were between 6 and 12 weeks old. Transgenic mouse strains OT-I (12) and OT-II (18) were bred in our facility from stocks generously provided by A. Schimpl (University of Würzburg, Würzburg, Germany). OT-I mice express a Vα2+ Vβ5+ T-cell receptor (TCR) that recognizes the OVA peptide from amino acids (aa) 257 to 264 complexed with the MHC class I molecule H-2Kb. OT-II mice contain CD4+ T cells that express a TCR that recognizes OVA peptide from aa 323 to 339 complexed with the MHC class II molecule I-A2. All animals were housed under specific-pathogen-free conditions at the Biocenter of the University of Würzburg. All animal experiments were approved by the government of Unterfranken and were performed according to the German animal protection guidelines.
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli DH10b was used as the host for all DNA manipulations. Competent L. monocytogenes cells were prepared and transformed by electroporation as described by Park and Stewart (25). After transformation of L. monocytogenes ΔtrpS with the plasmids that were constructed, the resulting L. monocytogenes strains were cultivated in an erythromycin-containing medium without tetracycline for removal of plasmid pTRPS. For infection of host cells, L. monocytogenes strains were grown to the late logarithmic phase (optical density at 600 nm [OD600], 1.0) at 37°C in brain heart infusion (BHI) medium, washed two times with endotoxin-free isotonic saline (0.9% NaCl), resuspended in 20% (vol/vol) glycerol in 0.9% NaCl, and stored at −80°C.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Listeria monocytogenes EGDe strains | ||
| WL-140 | ΔtrpS/pTRPS | 27 |
| ΔtrpS/pSP0 | 27 | |
| ΔtrpS/pSP118 | 27 | |
| ΔtrpS/pSP118-PCMV | This study | |
| WL-150 | Δ(trpS aroA/B)/pTRPS | 36 |
| ΔtrpS/pSP118-PSactAOVA | This study | |
| ΔtrpS/pSP118-PCMVOVA | This study | |
| S15 | ΔtrpS/pCSA1/pCSB1 | 31 |
| S62N | ΔtrpS/pCSA1/pCSB-OVA | 31 |
| Δ(trpS aroA/B)/pSP118 | This study | |
| Δ(trpS aroA/B)/pSP118-PSactAOVA | This study | |
| Δ(trpS aroA/B)/pSP118-PCMVOVA | This study | |
| Plasmids | ||
| pCI-OVA | Source of chicken albumin gene ova | J. Fensterle |
| pSP0 | Emr, trpS | 27 |
| pSP0 PSactAOVA | Emr, trpS, (PS)actA-ova-TinlA | This study |
| pSP118 | Emr, trpS, PactA-ply118 | 27 |
| pSP118 PSactAOVA | Emr, trpS, PactA-ply118, (PS)actA-ova-TinlA | This study |
| pSP118 PCMVOVA | Emr, trpS, PactA-ply118, PCMV-ova | This study |
| pCSA1 | Emr, trpS, PactA-ply118, PactA-polT7 | 31 |
| pCSB1 | Tcr, PT7-IRESEMCV-TT7 | 31 |
| pCSB-OVAa | Tcr, PT7-IRESEMCV-ova-TT7 | 31 |
Synonym, pCSB7.
Plasmid construction.
The plasmid DNA encoding full-length OVA (pCI-Ova) was obtained from J. Fensterle (University of Würzburg, Würzburg, Germany). Three different OVA antigen expression cassettes were constructed. First, expression plasmid pSP118-PCMVOVA was designed for expression of OVA in which a central internal fragment (encoding aa 19 to 144) of the native OVA cDNA was absent. OVA aa 1 to 18 were fused to OVA aa 145 to 386 by a recombinant PCR technique (10). The following specific primers were used: Ova2-F sense (5′-AAAAAAGCGGCCGCGCCACCATGGGCTCCATCGGCGCAGCAAGCATGGAATTT-3′), Ova-a antisense (5′-CCCAGGAATTGATGAGCTCCTTGAATACATCA-3′), Ova-b sense (5′-TGATGTATTCAAGGAGCTCATCAATTCCTGGG-3′), and Ova-R antisense (5′-AAAAAAGCGGCCGCTTAAGGGGAAACACATCTGC-3′). Using pCI-Ova as the template, a 37-bp fragment was amplified with primers Ova2-F sense and Ova-a antisense and a 727-bp fragment was amplified with primers Ova-b sense and Ova-R antisense. The two fragments were fused by performing a second PCR and were then amplified with primers Ova2-F sense and Ova-R antisense. The resulting 764-bp fragment was cloned into pSP118-PCMV using the NotI restriction site, resulting in plasmid pSP118-PCMVOVA.
Second, the expression cassette for listerial expression of secreted OVA (encoding aa 145 to 386 of the native OVA cDNA) was constructed. This prokaryotic expression cassette was under control of the listerial promoter and signal sequence of actA. The antigen expression cassette was obtained by PstI and SacI digestion of plasmid pUC18-(PS)actAOVATinlA (Schoen, unpublished). The resulting fragment, (PS)actA-ova-TinlA, was ligated into pSP118 using the PstI and SacI restriction sites, resulting in expression plasmid pSP118-PSactAOVA.
Construction of the third OVA expression plasmid, pCSB-OVA, has been described previously (31). All OVA expression plasmids were transformed into L. monocytogenes strains ΔtrpS and Δ(trpS aroA/B).
Preparation of protein extracts for Western blot analysis.
For preparation of protein extracts of L. monocytogenes, all strains were grown to the late logarithmic phase (OD600, 1.0) in BHI medium. Each supernatant was precipitated on ice with 12% trichloroacetic acid, pelleted by centrifugation (5,000 × g at 4°C), washed in acetone, and resuspended in 2× Laemmli buffer (16) to obtain a volume that was 0.2% of the original culture volume (OD600, 1.0). The culture pellet was washed with phosphate-buffered saline (PBS) and resuspended in 2× Laemmli buffer to obtain a volume that was 0.25% of the original culture volume (OD600, 1.0). The proteins were heated to 110°C for 5 min (supernatant proteins) or for 20 min (pellet proteins) before they were loaded on sodium dodecyl sulfate (SDS) gels.
About 2 × 106 COS-1 cells transfected with the plasmids using the Lipofectamine Plus reagent according to the manufacturer's instructions were washed with PBS and lysed in 400 μl of 2× Laemmli buffer. Cell extracts were then forced through a 23-gauge needle several times, frozen in liquid nitrogen, and boiled for 7 min before they were loaded on SDS gels.
Immunoprecipitation.
For immunoprecipitation of ovalbumin protein expressed in Caco-2 cells following infection with carrier strains for 24 h, cell extracts of 107 cells were obtained by suspending the infected cell layers in 1 ml of modified RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) containing the protease inhibitors phenylmethylsulfonyl fluoride (1 mM), aprotonin (1 μg/ml), EDTA (1 mM), leupeptin (1 μg/ml), and pepstatin (1 μg/ml) and also the phosphatase inhibitors Na3VO4 (1 mM) and NaF (1 mM). The cell suspensions were gently mixed on a vertical rotator at 4°C for 15 min to lyse the cells. After this, the suspensions were spun at 14,000 rpm for 15 min at 4°C, and the supernatants were transferred into fresh tubes for further use. The cell lysates were then diluted with PBS to obtain a total cell protein concentration of 1 mg/ml, and 25 μg of the immunoprecipitating rabbit anti-OVA antibody was added to 1-ml portions of the cell lysates. The cell lysate-antibody mixtures were gently mixed overnight at 4°C on a vertical rotator. The next day, immunocomplexes were captured by adding 100 μl of a 50% protein A-agarose bead slurry, and each mixture was again rotated overnight at 4°C. The agarose beads were then collected by centrifugation, washed three times with ice-cold PBS, resuspended in 2× Laemmli buffer, and boiled for 5 min. Finally, the protein A-agarose beads were removed by centrifugation, and SDS-polyacrylamide gel electrophoresis was performed with supernatant fractions.
Western blot analysis.
Protein samples (20 μl) were separated on 12% SDS-polyacrylamide gels by the method of Laemmli (16) and were analyzed by Western blotting. Fractionated proteins were then transferred to nitrocellulose membranes (Hybond ECL; Amersham Biosciences, Germany) using transfer buffer (48 mM Tris, 39 mM glycine, 1.3 mM SDS, 20% [vol/vol] ethanol) and a semidry electrotransfer unit. The membranes were blocked with 5% low-fat milk, washed, and incubated with polyclonal rabbit antibodies directed against ovalbumin that were specifically purified from antiserum (C6534; Sigma, Germany). Peroxidase-conjugated goat-anti-rabbit immunoglobulin G(H+L) (Dianova, Germany) was used as the second antibody. Bands were visualized by incubation of each membrane with ECL Western blot detection reagents (Amersham Biosciences) and exposure to Kodak XAR film.
Cell culture and infection experiments.
P388.D1 cells (murine lymphoid macrophages) were cultured in RPMI 1640 medium supplemented with 2 mM l-glutamine (Gibco) and 10% fetal calf serum (Biochrom, Berlin, Germany). Bactofection experiments were performed as described previously (27). Briefly, cells were seeded into 24-well plates 1 day prior to infection. After a washing step macrophages were infected at a multiplicity of infection (MOI) of 5 bacteria per cell for 1 h. The cells were washed three times and cultivated with medium containing 100 μg/ml gentamicin, which was replaced with medium containing 15 μg/ml gentamicin after 1 h. Viable bacterial counts of intracellular bacteria were determined at different times by plating serial dilutions of mechanically lysed cell suspensions on BHI agar.
Adoptive transfer.
In adoptive transfer experiments, total spleen cells were isolated from OT-I or OT-II donor mice as described previously (30). Spleens were removed, and single-cell suspensions were prepared in filter-sterilized Hanks balance salt solution medium using a 70-μm nylon cell strainer. After lysis of erythrocytes, cell suspensions were washed twice and collected in endotoxin-free PBS for adoptive transfer. Approximately 1 × 107 spleen cells resuspended in 0.2 ml were transferred intravenously (i.v.) into C57BL/6 recipient mice 1 day before immunization.
Infection of animals.
In adoptive transfer experiments, C57BL/6 recipient mice (groups of three animals) were intravenously infected with 5 × 107 bacteria resuspended in 0.1 ml endotoxin-free 0.9% NaCl 1 day after adoptive transfer. Three days postinfection, spleens were collected for flow cytometric analysis. In immunization experiments, groups of five mice were intravenously immunized three times with bacteria in endotoxin-free 0.9% NaCl at 2-week intervals. Control mice received 0.9% NaCl. Spleens of immunized mice were harvested on day 49 for enzyme-linked immunospot (ELISPOT) assay analysis 1 week after the last immunization.
Flow cytometric analysis of spleen cell suspensions.
Spleens were removed 3 days after infection with L. monocytogenes strains. Single-cell suspensions were prepared, and clonal expansion of OT-I or OT-II T cells was analyzed following labeling of total spleen cells with a phycoerythrin (PE)-conjugated iTAg MHC tetramer recognizing H-2Kb-SIINFEKL (Immunomics, France), Cy-Chrome-conjugated rat anti-mouse CD44 (IM7), and fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD8a (53-6.7) in OT-I transfer experiments or with FITC-conjugated anti-mouse Vα2 TCR (B20.1), PE-conjugated anti-mouse Vβ5.1, 5.2 TCR (MR9-4), and Cy-Chrome-conjugated anti-mouse CD4 (H129.19) in OT-II experiments (all obtained from BD Biosciences Pharmingen, Germany). In some OT-I transfer experiments, spleen cells were labeled with a combination of FITC-conjugated anti-mouse Vα2 TCR (B20.1), PE-conjugated anti-mouse Vβ5.1, 5.2 TCR (MR9-4), and Cy-Chrome-conjugated anti-mouse CD8 (53-6.7). The corresponding isotype controls were also used. The extent of clonal expansion of OT-I or OT-II T cells was assessed by measuring a minimum of 5 × 105 spleen cells using an Epics XL flow cytometer (Beckman Coulter).
Purification of lymphocytes.
Prior to ELISPOT analysis, T lymphocytes from spleens of immunized mice were purified by immunomagnetic separation using a DM mouse T-lymphocyte enrichment set (BD Biosciences Pharmingen) according to the manufacturer's protocol for negative selection of T lymphocytes from mouse spleens.
IFN-γ ELISPOT assays.
ELISPOT assays were performed 1 week after the final immunization with L. monocytogenes as described previously (7). To do this, 96-well nitrocellulose plates (Multiscreen-HA; Millipore) were coated overnight under sterile conditions with 5 μg/ml of purified anti-mouse gamma interferon (IFN-γ) monoclonal antibody (R4-6A2; BD Biosciences Pharmingen) in 100 μl of freshly prepared filter-sterilized carbonate buffer (pH 9.6 at 4°C), washed twice, and blocked for 2 h at 37°C with 200 μl of 1% bovine serum albumin in PBS. To determine the in vivo frequencies of CD8+ T cells specific for OVA epitope 257-264 in association with H-2Kb, 1 × 105 purified lymphocytes from immunized mice and OVA peptide 257-264 (SIINFEKL) at a final concentration of 2 μg/ml were added to 200 μl (total volume) of complete medium in each well containing 30 U/ml murine interleukin-2 (Strathmann Biotec AG, Germany). For each stimulant, measurements were obtained in triplicate, and unstimulated lymphocytes were used as controls. Cells were incubated for 24 h at 37°C. After this, the plates were washed with 0.05% Tween 20 in PBS, incubated with 0.25 μg/ml biotinylated rat anti-mouse IFN-γ monoclonal antibody (XMB1.2; BD Biosciences Pharmingen), washed, incubated with streptavidin-alkaline phosphatase (1:20,000; BD Biosciences Pharmingen), washed, and developed with the 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium substrate (Sigma). The areas in which a single cell was stimulated to secrete IFN-γ were detected as spots on the nitrocellulose membrane using a stereomicroscope (Zeiss). The frequency of peptide-specific T cells was expressed as the number of IFN-γ-secreting cells per 1 × 105 purified lymphocytes.
To determine the in vivo frequencies of I-A2 epitope OVA 323-339-specific CD4+ T cells, 4 × 105 spleen cells were added to nitrocellulose plates without purification and incubated with ovalbumin (final concentration, 100 mg/ml; Sigma) as the stimulant.
Statistics.
A two-tailed Student's t test was used for statistical analysis. P values of <0.05 were considered statistically significant.
RESULTS
Expression of ovalbumin by prokaryotic or eukaryotic mechanisms.
The main focus of the present study was to investigate whether infection of mice with L. monocytogenes strains transferring OVA with different delivery strategies results in efficient MHC class I-restricted presentation and activation of OVA-specific CD8 T cells. OVA is a glycoprotein (45 kDa) with an internal secretion sequence that directs the protein to the secretion pathway of eukaryotic cells (37). Secreted proteins, however, have limited access to the MHC class I molecules of the cells expressing the protein. Therefore, we used N-terminally truncated forms of OVA lacking the atypical internal secretion sequence. Such a truncated OVA protein remains within the cell and exhibits a higher degree of CD8 T-cell activation than OVA expressed in its native form exhibits (30). However, N-terminally truncated OVA, in turn, seems to have reduced access to the MHC class II presentation pathway (30).
The truncated OVA protein was expressed by L. monocytogenes by genetically fusing the ova gene (encoding aa 145 to 386) N terminally to the promoter and signal sequence of the listerial actA gene (Fig. 1A). For eukaryotic transcription and translation, the ova gene (encoding aa 1 to 18 and aa 145 to 386) was placed under control of the immediate early cytomegalovirus (CMV) promoter (Fig. 1B). In order to allow efficient prokaryotic transcription of mRNA and subsequent translation by host cell mechanisms, the ova gene (encoding aa 145 to 386) was N terminally linked to an internal ribosome entry site element and placed under control of a strong T7 promoter (Fig. 1C). The three expression cassettes were introduced into Listeria-E. coli shuttle plasmids carrying tetracycline resistance markers (pCSB-OVA) or erythromycin resistance markers (pSP derivatives) (Fig. 1A to C). The pSP derivatives (pSP118-PSactAOVA, pSP118-PCMVOVA, and pCSA1) harbored the essential listerial trpS gene for stable in vivo plasmid maintenance in trpS-deficient L. monocytogenes carrier strains (Table 1). For transcription of ova mRNA from the T7 promoter, a second plasmid (pCSA1) encoding the T7 RNA polymerase gene controlled by the actA promoter was transformed into the mRNA carrier strains (Fig. 1C).
FIG. 1.
Schematic maps of different antigen expression plasmids for delivery by L. monocytogenes. (A) Shuttle plasmid for expression and secretion of OVA by L. monocytogenes. (B) Plasmid for expression of OVA under control of the CMV promoter (PCMV). (C) Two-plasmid system for transcription of ova mRNA by L. monocytogenes under control of a T7 promoter (PT7) and for translation of OVA protein by host cells via an internal ribosomal entry site (IRES). In contrast to plasmid pSP0-PSactAOVA, plasmid pSP118-PSactAOVA contains the phage lysin 118 gene (ply118) under control of the promoter of the L. monocytogenes actA gene (PactA). (Panel i) Plasmid pCSA1 encodes the expression cassettes for T7 polymerase (T7 pol) and phage lysin 118 (ply118), both under control of PactA. (Panel ii) Plasmid pCSB-OVA encodes the ova mRNA expression cassette under control of the T7 promoter. trpS, gene encoding L. monocytogenes tryptophanyl tRNA synthetase; REm, erythromycin resistance marker; RTc, tetracycline resistance marker; ori Lm, origin of replication for L. monocytogenes; oriE1, origin of replication for E. coli; PSactA, promoter and secretion signal of actA; ova, partial ovalbumin gene; poly A, polyadenylation site.
Efficient transfer of DNA or mRNA but not secreted protein antigen by L. monocytogenes requires disruption of the listerial cell wall within the host cell cytosol (6, 27, 31). For this, plasmids pSP118-PCMVOVA and pCSA1 contain the endolysin gene ply118 from phage A118 under control of the listerial actA promoter, which turns L. monocytogenes strains into self-destructing DNA or mRNA carriers (Fig. 1B and 1C, panel i). As shown previously, the expression of this endolysin gene results in a marked reduction in the bacterial load in livers and spleens of orogastrically infected mice, leading to strong attenuation of the virulence of these carrier strains (27). In order to perform a comparative study, we had to use identical L. monocytogenes carrier strains that exhibited reasonably comparable virulence attenuation levels for all three strategies. To guarantee comparable virulence attenuation levels, the Ply118 expression cassette was also introduced into plasmid pSP0-PSactAOVA (Fig. 1A), and the resulting plasmid, pSP118-PSactAOVA, was used in the experiments.
Expression of the truncated OVA protein (∼27 kDa) was detected by Western blot analysis in pellet fractions, as well as in supernatants of L. monocytogenes cultures grown in culture broth. No OVA expression was detected in strains harboring plasmids for the delivery of ova DNA (ΔtrpS/pSP118-PCMVOVA) or in strains producing ova mRNA (ΔtrpS/pCSA1/pCSB-OVA) (Fig. 2A).
FIG. 2.
Expression of truncated OVA protein (∼27 kDa) determined by Western blot analysis using a purified rabbit anti-ovalbumin antiserum. (A) Expression of OVA by L. monocytogenes ΔtrpS containing the expression plasmids indicated, grown to the late logarithmic phase in BHI medium. Lanes 1 to 5, proteins from culture supernatants; lanes 6 to 10, total cellular proteins. (B) Expression of OVA by COS-1 cells 3 days after transfection with the expression plasmids indicated. (C) Expression of OVA by Caco-2 cells 24 h after infection with 50 CFU/cell of L. monocytogenes ΔtrpS containing different expression plasmids. Proteins were obtained by immunoprecipitation prior to Western blot analysis. The arrowheads indicate bands representing truncated OVA protein. Lm, L. monocytogenes.
To investigate the expression of OVA by the eukaryotic machinery, we transfected COS-1 cells with the OVA expression plasmids. As expected, truncated OVA protein was detected only in the cell extracts prepared from cells transfected with pSP118-PCMVOVA, the only plasmid that allowed transcription and translation of OVA from PCMV by host cells (Fig. 2B).
Expression of truncated OVA protein by host cells after bacterial delivery of ova mRNA transcribed by L. monocytogenes could be detected only in Caco-2 cells that had been infected with the corresponding carrier strain (Fig. 2C). Interestingly, we had to perform immunoprecipitation prior to Western blot analysis to visualize OVA in these infection experiments, suggesting that the level of expression was at the limit of detection. However, using the same experimental setup, we were not able to detect any OVA protein in cells infected with carriers that either secreted OVA protein or delivered ova DNA (Fig. 2C).
Self-destructing carrier strains have distinct extracellular and intracellular growth behavior.
L. monocytogenes carrier strains with plasmids pSP118-PSactAOVA and pSP118-PCMVOVA for delivery of the OVA protein and ova DNA, respectively, exhibited similar growth rates when they were cultured in BHI medium, whereas the ova mRNA delivery strain harboring plasmids pCSA1 and pCSB-OVA showed substantial growth inhibition (Fig. 3A). This was due to the fact that large amounts of candidate mRNA had to be synthesized at the cost of the total biosynthesis by this strain. Consequently, for in vivo experiments with DNA or protein delivery strains an L. monocytogenes strain containing plasmid pSP118 was used as a negative control. For infection experiments with mRNA delivery strains, we used a second control strain that contained plasmids pCSA1 and pCSB1 for transcription of egfp mRNA (31), which resulted in growth inhibition comparable to that of the test strain transcribing ova mRNA (Fig. 3A).
FIG. 3.
Replication of the L. monocytogenes ΔtrpS carrier strains containing the expression plasmids indicated. (A) Growth in BHI medium. mRNA delivery strains, L. monocytogenes ΔtrpS/pCSA1/pCSB1 (control strain), and L. monocytogenes ΔtrpS/pCSA1/pCSB-OVA have increased generation times. The DNA delivery strain was L. monocytogenes ΔtrpS/pSP118-PCMVOVA, the protein delivery strain was L. monocytogenes ΔtrpS/pSP118-PSactAOVA, and the control strain was L. monocytogenes ΔtrpS/pSP118. The optical densities at 600 nm of cultures were determined, and the results are expressed as means ± standard deviations. The results of one of three experiments are shown. (B) Intracellular growth of the L. monocytogenes delivery strains in macrophage cell line P388.D1. The mRNA delivery strains L. monocytogenes ΔtrpS/pCSA1/pCSB1 and L. monocytogenes ΔtrpS/pCSA1/pCSB-OVA are significantly less invasive than the other delivery strains but are able to replicate within the host cell cytosol. The OVA-secreting strain L. monocytogenes ΔtrpS/pSP118-PSactAOVA is more invasive than the mRNA delivery strains but is not able to replicate intracellularly. The results of one of three experiments are shown.
Additionally, the invasion and replication capacities of these strains were investigated. Macrophages (cell line P388.D1) were infected at an MOI of 5 with the corresponding carrier strains, and the numbers of intracellular bacteria were monitored for 7 h (Fig. 3B). The mRNA delivery strains harboring plasmids pCSA1 and pCSB-OVA or pCSB1 were taken up much less (about 10%) than the control strain with plasmid pSP118 and the DNA delivery strain containing plasmid pSP118-PCMVOVA were taken up. However, the intracellular replication of these mRNA-delivering strains was almost comparable to that of the control strains. In contrast, the L. monocytogenes strain secreting OVA protein was not able to replicate but was taken up more than the mRNA delivery strains.
Self-destructing carrier strains are virulence attenuated in vivo.
In an evaluation of the optimal intravenous infection dose of self-destructing (Ply118-expressing) L. monocytogenes strains, the 50% lethal doses (LD50) of these strains were determined to be 1 × 108 to 2 × 108 CFU per mouse when they were administered intravenously (not shown). In contrast, the LD50 of the isogenic wild-type L. monocytogenes strain was reported to be about 1 × 104 CFU per animal (6). To use bacterial doses that were significantly lower than the LD50, we injected 5 × 107 Ply118-expressing bacteria (0.25 to 0.5 LD50) i.v. into C57BL/6 mice and determined the bacterial loads in the spleens and livers of infected mice for 1 week.
At the beginning of the infection, about 1 × 106 CFU ply118+ L. monocytogenes was detected in total spleen homogenates. In mice that had received 5 × 103 CFU (0.5 LD50) of bacteria carrying control plasmid pSP0 (Fig. 4A), about 100 CFU was found in spleen tissue. During the course of the infection, the number of ply118+ L. monocytogenes in the spleen decreased continuously, whereas the number of control bacteria typically peaked 3 days after infection.
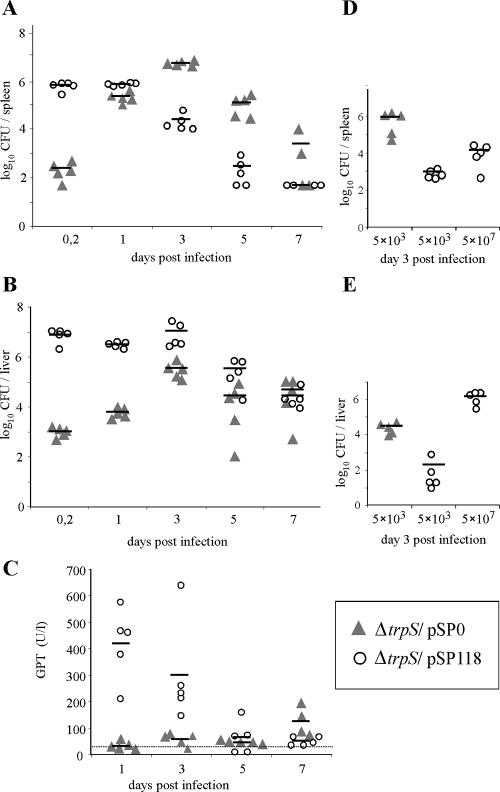
FIG. 4.
Viable bacterial counts in spleens and livers of C57BL/6 mice infected intravenously with 5 × 103 CFU (0.5 LD50) of L. monocytogenes ΔtrpS/pSP0 or 5 × 107 CFU (0.25 to 0.5 LD50) of the self-destructing carrier strain L. monocytogenes ΔtrpS/pSP118. (A) Bacterial loads in spleens of infected animals. (B) Bacterial loads in livers of infected animals. (C) GPT levels of infected mice at different times after infection. The dotted line indicates the mean GPT level for mice that were inoculated with a saline solution as a control. (D) Bacterial loads in spleens of animals infected with equal doses (5 × 103 CFU each) of L. monocytogenes ΔtrpS/pSP0 and L. monocytogenes ΔtrpS/pSP118. In this experiment, a third group of mice was also infected with 5 × 107 CFU of L. monocytogenes ΔtrpS/pSP118. Each symbol represents a single animal. The lines indicate the means for the experimental groups (n = 5). The results of one of two experiments are shown.
In contrast, the bacterial loads in the livers of animals infected with ply118+ L. monocytogenes were about 4 logs higher at the beginning of the infection, remained constant during the first 3 days, and decreased later (Fig. 4B). Also, the glutamic-pyruvic transaminase (GPT) levels of these animals increased significantly during the first 3 days of infection (Fig. 4C). This indicates that there was liver cell damage by intracellularly replicating bacteria that were apparently not lysed by Ply118. Taken together, it seems that self-destruction of carrier bacteria by Ply118-mediated lysis works more efficiently in infected cells that are in the spleen than in infected cells that are in the liver tissue. However, infections of mice with 5 × 107 CFU of L. monocytogenes expressing Ply118 were cleared faster than infections with 5 × 103 CFU control bacteria.
We also determined the extent of virulence attenuation of Ply118-expressing L. monocytogenes strains administered in equal doses (5 × 103 CFU each) as control bacteria (Fig. 4D and E). In spleens of infected mice, the amounts of Ply118-expressing L. monocytogenes were 3 logs lower than the amounts of control bacteria 3 days after infection (Fig. 4D). In liver tissue, the attenuation of the virulence of the carrier strain was lower, and the bacterial loads were reduced by 2 logs compared to the loads in the livers of mice infected with control bacteria (Fig. 4E).
MHC class I- and class II-restricted antigen presentation following infection with L. monocytogenes strains delivering OVA as mRNA, DNA, or secreted protein.
Antigen presentation by MHC I or MHC II followed by specific T-cell expansion can be quantitatively assessed by adoptive transfer of naive TCR-transgenic T cells with a known peptide/MHC specificity into syngeneic mice that are then exposed to the antigen (24, 30). We used CD8 T cells derived from OT-I transgenic mice expressing a TCR specific for OVA peptide 257-264 in association with H-2Kb (12) or CD4 T cells derived from transgenic OT-II mice that were specific for OVA peptide 323-339 in association with I-Ab (18). In particular, quantitative analysis of in vivo presentation of OVA 257-264 by MHC I was very sensitive when as many as 1 × 107 OT-I or OT-II spleen cells were transferred 1 day before infection of recipient mice with L. monocytogenes carrier strains.
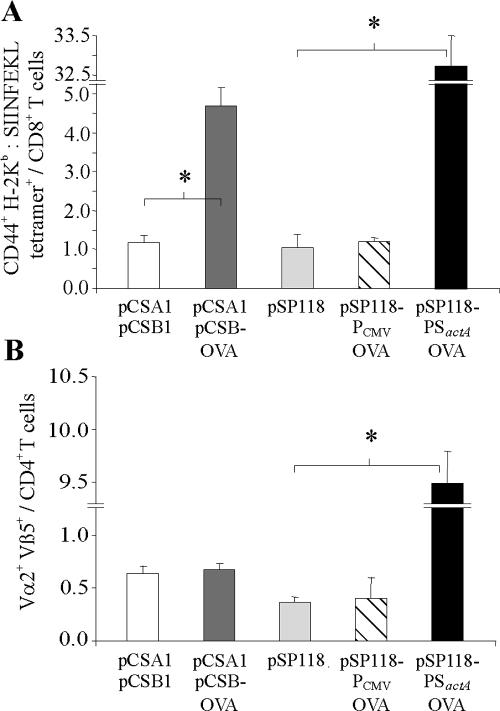
Therefore, groups of three recipient mice were infected with 5 × 107 bacteria delivering OVA protein, ova DNA, or ova mRNA, and spleen cells were analyzed for proliferating OT-I or OT-II cells by flow cytometry 3 days later. This experiment revealed that L. monocytogenes strain ΔtrpS/pCSA1/pCSB-OVA was able to deliver functional ova mRNA to appropriate MHC class I antigen presentation pathways in vivo, resulting in proliferation of specific OT-I cells so that they accounted for about 4.7% of the total CD8-T-cell population (Fig. 5A). These OT-I cells were also determined to be CD44high, representing an early marker of T-cell activation initiated by TCR signaling (5). Furthermore, the data indicated that while L. monocytogenes strain ΔtrpS/pSP118-PCMVOVA was rather inefficient in priming OT-I cells, strain ΔtrpS/pSP118-PSactAOVA exhibited comparably potent activation of OT-I cells so that they accounted for 33% of the total CD8 T-cell population (Fig. 5A). We also tested 10-fold-higher and -lower application doses of the strains delivering OVA protein or ova mRNA, but presentation of OVA 257-264 by MHC class I was most efficient when we used 5 × 107 carrier bacteria (0.25 to 0.5 LD50) in both cases (not shown). In addition, i.v. infection of mice with 5 × 103 CFU (0.5 LD50) of strain L. monocytogenes ΔtrpS/pSP0-PSactAOVA secreting amounts of OVA equal to the amounts secreted by ΔtrpS/pSP118-PSactAOVA but not expressing Ply118 resulted in clonal expansion of OT-I cells so that they accounted for only about 1% of the total CD8 T-cell population (not shown). These findings suggest that it may be advantageous to use as many bacteria as possible to deliver large amounts of secreted protein, at least in this system. However, infection with 5 × 103 CFU of strain ΔtrpS/pSP118-PSactAOVA also resulted in clonal expansion of OT-I cells so that they accounted for only about 1% of the cells, showing that Ply118-mediated lysis does not cause additional release of OVA protein from bacterial cells. Therefore, self-destruction of bacterial carriers has no advantage for the delivery of secreted proteins in terms of providing larger amounts of antigen from a single bacterium; rather, it facilitates the use of larger numbers of carrier bacteria per animal.
FIG. 5.
Clonal expansion of OVA-specific CD8 and CD4 T cells following infection with 5 × 107 CFU of L. monocytogenes ΔtrpS containing the plasmids indicated. (A) One day prior to infection, 107 spleen cells derived from OT-I mice were transferred into C57BL/6 mice. The frequencies of CD44+, H-2Kb-SIINFEKL tetramer-positive cells in all the CD8+ T cells in spleens were determined 3 days after infection, which indicated presentation of OVA epitopes via MHC class I. (B) To investigate presentation via MHC class II, 107 spleen cells derived from OT-II mice were transferred into C57BL/6 mice 1 day prior to infection. Three days later, the frequencies of Vα2+ Vβ5+ cells in all the CD4+ T cells in spleens of infected animals were determined. L. monocytogenes ΔtrpS/pCSA1/pCSB1 and L. monocytogenes ΔtrpS/pSP118 were the control strains. The results are expressed as means ± standard deviations (n = 3) and are representative of the results of at least two experiments. An asterisk indicates that there is a significant difference (P < 0.05, as determined two-tailed Student's t test) between experimental groups.
To see whether the truncated OVA variant also gains access to MHC class II molecules, we infected mice that had received OT-II spleen cells prior to i.v. infection with 5 × 107 CFU of the three carrier strains. As expected, only the L. monocytogenes strain secreting OVA protein was able to target OVA peptides to MHC class II presentation pathways, as shown by the proliferation of OT-II cells (Fig. 5B). In contrast, the L. monocytogenes strains delivering either ova DNA or ova mRNA failed to activate OT-II cells.
In line with these in vivo observations, when granulocyte-macrophage colony-stimulating factor-generated murine bone marrow-derived DC were infected with the carrier strain secreting OVA protein, a substantial proportion of cocultivated OT-I-derived CD8 T cells expressed IFN-γ (25 to 30%) after 6 h of coincubation (not shown). In contrast, DC infected with the L. monocytogenes carrier delivering ova mRNA activated only a small portion of OT-I CD8 T cells (1 to 2%), and the L. monocytogenes carrier delivering ova DNA was not successful at all in providing sufficient amounts of plasmid DNA to DC for subsequent OVA presentation via MHC class I.
These data provide further evidence that in vivo DC are probable target cells that are infected and express OVA for subsequent MHC class I-restricted presentation.
Virulence-attenuated L. monocytogenes carrier strains with additional deletions of aroA/B genes deliver amounts of secreted OVA protein and ova mRNA comparable to the amounts delivered by their parental strains.
Since intracellular Ply118 expression led to substantial attenuation of the virulence of L. monocytogenes carrier strains in vivo, we were able to administer as many as 5 × 107 Ply118-expressing cells per mouse without causing signs of disease. However, 3 days after infection the livers of the animals were colonized with about 107 bacteria (Fig. 4B), which was a considerable bacterial load for a vaccinee. Additionally, the serum transaminase levels of these mice increased during the first 3 days of infection, indicating that there was liver cell damage (Fig. 4C). From this it follows that additional attenuation of the virulence of self-destructing L. monocytogenes carrier strains is still required.
As shown recently, strain L. monocytogenes Δ(trpS aroA) WL-141 is a highly virulence-attenuated carrier of plasmid DNA that was able to transform several cell lines in vitro, and some of them were transformed with even higher efficiency than they were transformed with the corresponding wild-type strain (27). With regard to biosafety, this strain was improved by deletion of the aroB gene, whose gene product, as well as the gene product of the aroA gene, is involved in the biosynthesis pathway leading to aromatic compounds and to menaquinone, the lack of which results in a defect in oxidative respiration in these strains (36).
Therefore, we first investigated if deletion of the aroA/B genes results in greater attenuation of the virulence of the self-destructing carrier Δ(trpS)/pSP118. Mice that were infected with 5 × 107 CFU of L. monocytogenes strain Δ(trpS aroA/B)/pSP118 had 1-log fewer bacteria in their livers and spleens than mice infected with an equal dose of Δ(trpS)/pSP118 at the beginning of the infection (Fig. 6A). In livers of infected mice, however, the bacterial load was reduced 2 to 3 logs, starting on day 3 postinfection. Although the reduction in the bacterial loads of mice infected with strain Δ(trpS aroA/B)/pSP118 was not that impressive, we observed only slightly enhanced serum transaminase levels in single animals at the beginning of the infection (Fig. 6B). This reflects the fact that the aroA/B mutant strain destroyed little or no liver tissue, as shown previously for the aroA mutant WL-141 after in vitro infection of the HepG2 hepatocyte cell line (27).
FIG. 6.
Delivery properties and level of virulence attenuation for a self-destructing L. monocytogenes carrier strain with additional deletions of the aroA and aroB genes. (A) Viable bacterial counts in livers and spleens of C57BL/6 mice that were infected intravenously with 5 × 107 CFU of the self-destructing carrier strains L. monocytogenes ΔtrpS/pSP118 and L. monocytogenes Δ(trpS/aroA/B)/pSP118. (B) GPT levels of mice infected with 5 × 107 CFU of the self-destructing carrier strains L. monocytogenes ΔtrpS/pSP118 and L. monocytogenes Δ(trpS/aroA/B)/pSP118, determined at different times after infection. (C) Clonal expansion of OVA-specific CD8+ cells following infection with 1 × 108 CFU of L. monocytogenes Δ(trpS/aroA/B) containing the plasmids indicated. One day prior to infection, 107 OT-I spleen cells were transferred into C57BL/6 recipient mice. The frequencies of Vα2+ Vβ5+ cells in the total CD8+ T cells in spleens were determined 3 days after infection. Each symbol represents a single animal. The lines indicate the means for the experimental groups (n = 5). The results of one of two experiments are shown.
The aro mutant strains have longer generation times when they are cultured in rich medium (36). Similarly, transformation of L. monocytogenes strains with plasmids pCSA1 and pCBS-OVA enabling delivery of ova mRNA led to growth inhibition. Therefore, we did not investigate an mRNA delivery strain with the aroA/B genetic background as this strain grew very slowly on agar plates and not at all in broth cultures, and we were not able to determine whether the mutants could deliver ova mRNA into mice. In turn, we investigated whether the aroA/B mutant strain has a capacity to deliver OVA protein and ova-encoding DNA similar to that of its parental strain. Since this strain was shown to be more virulence attenuated than L. monocytogenes Δ(trpS)/pSP118, we used a slightly higher dose (1 × 108 CFU). Infection of mice that had received 1 × 107 OT-I spleen cells 1 day previously with L. monocytogenes Δ(trpS aroA/B)/pSP118-PSactAOVA resulted in proliferation of specific OT-I cells so that they accounted for about 49% of the total CD8 T cells (Fig. 6C), showing that this carrier provided large quantities of OVA for efficient MHC class I presentation. In spite of the high infectious doses, mice in this experiment did not show any signs of disease. Like the carrier ΔtrpS/pSP118-PCMVOVA, strain Δ(trpS aroA/B)/pSP118-PCMVOVA was not able to deliver sufficient amounts of ova DNA to mice for MHC class I presentation to occur at any measurable level.
L. monocytogenes strains delivering OVA protein or ova mRNA were able to induce T-cell responses in mice.
The experiments described above showed that after a single application, L. monocytogenes carrier strains ΔtrpS/pSP118-PSactAOVA and ΔtrpS/pCSA1/pCSB-OVA introduced ovalbumin and OVA-encoding mRNA, respectively, into antigen-presenting cells in vivo, leading to presentation of OVA 257-264 by MHC class I and activation of OT-I cells. The delivery of OVA protein resulted in activation of adoptively transferred antigen-specific CD4 T cells; however, the delivery of OVA-encoding mRNA did not result in activation of adoptively transferred antigen-specific CD4 T cells, which could have been due to the cytosolic retention of N-terminally truncated OVA expressed by the host cells.
These data also suggest that there is no exogenous source of OVA after bacterial mRNA delivery in vivo (e.g., by apoptosis of infected cells that have expressed OVA) that would result in uptake of OVA protein by APC for MHC class II-restricted antigen presentation. Alternatively, our experimental setup for the measurement of antigen presentation via MHC class II might not have been sensitive enough since activation of OVA-specific CD4 T cells was demonstrated to require about 10 times more OVA protein (cell associated or soluble) than activation of OVA-specific CD8 T cells requires (18).
To eliminate the latter possibility, we used a more sensitive immunization approach and investigated whether delivery of OVA protein, ova mRNA, or ova DNA by the L. monocytogenes carrier strains led to induction of OVA-specific CD8 or CD4 T-cell memory after repeated applications. To do this, C57BL/6 mice were immunized three times at 3-week intervals with 5 × 107 CFU of ΔtrpS/pSP118-PSactAOVA, ΔtrpS/pCSA1/pCSB-OVA, or ΔtrpS/pSP118-PCMVOVA. The frequency of OVA 257-264-specific CD8 T cells was calculated by determining the number of IFN-γ spots generated per 1 × 105 separated lymphocytes in the presence of the corresponding synthetic peptide. To determine the frequency of IFN-γ-secreting OVA-specific CD4 T cells, spleen cells were stimulated with purified soluble OVA. Mice immunized with ΔtrpS/pSP118-PSactAOVA, the L. monocytogenes strain secreting OVA protein, contained high numbers of IFN-γ-secreting CD8 T cells (Fig. 7A) and CD4 T cells (Fig. 7B). Interestingly, strain ΔtrpS/pCSA1/pCSB-OVA, the L. monocytogenes strain that delivered ova mRNA, was successful in eliciting only specific CD8 T cells at a comparatively lower frequency (Fig. 7A).
FIG. 7.
OVA-specific T-cell responses of mice infected three times at 3-week intervals with 5 × 107 CFU of the self-destructing L. monocytogenes carrier strain ΔtrpS containing the expression plasmids indicated. Spleen cells of infected mice were harvested at day 49 postinfection. Control mice received injections of a saline solution (0.9% NaCl). (A) CD8+ T-cell responses. For ELISPOT analysis, 105 lymphocytes purified from spleen cells were treated in triplicate for 24 h with OVA peptide 257-264 (SIINFEKL) and were assayed for IFN-γ-producing cells. (B) CD4+ T-cell responses. Frequencies of IFN-γ producing cells were determined following stimulation of total spleen cells (4 × 105 cells per well) for 24 h in triplicate with purified ovalbumin. Unstimulated cells (medium controls) had low levels of cytokine responses, and the values for these cells were not subtracted. The results are the results of one of two experiments and are expressed as means ± standard deviations for groups of five mice. An asterisk indicates that there is a significant difference (P < 0.05, as determined by two-tailed Student's t test) between experimental groups.
The low frequency could have been related to the fact that this carrier was not successful in eliciting specific CD4 helper T cells that are thought to be required for the expansion of CD8 T-cell memory (14). In accordance with our previous findings, L. monocytogenes strain ΔtrpS/pSP118-PCMVOVA delivering OVA-encoding plasmid DNA was not able to elicit CD8 or CD4 T-cell memory.
DISCUSSION
L. monocytogenes, a microorganism that replicates intracellularly, can be used to introduce heterologous self-expressed proteins, plasmid DNA, and (as a novel approach) mRNA into the cytoplasm of host cells (31). To do this, we developed a series of expression plasmids specific to L. monocytogenes that allow expression and secretion of antigens by the promoter and secretion signal sequence of the listerial actA gene, or expression of antigens by the host cell using the CMV promoter (27), or T7 polymerase-mediated transcription by the carrier bacteria of antigen-encoding mRNA that can subsequently be translated by the host cell (31). All three types of expression plasmids stably replicate in the carrier bacteria in vivo without the use of antibiotics, and release of DNA or mRNA occurs by disruption of the bacterial cells exclusively within the cytosol of host cells mediated by a phage lysin (27). It has been shown previously that all three strategies can be used to carry antigens or antigen-encoding nucleic acids into antigen-presenting cells in vitro (27, 31). However, the question of which strategy is the most efficient for generating strong CD8 and/or CD4 T-cell immune responses in vivo in order to guide future applications of these systems in novel vaccination approaches had not been addressed previously.
For this purpose, we used a truncated variant of OVA lacking its internal secretion sequence as a model antigen. This variant cannot be secreted by cells expressing the protein and seems to gain improved access to the proteasomal degradation and MHC class I presentation pathway (30). In addition, the adoptive transfer of antigen-specific CD8 and CD4 T cells derived from T-cell receptor transgenic mice prior to exposure to an antigen allows highly sensitive in vivo analysis of early antigen presentation events in the context of MHC classes I and II, respectively (30).
Self-destructing L. monocytogenes carrier strains containing mRNA expression plasmids were able to introduce antigen-encoding mRNA presumably directly into antigen-presenting cells in vivo, leading to activation of antigen-specific CD8 T cells (Fig. 5). Furthermore, delivery of OVA protein secreted by the bacterial carrier was most efficient for activating OVA-specific CD8 T cells, whereas delivery of ova DNA did not elicit any CD8 T-cell responses at all (Fig. 5).
OVA antigen was also presented in the context of MHC class II after infection of mice with L. monocytogenes secreting OVA. This could be explained by the fact that the actA promoter used for expression of OVA exhibits some extracellular background activity. However, MHC class II-restricted presentation was not observed after delivery of ova mRNA. In particular, this suggests that in vivo there might have been no source of extracellular OVA that could have been taken up by APC for MHC class II presentation, such as, e.g., apoptotic bodies that might have originated from OVA-expressing cells after infection with the OVA mRNA delivery strain.
Interestingly, the levels of OVA expression of in vitro infected cell lines were not consistent with the in vivo results. OVA secreted by L. monocytogenes was clearly detected in supernatants of broth cultures. When mice were infected with L. monocytogenes secreting OVA protein, there was strong activation of OT-I cells, indicating that OVA protein was in fact expressed in the cytosol of host cells. In contrast, OT-I cells were activated less in mice infected with L. monocytogenes delivering ova mRNA, implying that smaller amounts of OVA protein were provided by this strategy.
However, infection of Caco-2 epithelial cells with the same strain at a high MOI did not result in detectable amounts of OVA protein, but OVA protein could be detected in preparations of cells infected with L. monocytogenes delivering ova mRNA (Fig. 2) in the same experiments. Similar results were obtained when the infected cells were coincubated with lactacystin, a proteasome inhibitor. The differences might be attributable to different numbers of carrier bacteria during infection (Fig. 3B) since the number of intracellularly located L. monocytogenes cells secreting OVA protein decreased with time, whereas the number of L. monocytogenes cells delivering ova mRNA increased. However, one could also speculate that there might have been differences in the degradation and processing of OVA proteins that were produced by bacteria and mammalian cells.
The inability of the OVA-secreting L. monocytogenes strain to replicate intracellularly might have resulted from the fact that this delivery strain used the promoter of the actA gene fused to the signal peptide of actA to express OVA. This promoter construct was shown to be much more active than a comparable actA promoter construct without the signal peptide (unpublished results). This suggests that high rates of intracellular OVA expression, boosted by the transcriptional activator PrfA, interfere with the intracellular replication capacity of L. monocytogenes. Titration of PrfA by the actA promoter construct can be eliminated as a reason for the low replication capacity. The mRNA delivery strains replicated intracellularly nearly as efficiently as the control strains (Fig. 3B), even though they harbored a multicopy plasmid on which two actA promoter constructs (without a signal peptide) were inserted (Fig. 1).
Although there have been numerous studies describing the successful use of L. monocytogenes as a bacterial carrier for in vivo delivery of heterologous protein antigens (for a recent review see reference 26), until now there has been only one report, a report by Miki et al. (21), showing the delivery of a DNA vaccine plasmid by an attenuated L. monocytogenes strain in a murine model of tuberculosis. Whether the different results for the efficiency of DNA delivery in vivo using attenuated L. monocytogenes as a carrier between the study of Miki et al. and the work presented here are attributable to the different antigens used (mycobacterial antigens Ag85A, Ag85B, and MPB/MPT51 versus a truncated version of chicken ovalbumin), the different attenuated mutants used as carriers (the spreading-deficient Δ2 mutant lacking the entire lecithinase operon, including the virulence-associated genes actA, mpl, and plcB, versus the aroA/B trpS mutants used in this study), or the different experimental setups of the studies (protection against i.v. challenge with Mycobacterium tuberculosis versus analysis of the T-cell repertoire after infection using an adoptive transfer approach) awaits further experimental elucidation.
In contrast, the release of translation-competent mRNA from the bacteria directly into the host cell cytosol enables presentation of the antigen, particularly at early times after infection, which was shown to be crucial for efficient generation of antigen-specific T-cell responses in vivo (2, 4, 20). In addition, transformation of nondividing cells like DC with mRNA results in higher gene transfer efficiency and a higher expression rate than transformation with DNA results in, because DNA has to cross the nuclear membrane in order to be expressed, whereas mRNA only has to reach the cytosol (28, 29).
Therefore, the slow expression kinetics observed after bacterial DNA delivery (31), together with the much earlier onset of the cytotoxic side effects of the listerial carrier on the infected murine DC (9), might explain the lack of antigen presentation observed in vitro as well as in vivo.
In this study, we further characterized the use of a plasmid-encoded phage lysin (Ply118) expression cassette that strongly enhances release of DNA or mRNA from disrupted listerial cells (6, 27, 31). A self-destructing L. monocytogenes strain was shown to be more virulence attenuated in mice than wild-type L. monocytogenes was (Fig. 4). The extent of self-destruction in liver tissue, however, was not as great as that in spleen cells. Our hypothesis is that the phage lysin-mediated lysis of L. monocytogenes strongly depends on preceding disruption of some listerial cells within the cytosol by other mechanisms. Phage lysin 118 cleaves between l-alanine and d-glutamate residues of the L. monocytogenes peptidoglycan (19). This implies that the protein has to cross the bacterial cell membrane or has to be liberated from the bacterial cell to act as a lysin. In spleen cells of infected animals, however, there seem to be mechanisms that eventually assist phage lysin-mediated lysis of L. monocytogenes by bactericidal activities of cell populations like macrophages that act predominantly in spleen tissue.
With regard to safety issues, the expression of Ply118 does not lead to sufficient virulence attenuation of the carrier strains when it is used at doses necessary for efficient delivery of antigens. As indicated by the elevated GPT levels, the self-destructing carrier strains destroy some liver tissue. To study this, we also deleted the aroA and aroB genes in the self-destructing carrier strains, which encode proteins belonging to the basic branch of the aromatic amino acid pathway. aro mutants of L. monocytogenes were shown to have a defect in aerobic respiration, resulting in strongly reduced virulence compared to the virulence of the L. monocytogenes wild-type strain (36). Although the aroA/B deletion in the self-destructing carrier resulted in only a moderate decrease in the bacterial load in the liver, the strains did not destroy significant amounts of liver tissue (Fig. 6c). Delivery of secreted OVA by the self-destructing aroA/B carrier was shown to efficiently activate transferred OT-I cells, whose levels increased to levels comparable to those that were observed after infection with a self-destructing wild-type strain. These properties make the self-destructing aroA/B strain a promising new candidate for a vaccine carrier for the delivery of proteins.
In summary, the data presented in this paper demonstrate that virulence-attenuated L. monocytogenes strains, such as the aroA/B mutant, can be exploited for delivery of various types of molecules, such as proteins, plasmid DNA, or mRNA, in vivo. However, the strategy that is most appropriate for a particular therapeutic approach has to be determined in each case. Our data demonstrate that for vaccination purposes, antigens should be designed as proteins that are expressed and secreted by the carrier bacteria to gain access to both MHC class I and MHC II molecules. However, there are some limitations of this strategy. Not every protein is likely to be secreted by Listeria, and it might be difficult for prokaryotes to express sufficient amounts of proteins of eukaryotic origin without optimization of codon usage (39). Other therapeutic approaches, in which Listeria can be utilized as a carrier for the delivery of, e.g., cytokines in order to modulate immune responses (32), might be possible only by transfer of cytokine-encoding nucleic acids, since so far cytokines cannot be functionally expressed by prokaryotes. Accordingly, for the delivery of prodrug-converting enzymes by L. monocytogenes in the context of gene-directed enzyme prodrug therapy of tumor tissue (J. Stritzker, submitted for publication), the delivery of enzyme-encoding DNA was shown to be superior to the delivery of a secreted enzyme, at least in vitro, showing that there are challenges for every Listeria-based delivery strategy.
Acknowledgments
We thank J. Fensterle for plasmid pCI-OVA and helpful discussions, A. Schimpl for providing OT-I mice, S. Bauer for assistance with animal experiments, and B. Joseph for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through grants SPP 1089, GK 520 (to D.I.M.L.), and SP 479-B1 (to W.G.) and by grants from the Fonds der Chemischen Industrie (to W.G.).
Editor: D. L. Burns
REFERENCES
- 1.Angelakopoulos, H., K. Loock, D. M. Sisul, E. R. Jensen, J. F. Miller, and E. L. Hohmann. 2002. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect. Immun. 70:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badovinac, V. P., B. B. Porter, and J. T. Harty. 2002. Programmed contraction of CD8(+) T cells after infection. Nat. Immunol. 3:619-626. [DOI] [PubMed] [Google Scholar]
- 3.Brockstedt, D. G., M. A. Giedlin, M. L. Leong, K. S. Bahjat, Y. Gao, W. Luckett, W. Liu, D. N. Cook, D. A. Portnoy, and T. W. Dubensky, Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 101:13832-13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin, G. A., and J. T. Harty. 2004. Duration of infection and antigen display have minimal influence on the kinetics of the CD4+ T cell response to Listeria monocytogenes infection. J. Immunol. 173:5679-5687. [DOI] [PubMed] [Google Scholar]
- 5.DeGrendele, H. C., M. Kosfiszer, P. Estess, and M. H. Siegelman. 1997. CD44 activation and associated primary adhesion is inducible via T cell receptor stimulation. J. Immunol. 159:2549-2553. [PubMed] [Google Scholar]
- 6.Dietrich, G., A. Bubert, I. Gentschev, Z. Sokolovic, A. Simm, A. Catic, S. H. Kaufmann, J. Hess, A. A. Szalay, and W. Goebel. 1998. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat. Biotechnol. 16:181-185. [DOI] [PubMed] [Google Scholar]
- 7.Fensterle, J., L. Grode, J. Hess, and S. H. Kaufmann. 1999. Effective DNA vaccination against listeriosis by prime/boost inoculation with the gene gun. J. Immunol. 163:4510-4518. [PubMed] [Google Scholar]
- 8.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 9.Guzman, C. A., E. Domann, M. Rohde, D. Bruder, A. Darji, S. Weiss, J. Wehland, T. Chakraborty, and K. N. Timmis. 1996. Apoptosis of mouse dendritic cells is triggered by listeriolysin, the major virulence determinant of Listeria monocytogenes. Mol. Microbiol. 20:119-126. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. S. White (ed.), PCR protocols: a guide to methods and applications. Academic Press Inc., San Diego, Calif.
- 11.Hoebe, K., E. Janssen, and B. Beutler. 2004. The interface between innate and adaptive immunity. Nat. Immunol. 5:971-974. [DOI] [PubMed] [Google Scholar]
- 12.Hogquist, K. A., S. C. Jameson, W. R. Heath, J. L. Howard, M. J. Bevan, and F. R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell 76:17-27. [DOI] [PubMed] [Google Scholar]
- 13.Ikonomidis, G., Y. Paterson, F. J. Kos, and D. A. Portnoy. 1994. Delivery of a viral antigen to the class I processing and presentation pathway by Listeria monocytogenes. J. Exp. Med. 180:2209-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421:852-856. [DOI] [PubMed] [Google Scholar]
- 15.Kotton, C. N., and E. L. Hohmann. 2004. Enteric pathogens as vaccine vectors for foreign antigen delivery. Infect. Immun. 72:5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Levine, M. M., and M. B. Sztein. 2004. Vaccine development strategies for improving immunization: the role of modern immunology. Nat. Immunol. 5:460-464. [DOI] [PubMed] [Google Scholar]
- 18.Li, M., G. M. Davey, R. M. Sutherland, C. Kurts, A. M. Lew, C. Hirst, F. R. Carbone, and W. R. Heath. 2001. Cell-associated ovalbumin is cross-presented much more efficiently than soluble ovalbumin in vivo. J. Immunol. 166:6099-6103. [DOI] [PubMed] [Google Scholar]
- 19.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 20.Mercado, R., S. Vijh, S. E. Allen, K. Kerksiek, I. M. Pilip, and E. G. Pamer. 2000. Early programming of T cell populations responding to bacterial infection. J. Immunol. 165:6833-6839. [DOI] [PubMed] [Google Scholar]
- 21.Miki, K., T. Nagata, T. Tanaka, Y. H. Kim, M. Uchijima, N. Ohara, S. Nakamura, M. Okada, and Y. Koide. 2004. Induction of protective cellular immunity against Mycobacterium tuberculosis by recombinant attenuated self-destructing Listeria monocytogenes strains harboring eukaryotic expression plasmids for antigen 85 complex and MPB/MPT51. Infect. Immun. 72:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pamer, E. G. 2004. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 4:812-823. [DOI] [PubMed] [Google Scholar]
- 23.Pan, Z. K., G. Ikonomidis, A. Lazenby, D. Pardoll, and Y. Paterson. 1995. A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat. Med. 1:471-477. [DOI] [PubMed] [Google Scholar]
- 24.Pape, K. A., E. R. Kearney, A. Khoruts, A. Mondino, R. Merica, Z. M. Chen, E. Ingulli, J. White, J. G. Johnson, and M. K. Jenkins. 1997. Use of adoptive transfer of T-cell-antigen-receptor-transgenic T cell for the study of T-cell activation in vivo. Immunol. Rev. 156:67-78. [DOI] [PubMed] [Google Scholar]
- 25.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 26.Paterson, Y. 2003. Rational approaches to immune regulation. Immunol. Res. 27:451-462. [DOI] [PubMed] [Google Scholar]
- 27.Pilgrim, S., J. Stritzker, C. Schoen, A. Kolb-Maurer, G. Geginat, M. J. Loessner, I. Gentschev, and W. Goebel. 2003. Bactofection of mammalian cells by Listeria monocytogenes: improvement and mechanism of DNA delivery. Gene Ther. 10:2036-2045. [DOI] [PubMed] [Google Scholar]
- 28.Ponsaerts, P., V. F. Van Tendeloo, and Z. N. Berneman. 2003. Cancerimmunotherapy using RNA-loaded dendritic cells. Clin. Exp. Immunol. 134:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponsaerts, P., V. F. Van Tendeloo, N. Cools, A. Van Driessche, F. Lardon, G. Nijs, M. Lenjou, G. Mertens, C. Van Broeckhoven, D. R. Van Bockstaele, and Z. N. Berneman. 2002. mRNA-electroporated mature dendritic cells retain transgene expression, phenotypical properties and stimulatory capacity after cryopreservation. Leukemia 16:1324-1330. [DOI] [PubMed] [Google Scholar]
- 30.Rush, C., T. Mitchell, and P. Garside. 2002. Efficient priming of CD4+ and CD8+ T cells by DNA vaccination depends on appropriate targeting of sufficient levels of immunologically relevant antigen to appropriate processing pathways. J. Immunol. 169:4951-4960. [DOI] [PubMed] [Google Scholar]
- 31.Schoen, C., A. Kolb-Maurer, G. Geginat, D. Loffler, B. Bergmann, J. Stritzker, A. A. Szalay, S. Pilgrim, and W. Goebel. 2005. Bacterial delivery of functional messenger RNA to mammalian cells. Cell. Microbiol. 7:709-724. [DOI] [PubMed] [Google Scholar]
- 32.Shen, H., M. Kanoh, F. Liu, S. Maruyama, and Y. Asano. 2004. Modulation of the immune system by Listeria monocytogenes-mediated gene transfer into mammalian cells. Microbiol. Immunol. 48:329-337. [DOI] [PubMed] [Google Scholar]
- 33.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soussi, N., G. Milon, J. H. Colle, E. Mougneau, N. Glaichenhaus, and P. L. Goossens. 2000. Listeria monocytogenes as a short-lived delivery system for the induction of type 1 cell-mediated immunity against the p36/LACK antigen of Leishmania major. Infect. Immun. 68:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens, R., A. Lavoy, S. Nordone, M. Burkhard, and G. A. Dean. 2005. Pre-existing immunity to pathogenic Listeria monocytogenes does not prevent induction of immune responses to feline immunodeficiency virus by a novel recombinant Listeria monocytogenes vaccine. Vaccine 23:1479-1490. [DOI] [PubMed] [Google Scholar]
- 36.Stritzker, J., J. Janda, C. Schoen, M. Taupp, S. Pilgrim, I. Gentschev, P. Schreier, G. Geginat, and W. Goebel. 2004. Growth, virulence, and immunogenicity of Listeria monocytogenes aro mutants. Infect. Immun. 72:5622-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabe, L., P. Krieg, R. Strachan, D. Jackson, E. Wallis, and A. Colman. 1984. Segregation of mutant ovalbumins and ovalbumin-globin fusion proteins in Xenopus oocytes. Identification of an ovalbumin signal sequence. J. Mol. Biol. 180:645-666. [DOI] [PubMed] [Google Scholar]
- 38.Verch, T., Z. K. Pan, and Y. Paterson. 2004. Listeria monocytogenes-based antibiotic resistance gene-free antigen delivery system applicable to other bacterial vectors and DNA vaccines. Infect. Immun. 72:6418-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadava, A., and C. F. Ockenhouse. 2003. Effect of codon optimization on expression levels of a functionally folded malaria vaccine candidate in prokaryotic and eukaryotic expression systems. Infect. Immun. 71:4961-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]