Abstract
Gln synthetase (GS) is the key enzyme in N metabolism and it catalyzes the synthesis of Gln from glutamic acid, ATP, and NH4+. There are two major isoforms of GS in plants, a cytosolic form (GS1) and a chloroplastic form (GS2). In leaves, GS2 functions to assimilate ammonia produced by nitrate reduction and photorespiration, and GS1 is the major isoform assimilating NH3 produced by all other metabolic processes, including symbiotic N2 fixation in the nodules. GS1 is encoded by a small multigene family in soybean (Glycine max), and cDNA clones for the different members have been isolated. Based on sequence divergence in the 3′-untranslated region, three distinct classes of GS1 genes have been identified (α, β, and γ). Genomic Southern analysis and analysis of hybrid-select translation products suggest that each class has two distinct members. The α forms are the major isoforms in the cotyledons and young roots. The β forms, although constitutive in their expression pattern, are ammonia inducible and show high expression in N2-fixing nodules. The γ1 gene appears to be more nodule specific, whereas the γ2 gene member, although nodule enhanced, is also expressed in the cotyledons and flowers. The two members of the α and β class of GS1 genes show subtle differences in the expression pattern. Analysis of the promoter regions of the γ1 and γ2 genes show sequence conservation around the TATA box but complete divergence in the rest of the promoter region. We postulate that each member of the three GS1 gene classes may be derived from the two ancestral genomes from which the allotetraploid soybean was derived.
Gln synthetase (GS; EC 6.3.1.2) is responsible for the primary assimilation of ammonia in all living organisms (for reviews, see Reitzer and Magasanik, 1987; Forde and Cullimore, 1989; Cullimore and Bennett, 1992; Woods and Reid, 1993; Reitzer, 1996). Ammonia is assimilated into Gln and Glu through the combined actions of GS and Fd-GOGAT or NADH-GOGAT (Lea and Ireland, 1999). In plants, there are two major isoforms of GS: cytosolic GS (GS1), which occurs in the cytosol, and chloroplastic GS (GS2), which, although nuclear encoded, is located in the chloroplasts/plastids. In leaves, chloroplastic GS functions to assimilate primary ammonia reduced from nitrate and also to reassimilate ammonia released during photorespiration (Lam et al., 1996). In roots, GS1 assimilates ammonia (or NO3−) derived directly from the soil (Lea and Ireland, 1999), and in the cotyledons it reassimilates ammonia released by the breakdown of nitrogenous reserves during germination (Swarup et al., 1990). The GS1 in leaves and stems is localized in the phloem and functions to generate Gln for N transport (Brears et al., 1991; Kamachi et al., 1992). In the root nodules, the primary function of GS is the rapid assimilation of ammonia excreted into the plant cytosol of infected cells by the N-fixing bacteroids (Atkins, 1987).
Plant GS, like other eukaryotic GS, is made up of eight subunits (Meister, 1973) and is probably assembled as two tetramers stacked one upon the other. The native enzyme in plants has a molecular mass ranging from 320 to 380 kD, each subunit having a molecular mass of between 38 and 45 kD. The GS1 genes of several plants, especially legumes, have been cloned and sequenced (Tischer et al., 1986; Gebhardt et al., 1986; Tingey et al., 1987, 1988; Bennett et al., 1989; Boron and Legocki, 1993; Roche et al., 1993; Marsolier et al., 1995; Temple et al., 1995). The GS1 genes in all plants studied are members of small gene families, and the different members show a unique expression pattern suggesting that the gene members are differentially regulated (Gebhardt et al., 1986; Tingey et al., 1987; Bennett et al., 1989; Walker and Coruzzi, 1989; Peterman and Goodman, 1991; Marsolier et al., 1995; Temple et al., 1995).
The Phaseolus vulgaris (French bean) GS1 gene family contains three active GS1 genes (gln-α, gln-β, and gln-γ) and one psuedogene (gln-ε; Gebhardt et al., 1986). All of the three genes exhibit between 79% and 86% homology in the nucleotide sequence in the coding region. The gln-α gene is expressed in the cotyledons and embryonic axis of dry seeds (Swarup et al., 1990) and represents the most abundant GS mRNA in the first 2 d of germination. Functional analysis of the gln-α promoter showed that the gene is also expressed in the vascular tissues, and its expression is increased by mechanical wounding (Watson and Cullimore, 1996). gln-α mRNA appears to be a minor component in the nodules, and its level goes down during nodule development. The gln-β gene product was found to be the major form in the roots and in mature nodules (Gebhardt et al., 1986), and the expression of the gln-β gene appeared to be restricted to the vascular system (Forde et al., 1989). The major GS1 isoform in the nodules is the gln-γ gene product (Bennett et al., 1989), and based on the analysis of nodules from birdsfoot trefoil (Lotus corniculatus) plants containing a gln-γ promoter-β-glucuronidase (GUS) construct, it was concluded that the gln-γ gene is expressed only in the infected cells (Forde et al., 1989). The gln-γ gene also showed a low level of expression in the stems, petioles, and cotyledons of germinating seeds (Bennett et al., 1989; Swarup et al., 1990).
In pea (Pisum sativum), three active GS1 genes have been characterized: GS1, GS3A, and GS3B. All of the three genes are expressed in the nodules, and GS3A and GS3B are expressed strongly in the cotyledons (Tingey et al., 1987). In Medicago truncatula, two active genes (MtGSa, MtGSb) and one psuedogene (MtGSc) were characterized, MtGSa representing the nodule-enhanced form (Stanford et al., 1993). Two major classes of GS1 genes have been characterized in alfalfa (Medicago sativa; Temple et al., 1995), one of which (pGS13) appears to show nodule-enhanced expression and the other class (pGS100) appears to be more constitutive in its expression pattern.
Based on the characterization of GS1 cDNAs and their expression pattern, two classes of GS1 genes have so far been identified in soybean (Glycine max; Roche et al., 1993; Marsolier et al., 1995; Temple et al., 1996); however, the presence of multiple GS1 polypeptides in the roots and nodules of soybean, based on two-dimensional (2D) gel western analysis, suggests the presence of other members of GS1 genes that have not yet been characterized (Temple et al., 1996). The focus of this paper is the isolation, characterization, and regulation of all of the GS1 gene members in soybean. The study also involves a phylogenetic analysis of the different GS1 genes that have been described in the literature in relationship to the GS1 genes described in this paper. Based on genomic Southern analysis and analysis of hybrid-select translation (HST) products, it appears that there are three distinct classes of GS1 in soybean and each class has two members. Although the two members of each gene class show homology in the coding region and the 3′-untranslated region (UTR), there are differences in the expression pattern suggesting differential evolution of the promoters of orthologous GS1 gene members. In an attempt to understand the regulatory mechanism underlying the expression of the different GS1 genes, the promoter regions of the two gln-γ genes have been isolated and subjected to comparative sequence analysis with each other and with the promoter region of the gln-γ gene of French bean (Shen et al., 1992).
RESULTS
Analysis of GS1 Polypeptides in Different Plant Organs
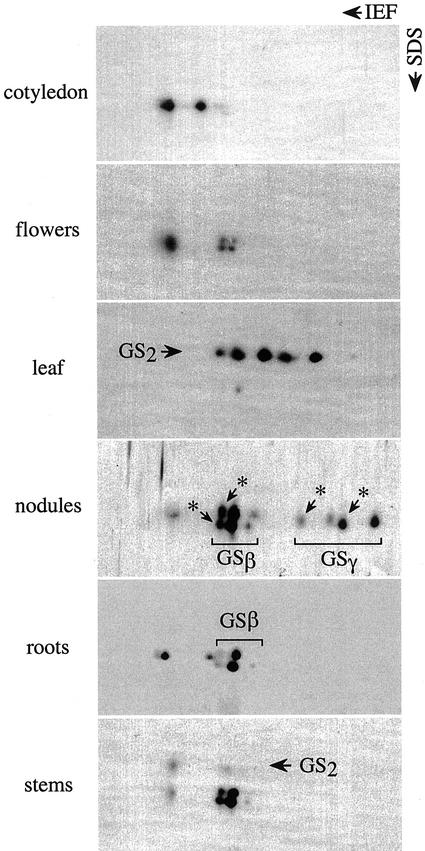
Protein extracts from different organs where GS1 is known to be the major isoform and is functionally significant were subjected to 2D gel electrophoresis followed by western analysis using GS antibodies. The GS polypeptide profile showed a unique organ-specific pattern (Fig. 1) with multiple spots in each case. Based on the analysis of HST products using two cDNA clones, one representing a constitutive form (β-form) and the other a nodule-specific form (γ-form), we had identified some of the spots in the nodule and root extracts as being the GS-β and GS-γ forms (Roche et al., 1993). Some of the spots associated with the GS-β form and the GS-γ form were identified to be their oxidatively modified forms (indicated with asterisks in Fig. 1; Ortega et al., 1999; data not shown). The cotyledons showed two unique spots that did not comigrate with either the two GS-β or the two GS-γ forms or their oxidatively modified counterparts. The unique GS1 polypeptides seen in the cotyledons are also present in the roots and nodules as minor components. The leaf and the stem, besides showing spots that corresponded to the GS1 forms, also showed spots that represented different forms of the GS2 polypeptides.
Figure 1.
The GS polypeptide profile in different organs of soybean. Protein extracts (10 μg) from different organs (as indicated) were subjected to 2D gel electrophoresis, followed by western analysis using GS antibodies. The identity of the GS1 and GS2 polypeptides is based on their Mr. The GS1 polypeptides previously identified as the β- and the γ-polypeptides (GSβ, GSγ) and their oxidized forms (*) are indicated.
Isolation and Characterization of cDNAs Representing the Different GS1 Gene Members
Genomic and cDNA clones for the gln-β and gln-γ genes from soybean have been described in the literature (Roche et al., 1993; Marsolier et al., 1995). However, the presence of multiple GS1 polypeptides in the different organs that cannot be accounted for by just the gln-β and gln-γ genes suggested that there may be other members. To characterize all of the functional genes representing the different members of the GS1 gene family and to obtain gene-specific probes for the different GS1 gene members, reverse transcriptase (RT)-PCR libraries were made separately from RNA isolated from the nodules and the cotyledons. The cotyledons were selected because they showed the presence of two GS1 polypeptides that did not belong to either the β or the γ class of GS1. The primers were designed to obtain the 3′-UTR and part of the 3′-coding region (see “Materials and Methods”). The RT-PCR libraries were subjected to differential hybridization using a 3′-coding region probe, a gln-β 3′-UTR gene probe, and the 3′-UTR of the gln-γ gene. The cDNA clones hybridizing to the coding region probe but not to the gln-β or gln-γ 3′-UTR probes were selected as cDNAs representing GS1 genes other than the gln-β and gln-γ forms. Several GS1 cDNAs representing the non-gln-β and -gln-γ forms were sequenced and found to be identical. The sequences were submitted to BLAST search and the 3′-coding region and the 3′-UTR of this new class of GS1 genes was found to share high sequence similarity to the gln-α gene of (French bean; Gebhardt et al., 1986; accession no. X04002) and probably represents the gln-α gene class of soybean (accession no. AF363020). To check whether there are multiple forms of the gln-α form, many cDNA clones from the cotyledon RT-PCR library representing gln-α were sequenced, but in all cases the 3′-UTR was identical. Several of the cDNA clones that hybridized to the gln-β and the gln-γ 3′-UTRs were also sequenced to check for any variation, and two different classes of 3′-UTR were identified in each case. The sequence of the 3′-UTR and the 3′-coding region for the members of the gln-β gene class and the gln-γ gene class were compared with each other. These genes, although showing a high degree of sequence similarity in the coding region, showed sequence divergence in the 3′-UTR. The two soybean gln-β genes (Gmgln-β1 and Gmgln-β2) showed about 70% sequence identity in the 276 bp of the 3′-UTR and a 90% identity in a stretch of 146 bp within the 3′-UTR. The Gmgln-β1 (accession no. AF301590) corresponds to the clone pGSGmD3′ in the study by Roche et al. (1993), to the pGS20 clone (Miao et al., 1991; accession no. S46513), and to the genomic clone λGS15 (Marsolier et al., 1995). There are no prior reports of any clones corresponding to the Gmgln-β2 (accession no. AF363021). The Gmgln-γ1 clone used in this paper was the same as the pGSGmE3′ reported by Roche et al. (1993; accession no. L20248). The sequence of the corresponding genomic clone was reported by Morey (1997; accession no. AF091456), and it is 99% identical at the 3′-UTR of the clone pGS34 (Marsolier et al., 1995; accession no. X81460). The Gmgln-γ2 clone described in this paper shows 98% identity to the cDNA clone pGS38 (Marsolier et al., 1995) and the genomic clone λGS21 (Marsolier et al., 1995; accession no. AF363022) in the 3′-UTR. There is only 48% identity in the 221 bp of the 3′-UTR of the Gmgln-γ1 and Gmgln-γ2 genes. A stretch of 64 bp in the 3′-UTR, however, shows 89% homology between the two gln-γ genes.
Expression Pattern of the Different GS1 Gene Members in the Different Plant Organs
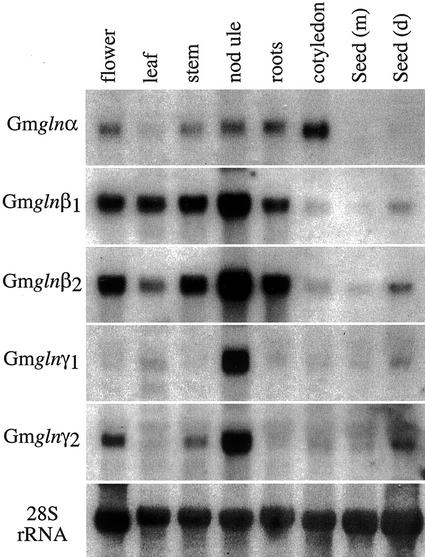
The expression pattern of the different GS1 gene members was analyzed by subjecting RNA from the different organs to northern-blot hybridization using the 3′-UTR of the different GS1 genes as probes (Fig. 2). The gln-α transcript was detected in all of the organs tested, with the highest level of accumulation in the cotyledons of germinating seeds and the lowest level in the seeds and the leaves. The gln-β1 and gln-β2 probes both showed strong hybridization with RNA from 14-d-old nodules. However, whereas the gln-β1 transcript was more abundant in the leaf compared with gln-β2, the gln-β2 probe showed a relatively stronger hybridization signal with RNA from roots. The gln-β2 probe also showed a higher level of hybridization to RNA from mature seeds compared with the gln-β1 probe. The gln-γ1 showed detectable hybridization only to RNA from the nodules, and gln-γ2, although showing the highest level of hybridization to nodule RNA, also showed significant levels of hybridization to RNA from flowers, stem, and to a smaller extent to RNA from the dry seeds.
Figure 2.
Expression pattern of the different GS1 gene members in the different organs of soybean. Total RNA (15 μg) isolated from the different organs (as indicated) was subjected to northern-blot hybridization sequentially with the 3′-UTR of the five different soybean GS1 genes as probes. The blot was also probed with a 28S rRNA gene probe as an indicator of RNA loads.
Relating the GS1 Polypeptides to the Different Classes of cDNAs
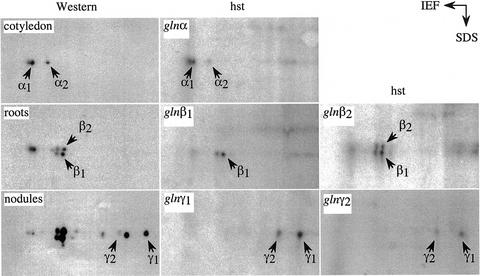
To relate the different GS1 gene members to the corresponding polypeptides, an HST experiment was done using the 3′-UTRs of the five different GS1 genes. The hybrid selected RNA was translated in vitro in the rabbit reticulocyte system, and the translation products combined with the partially purified proteins from the RNA source tissues were subjected to 2D gel electrophoresis. The proteins were transferred to a nitrocellulose membrane and subjected to western analysis using GS antibodies, followed by autoradiography. As shown in Figure 3, the HST products corresponding to the gln-α cDNA could be resolved into three spots, two of which comigrated with the two GS1 polypeptides seen in the cotyledon and the third one with the oxidized form of the GS1-α peptide. The identification of the oxidized form of the GS1-α polypeptide is based on previous oxidation studies performed on soybean root proteins (Ortega et al., 1999). The results would suggest that there are two forms of gln-α in soybean and that the 3′-UTRs of the two genes are highly homologous. The HST products for gln-β1 comigrated with the lower spot corresponding to the GS-β doublet and its oxidized form, whereas the gln-β2 HST product comigrated with both of the GS-β forms, suggesting that, whereas the former is the β1 form, the latter represents the β2 form. The gln-γ1 and the gln-γ2 3′-UTRs both selected mRNA for the GS-γ polypeptides, one of them being more heavily labeled than the other. Based on the relative abundance of the mRNA in the RT-PCR library, it may be concluded that the GS-γ polypeptide that is more basic is probably encoded by the Gmgln-γ1 gene member (Fig. 3). It is, however, interesting to point out that the gln-γ HST products when combined with the nodule extract for 2D gel electrophoresis did not show the oxidized forms of GS-γ, probably because the nodules have a very efficient oxygen radical-scavenging system (Dalton, 1995). In contrast, neither the root nor cotyledon extract protected the GS-α and GS-β translation products from becoming oxidized.
Figure 3.
Relating gene products to the different GS1 gene members. The 3′-UTR of the different GS1 gene members was used to hybrid select RNA from different tissues as indicated, and the selected RNA was translated in vitro in the rabbit reticulocyte system in the presence of [35S]Met. The translation products were then subjected to 2D gel electrophoresis along with protein extract from the same tissue as the source of RNA. The fractionated proteins were then electroblotted onto nitrocellulose and subjected to western analysis followed by autoradiography. The panels labeled western show the 2D GS polypeptide profiles of the different organs (as indicated) and the panels labeled hst represent the 2D profile of the HST products corresponding to the 3′-UTR of the different gene members (as indicated).
Analysis of Genomic DNA for the Hybridization Pattern with the Different GS1 Gene Members
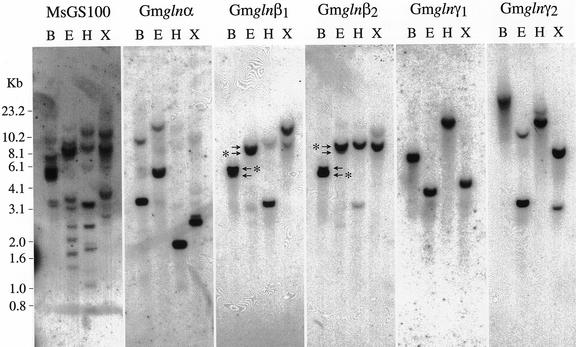
To determine the organization and complexity of the GS1 gene family in soybeans, genomic Southern blots were performed with the 3′-UTR of the different GS1 gene members. Soybean genomic DNA digested with XbaI, EcoRI, HindIII, and BamHI was fractionated on agarose gels and probed with the 3′-UTR of the five GS1 gene members (Fig. 4). The blot was also probed with the conserved coding region of an alfalfa GS1 gene member (MsGS100; Temple et al., 1995). The coding region probe hybridized to multiple bands in each lane, suggesting that GS1 is indeed encoded by a small gene family in soybeans. The 3′-UTRs of the gln-α gene showed one strong hybridizing band and a lighter band in all of the lanes, suggesting that the two bands probably represent two genes with homology in the 3′-UTR and may represent the two genes for gln-α. The gln-β1 and gln-β2 3′-UTRs each hybridized strongly to a unique band in each lane. The two probes, however, showed some degree of cross-hybridization. Thus, the gln-β1 probe hybridized to a small extent to the major β2-hybridizing band and vice versa. In the case of BamHI and EcoRI digests, the gln-β1 and gln-β2 probes hybridized to two closely migrating bands (indicated by arrows in Fig. 4, and the stronger hybridizing band in each case is indicated by an asterisk). The gln-γ1 3′-UTR hybridized exclusively to a single band in each lane, and the gln-γ2 3′-UTR hybridized strongly to one band and weakly to a second band in each lane. The hybridization pattern with the two 3′-UTRs of the gln-γ genes suggests that there is one gln-γ1 gene and two genes related to gln-γ2, and under our hybridization conditions the two probes did not cross-hybridize.
Figure 4.
Genomic Southern analysis. Total genomic DNA was digested to completion with four different restriction enzymes: BamHI (B), EcoRI (R), HindIII (H), and XbaI (X). Digested DNA (10 μg) with each of the restriction enzymes was subjected to electrophoresis followed by Southern-blot hybridization using the 3′-UTR of the different GS1 genes as probes. One of the blots was also probed with a conserved coding region probe. Mr standards were included in the gel during electrophoresis and the position of the different markers is indicated. (The arrows point to the two closely migrating bands in the B and E lanes for gln-β1 and gln-β2 probes and the asterisks point to the stronger hybridizing band in each lane.)
Expression Pattern of the Different GS1 Gene Members in Developing Roots
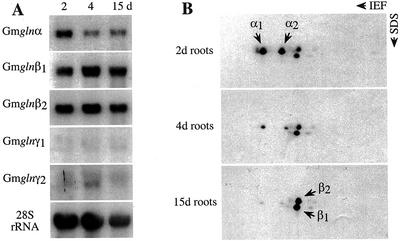
To check whether any of the GS1 gene members show developmental changes in the expression pattern in the roots, RNA from roots at 2, 4, and 9 d postgermination were subjected to northern analysis using the 3′-UTR of the five different GS1 gene members. As shown in Figure 5A, whereas the gln-α probe showed the highest level of hybridization to RNA from roots 2 d postgermination, the gln-β1 and gln-β2 probes showed an increase in the hybridization signal in roots 4 d postgermination compared with the signal in roots 2 d postgermination. Low-level hybridization signals were seen in the 4-d-old root RNA with the gln-γ2 probe. To check how the RNA expression pattern translates into the GS1 polypeptide pattern, protein extracts from roots of different ages were subjected to 2D gel electrophoresis, followed by western analysis with the GS antibodies (Fig. 5B). In the 2-d-old roots, the GS-α forms are predominant, and as the roots mature the GS-α forms are decreased to almost negligible levels and are taken over by the two GS-β forms. No polypeptides corresponding to the GS-γ forms are detectable in any of the root samples.
Figure 5.
Expression pattern of the different GS1 gene members in developing roots. A, RNA from roots at 2, 4, and 9 d postgermination was subjected to northern-blot analysis using the 3′-UTR of the five different GS1 gene members. The hybridization profile with the 28S rRNA gene probe is shown as an indicator of RNA loads. B, Protein extracts from roots of different ages as indicated were subjected to 2D gel electrophoresis followed by western-blot analysis using anti-GS antibodies. GS α and β forms are indicated.
Expression Pattern of the GS1 Gene Members in the Cotyledons of Germinating Seeds
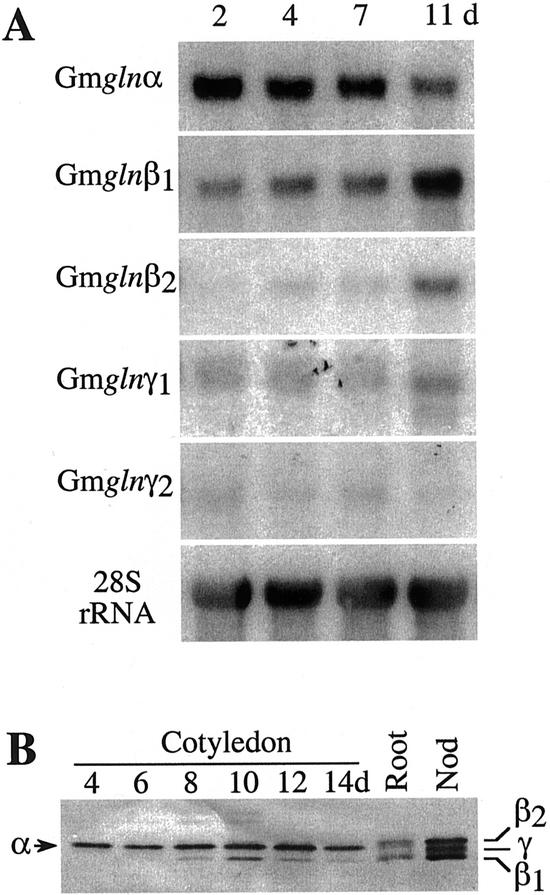
GS plays an important role in cotyledons during germination because it is a site of breakdown of protein reserves. To check which GS isoforms play the major role in assimilating NH3 released by the breakdown of nitrogenous reserves, RNA isolated from cotyledons at different times during germination (as indicated in Fig. 6A) were subjected to northern-blot analysis using the 3′-UTR of the different GS1 gene members. The gln-α probe showed the highest hybridization signal to RNA from cotyledons at 2 d postgermination, and there was a drop in the hybridization signal in the later stages of germination. The gln-β1 gene probe, on the other hand, showed a developmental increase in the level of hybridization, with the highest level being in RNA from cotyledons at 11 d postgermination. The gln-β2 probe did not show any significant hybridization to cotyledon RNA at early stages of germination (2–7 d). A hybridization signal was detectable in the cotyledons at 11 d postgermination. Low levels of gln-γ1 transcripts were also seen in cotyledons at 11 d postgermination. To check whether the RNA levels correlate with the GS1 polypeptide levels, soluble protein extracts from the cotyledons of the germinating seeds were subjected to SDS-PAGE, followed by western analysis using the GS antibodies. To differentiate between the different GS1 polypeptides on a one-dimensional gel, protein extracts from roots and nodules were also included in the gel. The GS polypeptides in the nodules separated out as three distinct bands, the two β forms with the γ form in between the two β forms. The roots, besides showing the two β forms, also showed a band migrating slightly faster than the β2 form. This band comigrates with the major GS isoform in the cotyledons and represents the α form. As shown in Figure 6B, the α form appears to be the predominant GS isoform at all stages of germination, and it is only approximately 8 d postgermination that the β1 form starts appearing.
Figure 6.
Expression pattern of the GS1 gene members in the cotyledons of germinating seeds. A, RNA from cotyledons of germinating seeds were subjected to northern-blot analysis using the 3′-UTR of the five different GS1 gene members. The hybridization profile with the 28S rRNA gene probe is shown as an indicator of RNA loads. B, Protein extracts (10 μg) from the cotyledons of seeds at different days following germination (as indicated) were subjected to SDS-PAGE followed by western-blot analysis using anti-GS antibodies. Root and nodule extracts were also included in the gel. GS α, β, and γ forms are indicated.
Expression Pattern of the Different GS1 Gene Members in Developing and Fix− Nodules
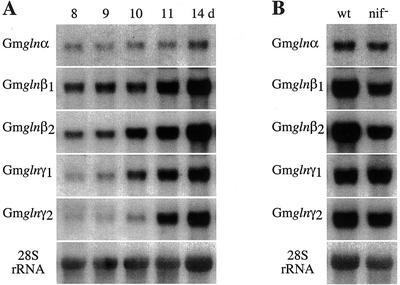
Nodules, besides going through developmental changes, also undergo physiological and biochemical changes associated with the onset of symbiotic N2 fixation and assimilation of fixed N. To check whether any of the GS1 gene members are affected by developmental changes in the nodules, RNA from nodules at different days following infection were subjected to northern-blot analysis using the 3′-UTR of the five different GS1 gene members (Fig. 7A). Except for the gln-α gene probe, all of the probes showed a developmental increase in the level of hybridization starting at d 10. Because the onset of N fixation occurs between d 9 and 10 (Sengupta-Gopalan and Pitas, 1986), the increase in the transcript level for the different GS1 gene members could be as much a developmental process as a physiological response to N2 fixation. To differentiate between the two possibilities, RNA from wild-type and Nif− nodules (14 d postinoculation) were subjected to northern-blot hybridization using all of the different GS1 gene probes (Fig. 7B). Whereas gln-α and the gln-γ genes showed no change in the level of the corresponding transcripts between the two types of nodules, both of the gln-β genes showed a dramatic drop in the level of hybridization in the Nif− nodules, suggesting that the gln-β genes are regulated by physiological changes rather than by developmental changes. The GS1 polypeptide profile in the nodules at different developmental stages and in the Nif− nodules match the expression profile of the GS1 genes at the RNA level (Temple et al., 1996).
Figure 7.
Expression pattern of the different GS1 gene members in developing nodules and in Fix− nodules. A, RNA from nodules at different days following infection was subjected to northern-blot analysis using the 3′-UTR of the five different GS1 gene members. The hybridization profile with the 28S rRNA gene probe is shown as an indicator of RNA loads. B, RNA from nodules formed by wild-type (wt) and Nif− bacteria at 11 d following infection were subjected to northern-blot analysis using the 3′-UTR of the five different GS1 gene members. The hybridization profile with the 28S rRNA gene probe is shown as an indicator of RNA loads.
Comparison of the Promoter Regions of the Gmgln-γ1, Gmgln-γ2, and Pvgln-γ Genes
The subtle differences in the expression pattern of the gln-γ1 and gln-γ2 genes prompted us to compare the promoter regions of the gln-γ genes. A genomic library of soybean (var. Resnik) in λ vector (EMBL3 SP6/T7) was screened with a full-length alfalfa GS1 cDNA clone, and the isolated clones were further characterized using the 3′-UTR of the different GS1 gene members and the conserved 5′-coding region of GS1. The 5′-flanking regions of the Gmgln-γ1 and the Gmgln-γ2 genes were subcloned and sequenced (accession nos. AF182214 and AF021456, respectively). The promoter sequences were submitted to BLAST and FASTA homology searches. Further homology searches were conducted using the Multiple Alignment Construction and Analysis (MACAW) program from the National Center for Biotechnology Information. Sequence analysis showed that there is high homology between the Gmgln-γ1 and Gmgln-γ2 promoters downstream of the putative TATA regions that is also shared with the promoter of the French bean gln-γ gene. The two soybean gln-γ promoters also share some homology with each other 50 bp upstream of the TATA box, upstream of which the two promoters share very little homology. The Gmgln-γ1 and Pvgln-γ promoters, however, share three regions of homology upstream of the TATA box that are conserved both spatially and in sequence. Forde et al. (1989) found a putative consensus sequence for NAT2/PNF-1 binding, TATTTWAT, which is derived from lining up the NAT2-binding sites of N23, leghemoglobin, and the PNF-1-binding region of the French bean gln-γ promoter. There was perfect homology between the first PNF-1-binding site from French bean and the Gmgln-γ1 promoter (−476), but in the second site the sequence is divergent in two of eight bases (−425). The putative NAT2-binding site was also found in the Gmgln-γ2 promoter at positions −426 and −229. A putative nodulin consensus sequence in Pvgln-γ (AAAGAT) at position −412 to −407 is found in the Gmgln-γ1 gene promoter at position −401 and at position −393 in the Gmgln-γ2 promoter. No extensive homology between the soybean gln-γ promoters and the promoter of the gln-γ of yellow lupin (Lupinus luteus (Llgln-γ; Boron and Legocki, 1993) was found, except that the Llgln-γ gene promoter, like the other gln-γ promoters, contain the nodulin consensus sequence and the NAT2-binding sites.
Phylogenetic Analysis of GS1 Genes in Legumes
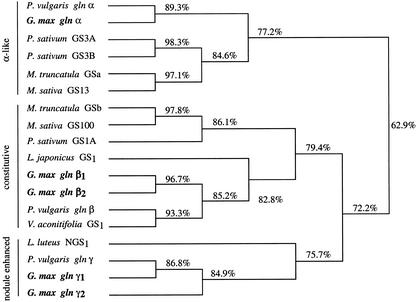
A phylogenetic tree of all of the legume GS1 sequences, including the soybean GS1 genes, was produced using the Mac DNASIS program (Higgins Algorithm with phylogenetic trees) from Hitachi Software (Tokyo). Only the available coding sequence was included in the analysis. As shown in Figure 8, all of the GS1 genes fall within three major groupings (gln-α, gln-β, and gln-γ) that have <80% homology to the other groups. The constitutively expressed members (pea GS1, Tingey et al., 1987, accession no. M20663; M. sativa GS100, Temple et al., 1995, accession no. X03931; M. truncatula GSb, Carvalho et al., 1997, accession no. Y10268; Lotus japonicus GS1 accession no. X94299; French bean gln-β, Gebhardt et al., 1986, accession no. X14605; moth bean [Vigna acontifolia] GS1, accession no. M94765; the Gmgln-β1, Roche et al., 1993, accession no. AF301590; and Gmgln-β2, this paper, accession no. AF363021) all appear to belong to the gln-β branch. The gln-α group contains four GS1 genes that are highly expressed in the nodules (GS3A and GS3B from pea, Tingey et al., 1987, accession no. U28924; GS13 from alfalfa, Temple et al., 1995, accession no. U15591; and MtGSa from M. truncatula, Carvalho et al., 1997, accession no. Y10267), whereas the orthologous members gln-α of French bean (Swarup et al., 1990; accession no. X04002) and soybean (this paper, accession no. AF363020) represent just a minor form in the nodules. The gln-γ group contains nodule-enhanced GS1 genes from soybean (Roche et al., 1993, accession no. L20248; Marsolier et al., 1995, accession no. AF363022), lupin (Boron and Legocki, 1993, accession no. X71399), and French bean (Bennett et al., 1989, accession no. X14605).
Figure 8.
Phylogenetic tree of GS1 genes in legumes. This was produced using the Mac DNASIS program from Hitachi Software. The GS1 sequences were obtained from GenBank and did not include the 5′- and 3′-UTRs. The percentages indicate the percentages of homology. The accession numbers for the reported sequences are: Pvgln-α (X04002); Gmgln-α (AF363020); PsGS3A (U28924); PsGS3B (U28924); MtGSa (Y10268); MsGS13 (U15591); MtGSb (Y10268); MsGS100 (X03931); PsGS1 (M20663); LjGS1 (X94299); Gmgln-β1 (AF301590); Gmgln-β2 (AF36021); Pvgln-β (X04001); VaGS1 (M94765); LINGS1 (X71399); Pvgln-γ (X14605); Gmgln-γ1 (L20248); Gmgln-γ2 (AF363022).
DISCUSSION
The data presented in this paper clearly establish the presence of three classes of active GS1 genes in soybean, each class being represented by at least two functional members. 2D gel profiles of the GS1 polypeptides from the different organs, however, showed that there are more GS1 polypeptides than can be accounted for by the number of GS1 genes in soybean. Based on the analysis of the oxidized GS polypeptides on 2D gels, we have identified six primary GS1 polypeptides; some of the remaining spots were identified as the oxidized forms of the primary GS1 polypeptides (Ortega et al., 1999). More recently, it has been shown that GS1 is also subject to phosphorylation (Finnemann and Schjoerring, 2000). It is possible that some of the unaccounted for spots on 2D gels are phosphorylated forms of some of the GS1 polypeptides. We have used the technique of RT-PCR and a set of nested primers in a conserved region in the 3′-coding region of the gene and an oligo(dT) primer to obtain DNA sequences of the 3′-end of GS1 genes that are expressed in the different organs. Based on the sequence comparison of the 3′-UTR of the amplified cDNAs with the 3′-UTR of the different GS1 gene classes in French bean, two gene members of the GS-β and GS-γ classes were identified, but only one of the members of the GS-α class was identified. Extensive screening of the RT-PCR library for the GS1 genes from the cotyledon library did not result in the identification of the second form of the gln-α gene. The two gln-α genes must have a 3′-UTR that is not identical, because genomic Southern-blot analysis with the 3′-UTR isolated for one of the gln-α genes showed hybridization to one major band and weaker hybridization to a second band in each lane. Moreover, the 3′-UTR of the isolated gln-α gene hybrid selected mRNA that translated into two products of identical molecular masses but different isoelectric focusing (IEF) values. The inability to get a gene for the second GS-α form probably is a reflection of the relative low abundance of the transcript for the second α-form. In fact, the 2D western analysis of cotyledon proteins and the HST products for the 3′-UTR of the GS-α gene does show that one of the products is more abundant than the other (Figs. 1 and 3). The genomic Southern data along with the results of the HST experiment establish the presence of two functional gln-β genes in soybean. The 3′-UTR of the gln-γ1 and gln-γ2 genes each hybridized strongly to unique bands in each lane and did not show cross-hybridization to each other, as was the case with gln-β1 and gln-β2 gene-specific probes. The gln-γ2 gene 3′-UTR, however, showed weak hybridization to an additional band in each lane. Because both the gln-γ1 and gln-γ2 3′-UTR and the coding region (Roche et al., 1993) selected mRNA from the nodule that translated into two primary GS-γ polypeptides, it would follow that the weaker hybridizing band on genomic Southern blot with the gln-γ2 probe probably represents a pseudogene. Pseudogenes for the different GS1 gene members have been reported for both in French bean (Forde and Cullimore, 1989) and M. truncatula (Stanford et al., 1993).
The gln-α gene(s) in soybean appears to show the same expression pattern as the gln-α gene of French bean, in that it is expressed in the cotyledons in the early stages of germination (Swarup et al., 1990), in the young roots (Ortega et al., 1986) and developing nodules (Bennett et al., 1989). Gmgln-α is active in the flowers, and based on promoter GUS fusions in transgenic tobacco, it was shown that the French bean gln-α gene promoter is also active in the flowers (Watson and Cullimore, 1996). Although we cannot distinguish between the transcripts of the two gln-α genes with the single 3′-UTR probe for gln-α, the two can be differentiated by analysis of the protein extract by 2D gel western analysis. Based on the analysis of protein extracts, it appears that the two gln-α genes may be differentially expressed because flowers and stems show only one of the gln-α products or it could be attributed to the relative promoter strength of the two genes.
The gln-β genes in soybean are the homologs of the gln-β gene of French bean, based not only on the sequence similarity in the 3′-UTR but also on the general expression pattern, in that they are expressed more or less constitutively but expressed at relatively high levels in the nodules. In fact, comparison of the promoter sequence of the gln-β1 gene promoter of soybean with the gln-β gene promoter of French bean shows blocks of similarity in a region of about 500 bp near the TATA box and another block of similarity further upstream in the region around −680 to −765 bp (Morey, 1997). The major difference between the gln-β genes of soybean and French bean is that the gln-β genes in soybean are induced in the nodules by reduced N or its assimilation product, whereas in French bean the gln-β gene is not NH3 inducible (Cock et al., 1990). Based on a promoter deletion analysis, Marsolier et al. (1995) showed that the NH3-inducible region is between −1.3 and −3.5 kb in the gln-β1 gene promoter of soybean. The exact cis element, however, has not been identified. It is, however, important to point out that when the soybean gln-β promoter GUS fusion was tested in transgenic plants it was found to be ammonia inducible in birdsfoot trefoil (Miao et al., 1991) but not in alfalfa or tobacco (Nicotiana tabacum; Carrayol et al., 1997). This would suggest that the differences in the NH3 inducibility of the gln-β gene promoters in soybean and French bean could be an attribute of the availability of trans factors rather than major differences in the promoters. This would become clear only after the NH3-inducible cis element has been identified in the promoter of the soybean gln-β genes. It is interesting to note that the gln-β1 and gln-β2 show some differences in their expression pattern in that the gln-β1 transcript is more abundant in the leaves and germinating cotyledons compared with gln-β2 transcripts, which are more abundant in the roots and flowers.
The 3′-UTR of the gln-γ1 and gln-γ2 genes of soybean share the same sequence similarity with each other as they do with the 3′-UTR of the Pvgln-γ gene, suggesting that these conserved sequences might have some functional significance or relatedness in ancestry. It is, however, interesting to note that all of the three gln-γ genes show nodule-specific/-enhanced expression and also some differences in the expression pattern. The Pvgln-γ gene, besides being expressed in the nodules, is also expressed in green cotyledons and stem, the gln-γ2 of soybean is expressed in the flowers, stem, and dry seeds, and the gln-γ1 gene of soybean appears to be primarily nodule specific. It is interesting to note that the two soybean gln-γ promoters are similar only in the vicinity of the TATA box and are completely divergent in the rest of the 5′-flanking regions and the soybean gln-γ1 gene promoter shares more sequence homology with the promoter of the French bean gln-γ gene promoter. A putative nodulin consensus sequence and a NAT2-binding site was found in all of the gln-γ promoters. These sites probably determine the nodule-enhanced expression of the gln-γ genes. This is interesting because it means that not only is there a wide divergence between the GS1 gene promoters but there is promoter divergence between similar GS1 gene members in legumes. There could be three possible explanations for the differential expression pattern of the two gene members for the gln-γ genes in soybean. The first possible explanation for the differential expression pattern is that the two genes are not in fact allelic but resulted from a recent gene duplication event. The loss of gene expression in tissues other than nodules for gln-γ1 could be due to possible loss of promoter sequences needed for expression in tissues other than nodules. The second possible explanation assumes that both of the gln-γ genes are allelic. Sometime during the recent evolution of soybean, the gln-γ1 gene could have lost the necessary cis-acting elements for expression in tissues other than nodules because of deletion or point mutations. A third possible explanation also assumes that the two genes are allelic. Soybean is an allotetraploid, and it is possible that there was some independent promoter evolution of the two alleles in the diploid ancestors of soybean, which was retained after the genetic event that created the allotetraploid soybean. The presence of two members for the gln-β and gln-α gene classes, which also show some differences in expression pattern, would support the third possibility.
The phylogenetic analysis of the legume GS1 genes (Fig. 8) would suggest that the tissue specificity does not necessarily correlate with orthologous relationships defined by sequence homology. For example, the gln-α GS1 isoform of French bean and soybean are not highly expressed in nodules, but their coding region is most like pGS13 from alfalfa (Temple et al., 1995) and GS3A and GS3B from pea (Tingey et al., 1987), which are highly expressed in nodules. In soybean, the gln-β genes are up-regulated when the plant is fed ammonia (Miao et al., 1991); however, the homologous gln-β gene from French bean (gln-β) is not (Cock et al., 1990). The differential expression pattern of the two gln-γ genes of soybean and the gln-γ gene of French bean would also support the notion that differential evolution of the promoters of orthologous GS1 gene family members is an important determinant of the differential expression of the GS1 gene family in legumes. It is interesting to note that only in the tropical legumes has a nodule-specific/-enhanced form of GS (the γ-form) been reported, whereas in the temperate legumes the α-form is the predominant form in the nodules.
MATERIALS AND METHODS
RT-PCR and DNA Manipulations
Standard techniques (Sambrook and Russel, 2001) were used unless otherwise stated. RT-PCR libraries of the cotyledon and nodule RNA were made by reverse transcribing RNA (5 μg) using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and an oligo(dT)18-anchor 20 primer, followed by PCR using the anchored primer and a conserved GS1 primer located 338 bases upstream from the stop codon. A second round of PCR was performed using the anchored primer and a nested GS1 primer 243 bases upstream from the stop codon. The sequence of the dT 18-anchor 20 primer was 5′-GTGAACTTAGGTGACTGACGT18 and those of the nested primers were, 5′-GTGCTGGTGCTCACACA-3′ (outer) and 5′-CACAAGGAGCACATTGC-3′ (inner). The PCR product was gel purified and subcloned into the pGEM-T vector (Promega, Madison, WI) and transformed into Escherichia coli strain DH5-α. The transformed E. coli was transferred to a duplicate gridded nitrocellulose membrane and was hybridized either to the coding region GS1 probe or the 3′-UTR of the gln-β and the gln-γ genes (Roche et al., 1993). The clones hybridizing to the coding region probe and not the gln-β or the gln-γ 3′-UTRs were selected to represent the unidentified GS1 gene members. These clones along with some representative clones hybridizing to the gln-β and gln-γ 3′-UTR probes were subjected to DNA sequencing. The sequences were subjected to homology searches using the BLAST, FASTA, and the MACAW program from National Center for Biotechnology Information.
Nucleic Acid Isolation and Analysis
Total RNA was isolated using the LiCl precipitation procedure (DeVries et al., 1982). RNA was fractionated in 1% (w/v) agarose/15% (v/v) formaldehyde gels and blotted onto nitrocellulose. Genomic DNA was isolated from young expanding leaves of soybean (Glycine max) by a modified hexadecytrimethyl ammonium bromide procedure (Richter et al., 1991). Restricted DNA was fractionated on a 1% (w/v) agarose gel, blotted onto nitrocellulose, and probed with 32P-labeled DNA probes. DNA probes were prepared from plasmid inserts isolated from agarose using the Wizard prep columns (Promega) and labeled by the random primer method as described by Temple et al. (1998). All filters were prehybridized for a minimum of 4 h and hybridized for 20 to 24 h in 50% (v/v) formamide, 5× SSC (1× SSC is 0.15 m NaCl, 0.015 m sodium citrate), 5× Denhardt's solution, 5 mm sodium phosphate (pH 7.0), 0.1% (w/v) SDS, 0.1 mg/mL denatured calf thymus DNA, and 0.04 mg/mL poly(A) at 42°C. Following hybridization, the filters were washed three times with 2× SSC, 0.5% SDS at 42°C for 15 min each, followed by one wash with 0.5× SSC, 0.5% (w/v) SDS at 42°C for 20 min, and exposed to x-ray film. The DNA blot and the different RNA blots were done several times, and only representative experiments are presented here.
Protein Extraction and PAGE
All procedures were carried out at 4°C. The different tissues were ground in liquid N with 15% (w/w) insoluble polyvinylpolypyrrolidone and homogenized with 2 (roots) or 5 (leaves, cotyledons, and nodules) volumes of extraction buffer (50 mm Tris-Cl, pH 8.0, 5 mm EDTA, 5% [v/v] ethylene glycol, 20% [v/v] glycerol, 1 mm magnesium acetate, 1 mm DTT, and a mixture of protease inhibitors: 50 μg/mL antipain, 1 μg/mL cystatin, 10 μg/mL chymostatin, 2 μg/mL leupeptin, and 1 mm phenylmethylsulfonyl fluoride). The homogenate was centrifuged for 15 min at 20,000g. For 2D SDS-PAGE analysis, the tissue extracts were desalted in Sephadex G25 columns against the same buffer as described above.
Protein concentration was measured by the Bio-Rad Protein assay (Bio-Rad, Hercules, CA), using bovine serum albumin as the protein standard. Two different PAGE systems were used: SDS-PAGE using 12% slab mini-gels and 2D SDS-PAGE carried out essentially as described by O'Farrell (1975) and modified by Ortega et al. (1999). Western analysis was carried out essentially as described previously (Ortega et al., 1999). The western analysis described in this paper is a representative example of many experiments.
HST
This was carried out essentially as described by us in an earlier paper (Roche et al., 1993). Plasmid DNA containing the 3′-UTR of the different GS1 gene members (Fig. 2) was immobilized on nitrocellulose discs and hybridized with 250 μg of target total RNA. The hybrid-selected RNA was translated in vitro using the rabbit reticulocyte system (Promega) with [35S]Met as the tracer amino acid. Prior to sample preparation for 2D SDS-PAGE, aliquots of the in vitro translations were mixed with an aliquot of soluble proteins from the appropriate tissues of soybean, and the samples were denatured by boiling in 2% SDS before the addition of 9.5 m urea. Following 2D SDS-PAGE, the polypeptides were transferred to nitrocellulose as described above and subjected to western analysis using GS antibody, and then the filter was air dried and exposed to x-ray film, allowing the detection of the radiolabeled HST products. The HST experiment was done twice and gave similar results.
Screening of Soybean Genomic Library
A soybean genomic library in l vector EMBL3 SP6/T7 from CLONTECH (Palo Alto, CA), was screened with the coding region of an alfalfa (Medicago sativa) GS1 cDNA (pGS100, Temple et al., 1995). The selected clones were further characterized by hybridizing the digested DNA to the 3′-UTR of the different classes of GS1 genes and the 5′-coding region probe. Genomic fragments containing the promoter regions were subcloned and sequenced with Sequenase version 2.0 (Amersham, Piscataway, NJ) kit according to the manufacturer's protocol. The sequences were subjected to homology searches using the BLAST, FASTA, and the MACAW program from the National Center for Biotechnology Information.
Footnotes
This research was supported by the U.S. Department of Agriculture (grant no. 92.37305–7941) and the Agricultural Experiment Station at New Mexico State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010380.
LITERATURE CITED
- Atkins CA. Metabolism and translocation of fixed nitrogen in the nodulated legume. Plant Sci. 1987;100:157–169. [Google Scholar]
- Bennett MJ, Lightfoot DA, Cullimore JV. cDNA sequence and differential expression of the gene encoding the glutamine synthetase γ polypeptide of Phaseolus vulgaris L. Plant Mol Biol. 1989;12:553–565. doi: 10.1007/BF00036969. [DOI] [PubMed] [Google Scholar]
- Boron LJ, Legocki AB. Cloning and characterization of a nodule-enhanced glutamine synthetase encoding gene from Lupinus luteus. Gene. 1993;136:95–102. doi: 10.1016/0378-1119(93)90452-9. [DOI] [PubMed] [Google Scholar]
- Brears T, Walker EL, Coruzzi GM. A promoter sequence involved in cell-specific expression of the pea glutamine synthetase GS3A gene in organs of transgenic tobacco and alfalfa. Plant J. 1991;1:235–244. doi: 10.1111/j.1365-313x.1991.00235.x. [DOI] [PubMed] [Google Scholar]
- Carrayol E, Tercé-Laforgue T, Desbrosses G, Pruvot-Maschio G, Poirier S, Ratet P, Hirel B. Ammonia regulated expression of a soybean gene encoding cytosolic glutamine synthetase is not conserved in two heterologous plant systems. Plant Sci. 1997;125:75–85. [Google Scholar]
- Carvalho H, Sunkel C, Salema R, Cullimore JV. Heteromeric assembly of the cytosolic glutamine synthetase polypeptides of Medicago truncatula: complementation of Escherichia coli mutant with a plant domain-swapped enzyme. Plant Mol Biol. 1997;35:623–632. doi: 10.1023/a:1005884304303. [DOI] [PubMed] [Google Scholar]
- Cock JM, Mold JM, Bennett MJ, Cullimore JV. Expression of glutamine synthetase genes in roots and nodules of Phaseolus vulgaris following changes in the ammonium supply and infection with various Rhizobium mutants. Plant Mol Biol. 1990;14:549–560. doi: 10.1007/BF00027500. [DOI] [PubMed] [Google Scholar]
- Cullimore JV, Bennett MJ. Nitrogen assimilation in the legume root nodule: current status of the molecular biology of the plant enzymes. Can J Microbiol. 1992;38:461–466. [Google Scholar]
- Dalton DA. Antioxidant defenses of plants and fungi. In: Ahmad S, editor. Oxidative Stress and Antioxidant Defenses in Biology. New York: Chapman & Hall; 1995. pp. 298–355. [Google Scholar]
- DeVries SC, Springer J, Wessels JGH. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta. 1982;156:120–135. doi: 10.1007/BF00395427. [DOI] [PubMed] [Google Scholar]
- Finnemann J, Schjoerring JK. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14–3-3 protein interaction. Plant J. 2000;24:171–181. doi: 10.1046/j.1365-313x.2000.00863.x. [DOI] [PubMed] [Google Scholar]
- Forde BG, Cullimore JV. The molecular biology of glutamine synthetase in higher plants. Oxford Surv Plant Mol Cell Biol. 1989;6:247–296. [Google Scholar]
- Forde BG, Day HM, Turton JF, Wen-jun S, Cullimore JV, Oliver JE. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1989;1:392–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Oliver JE, Forde BG, Saarelainen R, Miflin BJ. Primary structure and differential expression of glutamine synthetase genes in nodules, roots and leaves of Phaseolus vulgaris. EMBO J. 1986;5:1429–1435. doi: 10.1002/j.1460-2075.1986.tb04379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiol. 1992;99:1481–1486. doi: 10.1104/pp.99.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi G. The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Ireland RJ. Nitrogen metabolism in higher plants. In: Singh BK, editor. Plant Amino Acids, Biochemistry and Biotechnology. New York: Marcel Dekker; 1999. pp. 1–47. [Google Scholar]
- Marsolier MC, Debrosses G, Hirel B. Identification of several soybean cytosolic glutamine synthetase transcripts highly or specifically expressed in nodules: expression studies using one of the corresponding genes in transgenic Lotus corniculatus. Plant Mol Biol. 1995;27:1–15. doi: 10.1007/BF00019174. [DOI] [PubMed] [Google Scholar]
- Meister A. Glutamine synthetase enzymes of mammals. In: Boyer PD, editor. The Enzymes. X. New York: Academic Press; 1973. pp. 699–754. [Google Scholar]
- Miao GH, Hirel B, Marsolier MC, Ridge RW, Verma DPS. Ammonia-regulated expression of a soybean gene encoding cytosolic glutamine synthetase in transgenic Lotus corniculatus. Plant Cell. 1991;3:11–22. doi: 10.1105/tpc.3.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey KJ. Transcriptional regulation of the glutamine synthetase gene family in soybean: characterization of cis-acting elements involved in the differential expression of individual GS gene family members. PhD thesis. Las Cruces, NM: New Mexico State University; 1997. [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Campos F, Sanchez F, Lara M. Expression of two different glutamine synthetase polypeptides during root development in Phaseolus vulgaris L. Plant Physiol. 1986;80:1051–1054. doi: 10.1104/pp.80.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Roche D, Sengupta-Gopalan C. Oxidative turnover of soybean root glutamine synthetase: in vitro and in vivo studies. Plant Physiol. 1999;119:1483–1495. doi: 10.1104/pp.119.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman TM, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;330:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Reitzer LJ. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate and asparagine, l-alanine and d-alanine. In: Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Ed 2. Washington, DC: American Society of Microbiology Press; 1996. pp. 391–407. [Google Scholar]
- Reitzer LJ, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate and asparagine, l-alanine and d-alanine. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: American Society of Microbiology Press; 1987. pp. 302–320. [Google Scholar]
- Richter HE, Sandal NN, Marker KA, Sengupta-Gopalan C. Characterization and genomic organization of a highly expressed late nodulin gene subfamily in soybeans. Mol Gen Genet. 1991;229:445–452. doi: 10.1007/BF00267468. [DOI] [PubMed] [Google Scholar]
- Roche D, Temple SJ, Sengupta-Gopalan C. Two classes of differentially regulated glutamine synthetase genes are expressed in the soybean nodule: a nodule-specific class and a constitutively expressed class. Plant Mol Biol. 1993;22:971–983. doi: 10.1007/BF00028970. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Ed 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sengupta-Gopalan C, Pitas JW. Expression of nodule-specific glutamine synthetase genes during nodule development in soybeans. Plant Mol Biol. 1986;7:189–199. doi: 10.1007/BF00021330. [DOI] [PubMed] [Google Scholar]
- Shen W-J, Williamson MS, Forde BG. Functional analysis of the promoter region of a nodule-enhanced glutamine synthetase gene from Phaseolus vulgaris L. Plant Mol Biol. 1992;19:837–846. doi: 10.1007/BF00027079. [DOI] [PubMed] [Google Scholar]
- Stanford AC, Larsen K, Barker DG, Cullimore JV. Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol. 1993;103:73–81. doi: 10.1104/pp.103.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Bennett MJ, Cullimore JV. Expression of glutamine-synthetase genes in cotyledons of germinating Phaseolus vulgaris L. Planta. 1990;183:51–56. doi: 10.1007/BF00197566. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C. Down-regulation of specific members of the glutamine synthetase gene family in alfalfa by antisense RNA technology. Plant Mol Biol. 1998;37:535–547. doi: 10.1023/a:1006099512706. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Heard J, Ganter G, Dunn K, Sengupta-Gopalan C. Characterization of a nodule-enhanced glutamine synthetase from alfalfa: nucleotide sequence, in situ localization and transcript analysis. Mol Plant-Microbe Interact. 1995;8:218–227. doi: 10.1094/mpmi-8-0218. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Kunjibettu S, Roche D, Sengupta-Gopalan C. Total glutamine synthetase activity during soybean nodule development is controlled at the level of transcription and holoprotein turnover. Plant Physiol. 1996;112:1723–1733. doi: 10.1104/pp.112.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingey SV, Tsai F-Y, Edwards JW, Walker EL, Coruzzi GM. Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J Biol Chem. 1988;263:9651–9657. [PubMed] [Google Scholar]
- Tingey SV, Walker EL, Coruzzi GM. Glutamine synthetase genes of pea encode distinct polypeptides which are differentially expressed in leaves, roots and nodules. EMBO J. 1987;6:1–9. doi: 10.1002/j.1460-2075.1987.tb04710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer E, DasSarma S, Goodman HM. Nucleotide sequence of an alfalfa glutamine synthetase gene. Mol Gen Genet. 1986;203:221–229. [Google Scholar]
- Walker EL, Coruzzi GM. Developmentally regulated expression of the gene family for cytosolic glutamine synthetase in Pisum sativum. Plant Physiol. 1989;91:702–708. doi: 10.1104/pp.91.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AT, Cullimore JV. Characterization of the expression of the glutamine synthetase gln-α gene of Phaseolus vulgaris using promoter-reporter gene fusions in transgenic plants. Plant Sci. 1996;120:139–151. [Google Scholar]
- Woods DR, Reid SJ. Recent developments on the regulation and structure of glutamine synthetase enzymes from selected bacterial groups. FEMS Microbiol Rev. 1993;11:273–284. doi: 10.1111/j.1574-6976.1993.tb00001.x. [DOI] [PubMed] [Google Scholar]