Abstract
Human diseases caused by species of Aeromonas have been classified into two major groups: septicemia and gastroenteritis. In this study, we reported the molecular and functional characterization of a new virulence factor, ToxR-regulated lipoprotein, or TagA, from a diarrheal isolate, SSU, of Aeromonas hydrophila. The tagA gene of A. hydrophila exhibited 60% identity with that of a recently identified stcE gene from Escherichia coli O157:H7, which encoded a protein (StcE) that provided serum resistance to the bacterium and prevented erythrocyte lysis by controlling classical pathway of complement activation by cleaving the complement C1-esterase inhibitor (C1-INH). We purified A. hydrophila TagA as a histidine-tagged fusion protein (rTagA) from E. coli DE3 strain using a T7 promoter-based pET30 expression vector and nickel affinity column chromatography. rTagA cleaved C1-INH in a time-dependent manner. The tagA isogenic mutant of A. hydrophila, unlike its corresponding wild-type (WT) or the complemented strain, was unable to cleave C1-INH, which is required to potentiate the C1-INH-mediated lysis of host and bacterial cells. We indeed demonstrated colocalization of C1-INH and TagA on the bacterial surface by confocal fluorescence microscopy, which ultimately resulted in increased serum resistance of the WT bacterium. Likewise, we delineated the role of TagA in contributing to the enhanced ability of C1-INH to inhibit the classical complement-mediated lysis of erythrocytes. Importantly, we provided evidence that the tagA mutant was significantly less virulent in a mouse model of infection (60%) than the WT bacterium at two 50% lethal doses, which resulted in 100% mortality within 48 h. Taken together, our data provided new information on the role of TagA as a virulence factor in bacterial pathogenesis. This is the first report of TagA characterization from any species of Aeromonas.
Aeromonas hydrophila represents one of the most predominant species within the family Aeromonadaceae that leads to human diseases, such as gastroenteritis, wound infections, and septicemia (21). These pathogens have been isolated from a wide variety of food and water sources and are being recovered with increasing frequency from patients with traveler's diarrhea (9, 19, 21). Resistance of Aeromonas spp. to water chlorination and to multiple antibiotics has resulted in this organism being placed on the “Contaminant Candidate List” by the Environmental Protection Agency (9). These ubiquitous bacteria produce a large number of virulence factors, many of which have been linked to Aeromonas-associated pathogenesis. Among them are matrix-binding proteins, elastases, proteases, cytotonic and cytotoxic enterotoxins, hemolysins, aldolase, chitinase, lipases/phospholipases, and the ability to form a capsule-like outer layer (10, 30, 35, 45). Aeromonas spp. also possess type IV pili and bundle-forming pili, the latter of which has been shown to be associated with gastroenteritis (23, 29).
Our laboratory purified and extensively characterized the cytotoxic enterotoxin Act and two cytotonic enterotoxins, designated Alt (heat labile) and Ast (heat stable), from a diarrheal isolate, SSU, of A. hydrophila (44). Deletion of these enterotoxin genes from the wild-type (WT) A. hydrophila SSU by marker exchange mutagenesis revealed that they all contributed to fluid secretion in ligated ileal loops in a mouse model, with Act contributing maximally, followed by Alt and Ast (3, 44). More recently, our laboratory characterized a type III secretion system (T3SS) from A. hydrophila SSU and showed that a mutant deleted for both act and the Aeromonas outer membrane protein B gene (aopB), involved in the formation of a needle complex in the T3SS, was avirulent in a mouse model (46). Further, we reported characterization of the DNA adenine methyltransferase gene from A. hydrophila SSU and its role in modulating the function of both T3SS- and type 2 secretion system-associated bacterial virulence (15).
In our attempt to identify new virulence factors in isolate SSU of A. hydrophila, we performed mass spectrometric analysis of several secreted proteins. One of the proteins we identified exhibited homology to a ToxR-regulated lipoprotein, or TagA, recently identified in the enteric pathogens Vibrio cholerae (20) and Escherichia coli O157:H7 (18, 24, 26). Based on recent studies of the mechanism of action of E. coli O157:H7 TagA (now designated StcE for secreted protease of complement C1-esterase inhibitor [C1-INH] from enterohemorrhagic E. coli), it was noted that this protein potentiated the activity of the C1-INH (24). Henceforth, the gene from E. coli O157:H7 will be referred to as stcE and its gene product as StcE. We use the designation of tagA to refer to the gene from A. hydrophila SSU, because this nomenclature was originally adopted for the gene in both V. cholerae and E. coli 0157:H7 (20, 37). The C1-INH belongs to the superfamily of serine protease inhibitors (also referred to as serpins) and is the only inhibitor of activated C1r and C1s of the classical complement cascade, the contact activation pathway, and the intrinsic pathway of coagulation (6). It is therefore endowed with anti-inflammatory properties. Serpins, such as C1-INH (circulating C1-INH has a molecular mass of 105 kDa), inhibit the actions of their respective serine proteases by mimicking the three-dimensional structure of the protease's normal substrate, thus blocking the enzyme's activity (13). Importantly, these proteases cleave within the serpins, leading to the formation of a covalent bond linking the two molecules. A massive allosteric change in the serpin's tertiary structure causes the attached protease to be moved to a site where it can be destroyed (13). Almost 20% of the proteins found in blood plasma are serpins, and their abundance reflects the fact that serpins inhibit excessive proteolysis, which affects host clotting and complement systems (7).
Metalloproteases are widely spread in many pathogenic bacteria, where they play crucial functions related to colonization and evasion of host immune defenses, acquisition of nutrients for growth and proliferation, facilitation of dissemination, or tissue damage during infection of the host (38, 48, 50). StcE is a metalloprotease with a catalytical metal ion, Zn2+, and hence is known as a zinc metalloprotease (26). The cleavage of C1-INH by StcE was initially believed to abrogate the activity of the esterase inhibitor, ultimately resulting in proinflammatory and coagulation responses culminating in localized tissue damage, intestinal edema, and thrombotic abnormalities due to the increased activation of complement (26). However, recent data indicated that StcE might increase and prolong survival of the bacterium within the host by preventing complement activation and the subsequent tissue damage (24). In addition to C1-INH, two other substrates were recently identified for StcE in E. coli O157:H7 (18). The substrates, gp340 and mucin 7, are heavily glycosylated proteins found in human saliva, and therefore, the cleavage of these substrates by StcE could potentially allow the bacterium to establish a successful infection in the host by evading its mucosal defenses (18).
Here, we reported the first identification and characterization of TagA in an Aeromonas species. We provided evidence that the tagA gene was functional in the clinical isolate SSU of A. hydrophila and that purified TagA interacted with and cleaved C1-INH. By cleaving C1-INH, TagA potentiated the activity of this serpin, resulting in more inhibition of complement. We also constructed a tagA isogenic mutant, which lost the ability to bind C1-INH and to cleave the serpin; these activities were restored after complementation of the tagA mutant. Finally, we demonstrated that TagA provided increased serum resistance to A. hydrophila SSU as well as to E. coli DH5α and contributed significantly to bacterial virulence in a mouse model.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. hydrophila SSU and its rifampin-resistant (Rifr) derivative have previously been described (44, 52). Vectors pBluescript and pBR322 were used for cloning, and plasmid pBRtagA, which contained the coding region of the A. hydrophila tagA gene, was used for complementation. A suicide vector, pDMS197, with a conditional R6K origin of replication, a levansucrase gene (sacB) from Bacillus subtilis, and a tetracycline resistance (Tcr) gene was used for homologous recombination (14). The plasmid pHP45Ω, containing a spectinomycin and streptomycin resistance (Sm/Spr) gene cassette, was employed as a selective marker for generating the tagA isogenic mutant. Ampicillin, tetracycline, kanamycin, and spectinomycin and streptomycin (Sm/Sp) antibiotics were used at concentrations of 15, 25, 50, and 100 μg/ml, respectively, in Luria-Bertani (LB) medium or agar plates. The antibiotic rifampin was utilized at a concentration of 40 μg/ml for bacterial growth and 300 μg/ml during conjugation experiments. Chromosomal DNA was isolated using a QIAamp DNA mini kit, and digested plasmid DNA or DNA fragments from agarose gels were purified using a QIAprep miniprep kit (QIAGEN, Inc., Valencia, CA).
Identification of the tagA gene on the chromosome of A. hydrophila SSU.
We identified the tagA gene in A. hydrophila while searching for potential T3SS-secreted effector proteins. One such effector protein (AexT) was recently identified in a fish isolate of Aeromonas salmonicida (5). To evaluate any obvious differences in the protein profiling of A. hydrophila SSU and of A. salmonicida ATCC 49385 strain (American Type Culture Collection, Manassas, VA), the culture filtrates (1 liter) from these bacteria, grown at their optimal temperatures of 37°C and 26°C, respectively, were concentrated by trichloroacetic acid (TCA) (10% final concentration) precipitation. The precipitated proteins were resuspended in 300 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) tracking buffer, and approximately 50 to 100 μg of total proteins was separated by SDS-12% PAGE (42). The gels were then stained with either Coomassie blue or SYPRO Ruby (Bio-Rad, Hercules, CA). A total of 15 bands that were unique in the supernatant of A. hydrophila were isolated from the stained gel, trypsin digested, and subjected to mass spectrometry (MS) and tandem MS analysis in the Proteomics Core Laboratory at the University of Texas Medical Branch (UTMB), Galveston, TX.
One of the proteins of 85 to 90 kDa in size from A. hydrophila SSU exhibited homology (with a score of 0.025 based on an MS analysis search using the Profound database [Genomic Solutions, Ann Arbor, MI]) to a T3SS effector protein homolog from the hrp (hypersensitivity and pathogenicity) T3SS gene cluster of Erwinia amylovora (AAF63400) (51). To better characterize this 85- to 90-kDa polypeptide, the TCA-precipitated proteins were separated by SDS-12% PAGE, transferred to a polyvinylidene difluoride membrane, and stained by Coomassie blue (11, 43). Subsequently, the 85- to 90-kDa band was trypsin digested, and the fragments were subjected to NH2-terminal and internal sequencing at UTMB's Protein Chemistry Core Facility. Based on the sequences of three major tryptic digest peaks of this 85- to 90-kDa protein, a significant homology (64%) was noted within amino acid (aa) residues 109 to 130 of an unknown environmental protein obtained from the Sargasso Sea (EAI65408) (49), based on a ClustalW (Supercomputer Laboratory, Institute for Chemical Research, Kyoto University, Kyoto, Japan) alignment program. The homology with the E. amylovora T3SS effector protein homolog, however, was not as significant (22%) and was limited to a very small region of the 393-aa protein.
With a view that the “unknown” protein could be of interest to us (as a potential T3SS effector protein from A. hydrophila), we designed specific 5′ and 3′ primers (AH5F, 5′-ACCGCCTACTACCTGGAAGGAACGCCGGAGGAGGGG-3′; and AH5R, 5′-CCCCCTGGCTCAGTCGCAGGG-3′). These primers were designed based on the region in the unknown environmental sequence of highest homology beginning from aa residue 115 (a position that corresponded to nucleotide position 353 within the DNA sequence [based on reported 808 bp] of the unknown environmental gene) and the amino acid sequence obtained from two of the tryptic digest peaks of the 85- to 90-kDa polypeptide. The 3′ primer represented the terminal 21 bases of the 808-bp sequence of the unknown environmental gene. This strategy allowed us to amplify, by PCR, a corresponding DNA fragment (455 bp) from the genomic DNA (gDNA) of A. hydrophila SSU, using conditions previously described (46). DNA sequencing of the amplified fragment was performed using an automated DNA sequencer 373XL (Applied Biosystems, Inc., Foster City, CA) in the Protein Chemistry Core Laboratory. Subsequently, our National Center for Biotechnology Information (NCBI) nucleic acid and protein BLAST searches demonstrated a significant identity and homology of this partial sequence to that of stcE of E. coli O157:H7 (AY714880).
Cloning and DNA sequence analysis of the A. hydrophila SSU tagA gene.
By PCR amplification, using gDNA of A. hydrophila and primers designed to the 5′ start and 3′ stop coding region of the E. coli O157:H7 stcE gene, we attempted to amplify the corresponding tagA gene from the gDNA of A. hydrophila. However, we were unsuccessful in amplifying a product, as we later discovered that the 3′ ends of A. hydrophila tagA and E. coli O157:H7 stcE genes were entirely different. Consequently, different strategies were used to obtain a full-length sequence of the tagA gene from A. hydrophila.
Within the 455 bp of the sequenced tagA gene, a single BamHI restriction enzyme site existed. We digested gDNA of A. hydrophila SSU with the BamHI enzyme and performed Southern blot analysis using a [α-32P]dCTP-labeled partial tagA gene (455 bp) as a probe under high stringency conditions (46). Briefly, the BamHI-digested gDNA from WT A. hydrophila SSU (10 μg) was subjected to 0.8% agarose gel electrophoresis and Southern blot analysis (44). Next, the digested DNA was transferred to a nylon membrane and baked at 80°C for 2 h. The conditions for prehybridization, hybridization, and washings of the filters have previously been described (44). Two bands, 2.3 kb and 4.4 kb in size, reacted with the tagA gene probe. Subsequently, DNA fragments within these size ranges were recovered from the agarose gel, purified, and cloned into a pBluescript vector at the BamHI restriction enzyme site. The resulting plasmid libraries in E. coli DH5α were plated (150 to 200 colonies per LB agar plate with 100 μg/ml ampicillin). Colonies from the plates were transferred to nylon filters (Gibco BRL, Gaithersburg, MD), which were then prehybridized and hybridized (46) using the [α-32P]dCTP-labeled tagA gene probe. After being washed, the filters were exposed to X-ray films at −70°C for 2 to 12 h. The plasmid DNA isolated from the pure positive clones (after two to three rounds of purification) was digested with the BamHI enzyme for correct identification of the clones. Both the 2.3- and 4.4-kb cloned DNA fragments were sequenced.
We also screened an existing fosmid library that was prepared using gDNA of A. hydrophila SSU for obtaining the entire sequence of the tagA gene (46). The fosmid library was screened with the labeled tagA gene probe (455 bp) of A. hydrophila, as described above. Finally, DNA isolated from the positive fosmid colonies was used as a template for DNA sequencing. The DNA sequence data were analyzed and compared with the databases using the online BCM Search Launcher (Baylor College of Medicine Human Genome Sequencing Center, Houston, TX) and the ClustalW program.
Generation and characterization of tagA mutant of A. hydrophila SSU.
Based on the DNA sequence of the tagA gene, a portion (1,022 bp from the ATG start codon) was PCR amplified using two primers (Tag5, 5′-ATGACCACATGCACCACACG-3′; and TagR1, 5′-TCCGGCAGCATCACTTCCGG-3′), and the amplified DNA fragment was cloned in the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA). Following DNA sequence analysis, we noted a unique restriction enzyme site, SmaI, in this partial tagA gene of A. hydrophila. By using SmaI digestion, the Sp/Smr gene cassette was removed from the plasmid pHP45Ω and inserted at the SmaI site of the partial tagA gene on the plasmid pCR2.1, thus truncating the tagA gene. By using KpnI-XbaI restriction enzyme digestion (at sites that existed in the pCR2.1 vector), the Sp/Sm-truncated tagA gene containing DNA fragment was isolated from the pCR2.1 vector and ligated to the pDMS197 suicide vector at the compatible restriction enzyme sites. The resulting plasmid (pDMS197TagAPSm/Sp) was transformed into E. coli SM10, which contained λpir (14). This strategy provided 401 and 621 bp of flanking upstream and downstream DNA sequences on each side of the Sp/Smr gene cassette for homologous recombination. The E. coli with recombinant plasmid pDMS197TagAPSm/Sp was conjugated with WT A. hydrophila SSU-R (44, 52). The transconjugants were selected based on resistance to appropriate antibiotics and sucrose and subjected to further Southern blot analysis (44).
Briefly, the gDNA from the tagA mutant as well as from WT A. hydrophila SSU was isolated, and an aliquot (10 μg) was digested with NotI-BglII restriction enzymes and subjected to Southern blot analysis as described above. Three DNA probes representing the partial coding region of the tagA gene (1,022 bp that was PCR amplified using specific primers Tag5/TagR1), a 2.0-kb Sm/Spr gene cassette from plasmid pHP45Ω, obtained by SmaI restriction enzyme digestion, and suicide vector pDMS197 (6.0 kb) were used for Southern blot analysis (44).
Purification of ToxR-regulated lipoprotein (TagA).
PCR amplified from A. hydrophila SSU gDNA, the tagA gene was cloned into a pET-30a(+) T7 promoter-based expression vector (Novagen, San Diego, CA) by using primers with NdeI and BglII restriction enzyme sites, respectively, with the following sequences: TagA5/NdeI, 5′-GGAATTCCATATGACCACATGCACCACACG-3′; and TagA3/BglII, 5′-GAAGATCTTGCGCGTCGCCAGCGGCATGC-3′. To overexpress the tagA gene in E. coli with a histidine tag, the recombinant plasmid was transformed into the E. coli DE3 strain which harbored the T7 RNA polymerase gene on the chromosome. The E. coli (pET-30a-tagA) culture was grown in 300 ml of the LB medium with kanamycin (30 μg/ml) to an optical density at 600 nm (OD600) of 0.5 before induction with a final concentration of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. As a majority of rTagA was present in the bacterial membrane fraction, a high concentration of urea was used to solubilize and purify the protein. Briefly, the bacterial cells were harvested, resuspended in 13 ml of appropriate buffer (8 M urea, 20 mM NaH2PO4, and 500 mM NaCl, pH 7.8), and disrupted by sonication. The cell lysates were passed through the nickel-charged resin column (ProBond; Invitrogen, Carlsbad, CA) (2-ml bed volume in a 10-ml column). The resin was washed with 3-column volumes of the wash buffer, and the TagA protein was eluted in 1-ml fractions (a total of five fractions) with a buffer containing 8 M urea, 20 mM NaH2PO4, and 500 mM NaCl, pH 4.0. The purity of the TagA protein from fractions 1 to 5 was examined by SDS-12% PAGE, followed by Coomassie blue or SYPRO Ruby staining of the gel. The eluted fractions 3 and 4, containing purified rTagA, were dialyzed separately against phosphate-buffered saline (PBS) and examined for enzymatic activity by testing for cleavage of C1-INH, which was obtained from Cortex Biochem, San Leandro, CA.
Complementation of the A. hydrophila SSU tagA mutant.
The tagA gene (2,379 bp) along with an upstream sequence (307 bp) was PCR amplified from the gDNA of A. hydrophila using primer sets (tagA-N/EcoRI, 5′CCGGAATTCACAACCAGCTGGTATGGCAGG-3′; and tagA-C/PvuI, 5′ ATCGATCGTCAGCGCGTCGCCAGCGGCATG-3′). The amplified DNA fragment (2,686 kb) was cloned into the pBR322 vector at the EcoRI-PvuI restriction enzyme sites and subsequently transformed into E. coli JM109 strain. The pBRtagA recombinant plasmid was isolated from the E. coli strain and electroporated (Bio-Rad) into the A. hydrophila tagA mutant strain. To verify the presence of the pBR322tagA plasmid in the A. hydrophila tagA mutant, the isolated plasmid DNA was digested with both EcoRI and PvuI restriction enzymes and the sizes of DNA fragments were compared from both E. coli and A. hydrophila strains. Furthermore, we also generated an A. hydrophila SSU tagA mutant strain containing only pBR322 vector (without an insertion) to be used as a control.
Proteolysis of C1-INH by TagA.
Purified C1-INH (12 μg) was incubated with 1 μg of purified rTagA fusion protein in 120 μl of AD buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, and 0.01% Tween 20) at room temperature. Subsequently, 20 μl of the reaction mixture was removed at various time points and subjected to Western blot analysis using polyclonal goat anti-human C1-INH antibody (Cedarlane, Ontario, Canada) diluted 1:5,000 in 5% bovine serum albumin. The blots were then treated with horseradish peroxidase (HRP)-conjugated secondary antibody (rabbit anti-goat) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) diluted 1:20,000 in 5% skim milk. Subsequently, the membranes were washed, and a chemiluminescence substrate (Pierce Biotechnology, Rockford, IL) was applied and allowed to incubate for 5 min at room temperature before the membranes were exposed to X-ray film (17). To test cleavage of C1-INH by native TagA, WT A. hydrophila SSU, its tagA mutant, and complemented strains were grown to an OD600 of 0.5, washed once with PBS, and resuspended in a final volume of 200 μl of AD buffer. Cells were either concentrated from 1 ml starting volume down to 200 μl of AD buffer (109 cells) or diluted in a volume of 200 μl having 106 bacterial cells. This mixture was then incubated with 20 μg of C1-INH for various time points (0 and 8 h), upon which 20 μl of the reaction mixture was removed and subjected to Western blot analysis.
Lysis of sheep erythrocytes by TagA.
Increasing concentrations of C1-INH (2, 8, and 16 μg) were mixed with 1 μg of rTagA, heated rTagA (80°C for 10 min to inactivate TagA), or elastase (as a negative control) from Pseudomonas aeruginosa (Calbiochem, San Diego, CA) in a total volume of 149 μl AD buffer overnight at room temperature. On the next day, sheep erythrocytes (Colorado Serum Co., Denver, CO) were opsonized with an anti-sheep red blood cell antibody (Rockland Immunochemicals, Inc., Gilbertsville, PA) for 10 min. Human serum (0.5%; Cambrex, Baltimore, MD) was mixed with the opsonized erythrocytes (107 cells) and added to the C1-INH/rTagA (or elastase) reaction mixture from overnight incubation in a total volume of 200 μl. The reaction was allowed to proceed for 1 h at 37°C. Next, 1 ml of AD buffer plus 10 mM EDTA was added to stop complement activity. Erythrocytes were pelleted, and the OD412 of the supernatant was measured using a VersaMax microplate reader (Molecular Devices Corporation, Sunnyvale, CA).
Protease activity.
To further test the protease activity of rTagA, a slight modification of the method described by Erova et al. was used (15). Briefly, increasing concentrations of rTagA (25 ng, 50 ng, 100 ng, and 1 μg), heated rTagA (1 μg heated to 80°C for 10 min), or rTagA (1 μg) neutralized with anti-StcE antibodies (1:20 dilution in PBS) were added to 6-ml snap-cap tubes which contained 500 μl of 1× Dulbecco's PBS and 5 mg of hide azure powder substrate (Calbiochem, La Jolla, CA). The tubes were incubated in a shaker incubator at 37°C for 2.5 to 18 h. Blue color was released as the substrate was catalyzed, which was quantified at OD595. The substrate incubated with 1 μg of elastase from P. aeruginosa (Calbiochem, San Diego, CA) served as a positive control, while substrate with PBS alone was used as negative control.
Serum resistance.
E. coli DH5α was grown to an OD600 of 0.5, washed once with PBS, and resuspended in an equivalent volume of AD buffer. An aliquot (20 μl) of bacteria was added to the overnight room temperature incubation reaction mixture of 8 μg C1-INH treated with or without 1 μg rTagA in a total volume of 176 μl of AD buffer. Human serum was then added to the bacteria and C1-INH/rTagA mixture to a final concentration of 2%. The reaction mixtures were incubated at 37°C for 1 h. The AD buffer was then added to 1 ml final volume plus 10 mM EDTA to stop complement activity. A 10-fold serial dilution of this reaction mixture was plated on LB agar plates, and the percentage of surviving bacteria was determined by dividing the number of CFU by the total number of bacteria after 1 h in the absence of serum. As A. hydrophila SSU, compared to E. coli, is naturally more serum resistant (2), a modification of this method was used to determine increased serum resistance of A. hydrophila due to the presence of TagA. Both WT A. hydrophila and its tagA mutant were grown to an OD600 of 0.5. The bacterial cells were washed and resuspended in AD buffer, and 20 μl of various bacterial strains was added to a reaction mixture of AD buffer and 50% human serum. The reaction mixtures were incubated at 37°C for 3 h. The AD buffer was again added to a final volume of 1 ml along with 10 mM EDTA. A 10-fold serial dilution was plated out on LB-rifampin agar plates, and the percentage of surviving bacteria was determined as described above.
Sandwich Western blot analysis.
Briefly, equal amounts (1 μg) of rTagA, C1-INH, or cholera toxin (negative control) were loaded and separated on SDS-10% polyacrylamide gels, and then these proteins were transferred to nitrocellulose membranes. Membranes were blocked and washed as recently described (17), treated with either rTagA (1 μg/ml in Tris-buffered saline with 0.1% Tween 20, pH 7.5 [TTBS]) or C1-INH (1 μg/ml) in TTBS with gentle shaking for 24 h at 4°C, and then washed. Primary antibodies to StcE, received from Rodney Welch (University of Wisconsin, Madison, WI) (1:5,000), or to C1-INH (1:5,000) were diluted in 5% bovine serum albumin (prepared in 1× TTBS) and allowed to incubate at room temperature for 1 h. After being washed, the membranes were treated with HRP-conjugated secondary antibody and the chemiluminescence substrate as described above.
Binding of C1-INH to TagA on bacterial cell surface.
Briefly, bacterial cells (WT A. hydrophila, its tagA mutant, and the complemented strain) grown to an OD600 of 0.5 were washed with PBS and treated with 2 μg/ml of C1-INH for 2 h. Cells were then washed thoroughly and resuspended in PBS to a final concentration of 107 cells/20 μl. A 20-μl drop of bacterial cells was placed on a glass slide and allowed to air dry. The cells were then fixed with 4% paraformaldehyde for 20 min, washed with PBS, and treated with the appropriate primary antibodies (anti-StcE [diluted 1:500 in PBS], anti-C1-INH [1:500 in PBS], or a combination of both) for 1 h. After being washed two times in PBS, the cells were incubated with fluorescein-conjugated secondary antibodies, purchased from Molecular Probes, Eugene, CA (Alexa Fluor anti-rabbit [1:100 in PBS], Texas Red anti-goat [1:100 in PBS], or a combination of both), for 1 h. Cells were washed again, and 5 μl of DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories Inc., Burlingame, CA) was added to stain the bacterial cell nuclei. Fluorescence labeling was visualized using a Zeiss 510 UV meta confocal microscope (Carl Zeiss, Inc., Thornwood, NY) with an objective lens providing a magnification of ×63 (total magnification, ×2,520).
Animal experiments.
Groups of 10 Swiss Webster mice (Taconic Farms, California) were infected by the intraperitoneal route with 4 × 107 CFU (WT or its tagA mutant) in accordance with approved animal care protocols. Deaths were recorded for 14 days postinfection. This bacterial dose used represented approximately two 50% lethal doses of WT A. hydrophila (52).
Statistics.
Wherever applicable, at least three independent experiments were performed and the data analyzed by Student's t test, P values of ≤0.05 were considered significant. The animal data were analyzed using Fisher's exact test.
Nucleotide sequence accession number.
The sequence of the A. hydrophila SSU tagA gene was deposited in the GenBank database under accession number DQ398103.
RESULTS
Identification and cloning of the tagA gene from A. hydrophila SSU.
The presence of a T3SS in the diarrheal isolate SSU of A. hydrophila (46) led us to postulate the functionality of the system by the secretion of effector proteins. Consequently, we concentrated culture supernatants of A. hydrophila by TCA precipitation and separated the resulting proteins by SDS-12% PAGE. Unique protein bands present in the supernatant of A. hydrophila, but not in A. salmonicida, were isolated from the stained gel, trypsin digested, and analyzed by MS and tandem MS analysis. One of the proteins (AH5; 85 to 90 kDa) yielded some homology to a T3SS-associated effector protein homolog from a plant pathogen, E. amylovora (51). Consequently, this protein was further subjected to NH2-terminal and internal sequencing. Three major tryptic digest peaks were sequenced, and ClustalW alignment of two tryptic digest sequences, a total of 22 aa residues identified by sequencing, also exhibited a 64% homology at the amino acid level within residues 109 to 130 of an unknown environmental protein from the Sargasso Sea (49).
Believing this protein could be of interest to us, we designed primers against the unknown gene sequence to determine the identity of our AH5 gene. Subsequent sequencing of a 455-bp fragment amplified from the gDNA of A. hydrophila SSU resulted in a match with the stcE gene of E. coli O157:H7 (26). Interestingly, updated NCBI BLAST homology searches with the unknown sequence also resulted in a significant match (52% at the amino acid level) with StcE. Consequently, we cloned and sequenced the entire tagA gene, along with its flanking upstream and downstream sequences, by screening two recombinant plasmid libraries of A. hydrophila SSU (for details, see Materials and Methods). We screened approximately 3,000 colonies of each library to obtain four to five positive clones.
We also used an A. hydrophila SSU fosmid library previously prepared in the laboratory to obtain and confirm the sequence of the tagA gene and its flanking sequences. Five positive fosmid clones that contained inserts of approximately 25 kb reacted with the tagA gene probe out of 500 screened colonies. The full-length tagA gene encoded a protein of 793 aa with a molecular mass of 89 kDa. The overall homology at the amino acid level of A. hydrophila SSU TagA with that of E. coli O157:H7 StcE was 64% (Fig. 1), and the identity at the nucleotide level was 60%. It is important to note that the A. hydrophila tagA gene was considerable shorter (by 285 bp) at the 3′ end than its homolog in E. coli O157:H7. However, the TagA of A. hydrophila possessed the unique ligand binding site (which is underlined) (HEVGHNYGLGH) common to all zinc metalloproteases (22), suggesting that this protein should be functional (Fig. 1). Furthermore, A. hydrophila SSU TagA also had a hydrophobic leader sequence (69 bp), an indication that it, like E. coli O157:H7 StcE, would be secreted out of the bacterial cell. Interestingly, TagA of A. hydrophila SSU shared limited identity (<40%, and in the region that spanned the metalloprotease active site) with the only other ToxR-regulated lipoprotein sequence in the GenBank database of unknown function from V. cholerae (U12265). This poses an interesting evolutionary perspective on how A. hydrophila acquired this gene, i.e., whether it was horizontally acquired from the pathogenic E. coli O157:H7 strain, transferred from V. cholerae, or acquired from a third, unidentified source.
FIG. 1.
Amino acid sequence comparison of TagA from A. hydrophila SSU and E. coli O157:H7 StcE. The amino acid sequence of A. hydrophila TagA was obtained after cloning the tagA gene in the pBluescript cloning vector and by sequencing the fosmid library clones. The sequence of 793 aa residues of TagA was aligned by ClustalW Protein Sequence Alignment with the published sequence of E. coli O157:H7 StcE. The conserved zinc metalloprotease-active site of the enzyme is in bold and underlined. -, not found; *, conserved aa residues; :, identical aa residues; ., functionally similar aa residues. Numbers on the right indicate the positions of the aa residues.
Purification of A. hydrophila TagA.
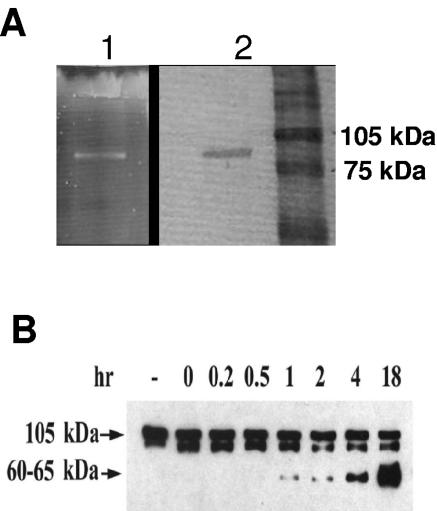
The tagA gene was overexpressed, and rTagA as a histidine-tagged fusion protein was purified using ProBond resin charged with nickel. Purified TagA was eluted from the column in 8 M urea buffer with 20 mM NaH2PO4 and 500 mM NaCl, pH 4.0. Based on SDS-12% PAGE and Coomassie blue or SYPRO Ruby staining, a single protein band with a molecular mass of ∼90 kDa was detected in eluted fractions 3 and 4 (Fig. 2A). The molecular mass of purified TagA was in agreement with the predicted size of 793 aa residues (89,006 Da) based on the DNA sequence plus the additional amino acid residues derived from the histidine tag region of the pET-30a(+) vector.
FIG. 2.
(A) Purification of recombinant TagA. TagA was produced as a fusion protein with a His tag in E. coli DE3 strain from pET30a vector, as described in Materials and Methods. After dialysis, the purified proteins from fractions 1 to 5 were examined by SDS-12% PAGE, followed by SYPRO Ruby (lane 1) or Coomassie blue (lane 2) staining of the gel. Data from fraction 3 (representing homogeneous preparation) are shown. The molecular masses of the protein markers are indicated. (B) Time-dependent digestion of C1-INH by purified recombinant TagA. C1-INH (12 μg) was mixed with rTagA (1 μg) in 120 μl AD buffer. An aliquot (20 μl) of the reaction mixture was removed at various time points (from 0 to 18 h) and subjected to Western blot analysis, as described in Materials and Methods. The primary antibodies used were to C1-INH, followed by HRP-conjugated rabbit anti-goat secondary antibody. The membranes were then treated with the chemiluminescence substrate. The leftmost lane represents untreated C1-INH (2 μg). Lanes designated 0 to 18 indicate hours of incubation of rTagA with C1-INH in order to observe cleavage of the native C1-INH from a 105-kDa to a 60- to 65-kDa polypeptide. The cleaved product of C1-INH (60 to 65 kDa) was observed after digestion with TagA. The presence of a doublet band for commercially available C1-INH was noted which could represent either the degradation product or different forms of C1-INH on a denaturing gel. This phenomenon was also evident in the Western blot analyses of C1-INH cleavage by rStcE′-His (26).
Cleavage of C1-INH by TagA.
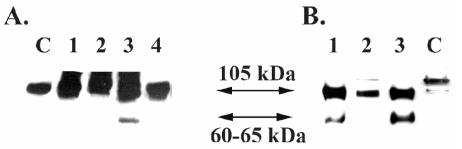
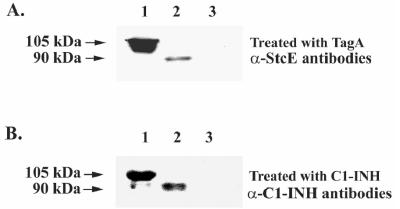
To determine functionality of A. hydrophila TagA, we examined cleavage of C1-INH with rTagA. We expected, based on previous studies (26), that TagA would cleave the native 105-kDa form of C1-INH to a 60- to 65-kDa product. It is known that proteases from several other bacteria, such as Serratia marcescens (28), play a role in cleaving the C1-INH component of the complement system. Purified human C1-INH was mixed with rTagA, and aliquots of the reaction mixture were removed at specific times and subjected to immunoblot analysis using antibodies to C1-INH (Fig. 2B). Over time, cleavage of the 105-kDa band, with the appearance of an ∼65-kDa cleavage product, was apparent. The intensity of the 65-kDa protein band increased significantly between 1 and 18 h. To determine whether this cleavage of C1-INH would be seen with whole cells of A. hydrophila, we treated purified C1-INH with log-phase-grown WT A. hydrophila, its tagA mutant, or its complemented strain (tagA/pBRtagA). As shown in Fig. 3, a specific cleavage pattern was evident only when C1-INH was treated with either the WT bacterium or its complemented strain (Fig. 3A, lane 3, and B, lanes 1 and 3) and not with the tagA mutant (Fig. 3A, lane 1, and B, lane 2). These data show functional protease activity of A. hydrophila TagA by using C1-INH as a substrate.
FIG. 3.
Cleavage of C1-INH by WT and tagA mutant of A. hydrophila SSU. (A). Whole WT A. hydrophila, tagA mutant, or E. coli DH5α bacterial cells (109) were mixed with purified human C1-INH (20 μg), and aliquots of the reaction mixture were removed at various time points (0 and 8 h) for analysis by immunoblotting as described in Materials and Methods. The whole cell of WT A. hydrophila SSU cleaved C1-INH from its native size of 105 kDa into a 60- to 65-kDa fragment (8 h) (lane 3). Lane 2 represents the absence of cleavage seen with WT A. hydrophila SSU at 0 h. The whole cell from the tagA mutant did not cleave C1-INH, even after 8 h of incubation (lane 1). E. coli DH5α cells not possessing the tagA gene were unable to cleave the inhibitor (lane 4), even after 8 h of incubation. A smearing pattern in this blot was due to the high load of bacterial proteins on the gel. (B) In this experiment, the WT, the tagA mutant, or the complemented strain (106 cells) was mixed with 20 μg of C1-INH and incubated for 8 h at room temperature. The absence of C1-INH cleavage noted with the tagA mutant was specific (lane 2), as this activity was restored in the complemented strain (lane 3) and was similar to the pattern seen with WT A. hydrophila SSU (lane 1). In both panels, lane C represents untreated C1-INH (2 μg). Once again, the presence of a doublet band for C1-INH was noted (26).
C1-INH-mediated inhibition of complement is potentiated by A. hydrophila TagA.
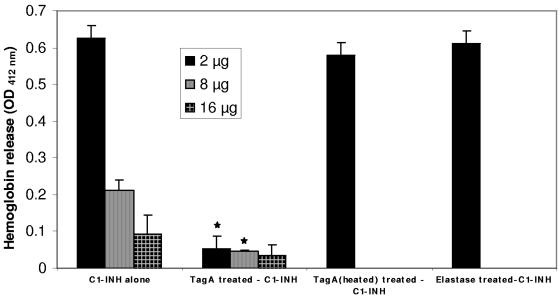
To evaluate A. hydrophila TagA's role in potentiating the inhibitory activity of C1-INH, we tested the effect of TagA-treated C1-INH on the lysis of erythrocytes in the presence of serum. Increasing concentrations of C1-INH (2, 8, and 16 μg) were treated either with or without 1 μg of rTagA overnight before we added opsonized sheep erythrocytes and human serum. As illustrated in Fig. 4, increasing concentrations of C1-INH resulted in a dose-dependent decrease in hemoglobin release or erythrocyte lysis. However, when the inhibitor was pretreated with 1 μg of rTagA, a significant reduction in the lysis of erythrocytes resulted due to increased complement inhibition (Fig. 4). These results were specific to TagA, as C1-INH treated with heated (inactivated) rTagA or with another metalloprotease, elastase from P. aeruginosa, did not reduce erythrocyte lysis at a concentration of 2 μg of C1-INH (the dose at which the greatest reduction in erythrocyte lysis was observed from untreated C1-INH to TagA-treated C1-INH). These data indicated the direct role of A. hydrophila SSU TagA in the potentiation of complement inhibition through its effect on C1-INH.
FIG. 4.
The C1-INH-mediated inhibition of the classical complement cascade is potentiated by TagA from A. hydrophila SSU. Increasing amounts of C1-INH (2, 8, and 16 μg) were treated with 1 μg of rTagA (or heated rTagA), treated with 1 μg of the metalloprotease elastase from P. aeruginosa, or left untreated overnight at room temperature. On the next day, opsonized sheep erythrocytes were mixed with the C1-INH overnight-incubated reaction mixture, as described in Materials and Methods. After incubation for 1 h at 37°C, the erythrocytes were pelleted and the OD412 of the supernatant was measured. * denotes statistically significant values (P ≤ 0.05).
Having illustrated an important function for TagA in contributing to complement inhibition, we examined whether rTagA demonstrated protease activity only against the substrate C1-INH or whether other substrates were also able to be hydrolyzed by this enzyme. We noted that rTagA (25 ng to 1 μg) was able to hydrolyze the chromogenic substrate hide azure powder in a dose-dependent manner. As a positive control, P. aeruginosa elastase demonstrated high protease activity. The protease activity of TagA against this additional substrate was shown to be specific, as recombinant protein heated to 80°C for 10 min or rTagA neutralized by anti-StcE antibodies was not able to hydrolyze the substrate. Further, the protease activity of rTagA was inhibited with 5 mM of EDTA, indicating the requirement of a metal ion (Zn2+) for the enzymatic activity.
TagA increases the serum resistance of E. coli and that of A. hydrophila SSU.
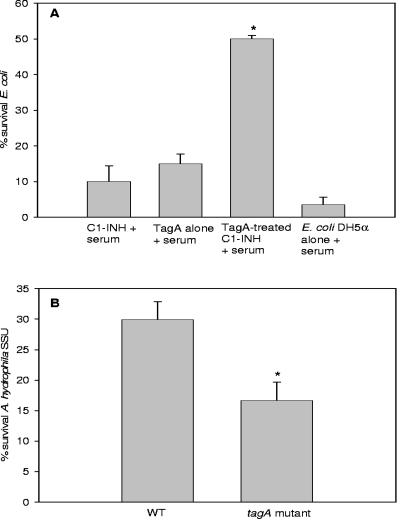
Many pathogenic bacteria have developed mechanisms to evade the host immune system by preventing killing by complement activation (16). As Aeromonas strains are naturally serum resistant due to the presence of either a capsule or surface layer and/or certain outer membrane proteins (2, 32), we first determined whether rTagA of A. hydrophila SSU could impart serum resistance to E. coli DH5α, which is serum sensitive. E. coli strain DH5α grown to mid-log phase was incubated with human serum (2%) and 1 μg of rTagA for 1 h at 37°C, serially diluted, and plated onto LB agar plates to determine the number of CFU and the percentage of survival. E. coli grown in the presence of serum alone had a survival rate of only 5%. However, the addition of TagA-treated C1-INH significantly increased survival of the bacteria to 50% in the presence of serum. The addition of TagA or C1-INH alone in the presence of serum did not significantly increase survival of the bacterium (Fig. 5A).
FIG. 5.
TagA provides increased serum resistance to E. coli DH5α and A. hydrophila SSU by increasing the inhibitory activity of C1-INH. (A) An aliquot (8 μg) of C1-INH was treated with or without 1 μg of rTagA and incubated overnight at room temperature and, on the next day, was added to an aliquot (20 μl) of mid-log-phase grown E. coli DH5α cells, as described in Materials and Methods. After incubation for 1 h at 37°C, the bacteria were serially diluted and plated onto LB agar plates. The percentage of surviving bacteria was calculated as described in Materials and Methods (*, P = 0.001 by unpaired t test). (B) TagA also contributes to the serum resistance of A. hydrophila SSU. WT bacterium or its tagA mutant was grown to mid-log phase, washed in PBS, and resuspended in an equivalent amount of PBS. An aliquot (20 μl) of the bacterial cells was added to a mixture of PBS and 50% serum and incubated at 37°C for 3 h (34). The percentage of surviving cells was calculated as described in Materials and Methods. (*, P = 0.04 by unpaired t test).
We then examined the role of TagA in the serum resistance of A. hydrophila SSU. For these experiments, we compared the survival of the WT bacterium with that of the tagA mutant in the presence of 50% human serum. A high concentration of human serum was required to kill A. hydrophila. Different human serum concentrations (5, 10, 15, 20, and 30%) were used initially to titrate the percentage of surviving WT cells (data not shown). However, only 50% human serum was shown to have any significant effect in killing WT A. hydrophila SSU and demonstrated a significant reduction in the survival of the tagA mutant. As shown in Fig. 5B, the tagA mutant was significantly less serum resistant than WT A. hydrophila SSU.
Binding of A. hydrophila SSU TagA to human C1-INH.
To further confirm the binding of A. hydrophila TagA to C1-INH, we performed sandwich Western blot analysis. In one set of experiments, the purified C1-INH was subjected to electrophoresis, followed by its transfer to a nitrocellulose membrane. This membrane was treated with rTagA before primary antibodies were added. When the membrane was probed with antibodies to StcE, a band of 105 kDa was detected corresponding to the size of purified human C1-INH (2 μg), which was run on the gel (Fig. 6A, lane 1). Therefore, detection of a band similar in size to C1-INH indicated binding of TagA to C1-INH on the membrane. rTagA was also run on the gel as a positive control, indicating the native size of the protein (Fig. 6A, lane 2). Similarly, when purified rTagA (1 μg) was first run on the gel and the membrane was treated with C1-INH, followed by antibodies to C1-INH, a band corresponding to the size of TagA (90 kDa) was detected (Fig. 6B, lane 2). As a positive control, C1-INH (1 μg) was also run on the gel and a band corresponding to 105 kDa reacted to the C1-INH antibodies (Fig. 6B, lane 1). No band was detected in either gel when cholera toxin (CT) was used (Fig. 6A and B, lane 3), indicating a specific interaction between C1-INH and TagA. It is important to note that antibodies to C1-INH specifically reacted with purified human C1-INH and did not react nonspecifically with rTagA and vice versa (data not shown). Sandwich Western blot analysis data further substantiated interaction of TagA with its substrate C1-INH in both their native and denatured forms.
FIG. 6.
Confirmation of the interaction of TagA with C1-INH by sandwich Western blot analysis. C1-INH (2 μg), rTagA (1 μg), or cholera toxin (1 μg) was loaded on SDS-12% polyacrylamide gels, and electrophoresis was performed before transfer to nitrocellulose membranes. The membranes were subsequently incubated with either TagA or C1-INH and probed with anti-StcE or anti-C1-INH antibody. (A) Results of a sandwich Western blot analysis using C1-INH (2 μg). A 105-kDa-sized band (lane 1) was detected when membranes were first treated with rTagA (1 μg/ml) overnight at 4°C and then probed with anti-StcE antibody. Lane 2 was used as a control to indicate the native size of TagA. No band was detected when CT was loaded on the gel instead of C1-INH (lane 3). (B) Results of a sandwich Western blot analysis in which TagA protein (1 μg) was loaded on the gel, incubated with C1-INH (1 μg/ml) overnight, and probed with anti-C1-INH antibody. Note the binding of C1-INH specifically to TagA (lane 2, a 90-kDa band). Lane 1 was used to visualize the native size of the C1-INH protein. No band was detected when CT was loaded on the gel instead of TagA (lane 3).
Binding of C1-INH and TagA on bacterial surface of A. hydrophila SSU.
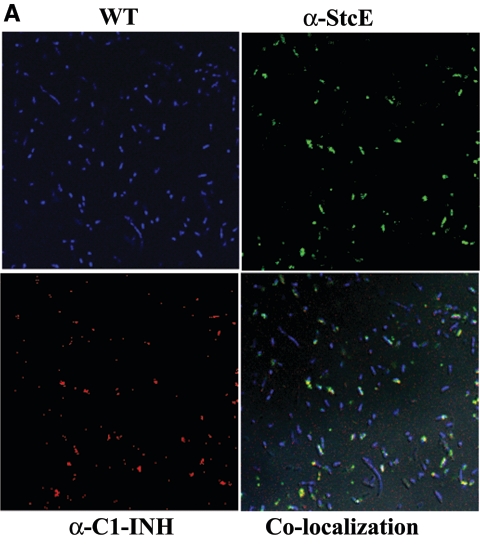
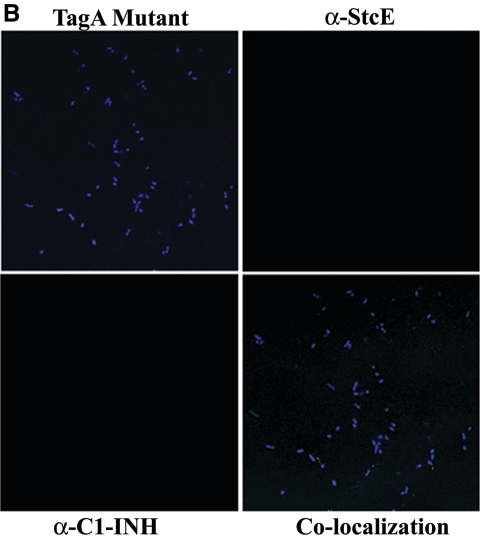
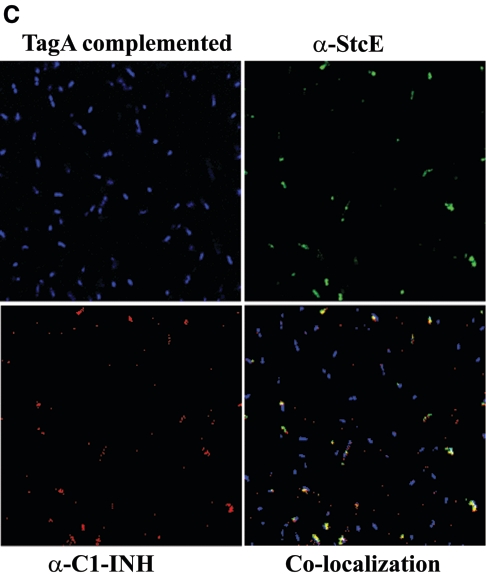
The cleavage of C1-INH by StcE has been reported previously (26). Further, to elucidate the functional interaction of C1-INH and TagA and the specificity of binding of C1-INH to the bacterial surface of A. hydrophila SSU, we performed confocal fluorescence microscropy. Colocalization experiments were performed using polyclonal antibodies to both C1-INH and StcE and using the WT bacterium, its tagA mutant, and the complemented strain. Treating WT bacteria with C1-INH and then staining the cells with antibodies to both C1-INH and StcE revealed a colocalization pattern consistent with the observed interaction between these two proteins (Fig. 7A). The binding specificity of C1-INH to TagA was demonstrated using the tagA mutant and the complemented strain. As shown in Fig. 7A and C, the WT and complemented strains bound individually to both of the fluorescently tagged antibodies. However, the tagA isogenic mutant did not express TagA on the surface, as seen from the lack of staining with anti-StcE antibodies and Alexa Fluor-labeled secondary antibodies, and hence did not show any colocalization of the metalloprotease and C1-INH (Fig. 7B). The complemented strain, however, displayed colocalization of TagA and C1-INH on the bacterial surface similar to that seen in the WT bacterium (Fig. 7C).
FIG.7.
Colocalization of C1-INH and TagA on the surface of A. hydrophila SSU and its tagA isogenic mutant. WT A. hydrophila and the tagA mutant were grown to an OD600 of 0.5, washed with PBS, and treated with 2 μg of C1-INH for 2 h. Cells were then washed thoroughly with PBS several times and resuspended in PBS to a final concentration of 107 cells/20 μl. The cells were then fixed with 4% paraformaldehyde, washed with PBS, and then incubated with primary and fluorescein-conjugated secondary antibodies to both C1-INH and StcE for 1 h, as described in Materials and Methods. After subsequent washes of cells in PBS, followed by DAPI staining of the cells, binding was inspected by confocal fluorescence microscopy. The panels indicated as WT cells, tagA mutant, and tagA-complemented strain show staining by DAPI. The panel indicated as α-StcE illustrates binding with anti-StcE antibody and Alexa Fluor-labeled secondary antibody, while the α-C1-INH panel indicates binding with anti-C1-INH antibody and Texas Red-labeled secondary antibody. The colocalization panel demonstrates binding of these two labeled proteins on the bacterial surface. (A) WT A. hydrophila. Approximately 70% of the total number of the cells in the field showed colocalization of TagA and C1-INH. (B) tagA mutant bacteria. No binding of TagA with C1-INH was seen. (C) pBRtagA-complemented strain. Approximately 60% of the total number of cells in the field showed colocalization of TagA with C1-INH. At least five fields were visualized to calculate the percentage of positive cells. Total magnification, ×2,520.
In vivo effects of TagA in A. hydrophila SSU.
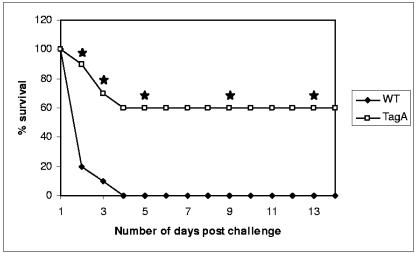
In our in vivo studies, we noted that 100% of the animals infected with the 4 × 107 dose of WT A. hydrophila died within 48 h (Fig. 8). However, only 40% (P = 0.0038 compared to WT bacteria) of the animals died when inoculated with the tagA mutant of A. hydrophila SSU at the same dose. These data indicated that TagA played a significant role in contributing to the overall survival of the bacteria within the host. We also investigated the colonization of the tagA mutant bacteria in comparison to that of WT A. hydrophila. The in vivo survival of WT A. hydrophila and its isogenic mutant was determined by counting the number of bacteria in the spleen of the mice on days 1 and 3 postinfection. We noted no significant difference in the numbers of WT and tagA mutant bacteria in the spleen on day 1 (data not shown). Further, the bacteria were quickly cleared from the host system, as day 3 showed almost no bacteria to be present in the tissue, with no significant difference between the WT and the mutant (data not shown). These data indicated that the observed difference in survival of mice challenged with the tagA mutant bacteria was not due to an impaired ability of these bacteria to colonize host tissue.
FIG. 8.
TagA of A. hydrophila SSU contributes to the virulence of the bacterium. Swiss Webster mice (n = 10 per group) were injected intraperitoneally with two 50% lethal doses of WT A. hydrophila SSU. The same dose was used to infect mice with the tagA mutant, and both groups were monitored for death over a 14-day period. The data were statistically analyzed using Fisher's exact test. Three independent experiments were performed, and data from a typical experiment are shown. * denotes statistically significant differences between the tagA mutant and WT bacteria (P ≤ 0.05).
DISCUSSION
The role of TagA in bacterial virulence is relatively new, with the presence of the tagA gene detected only in V. cholerae and E. coli O157:H7 (20, 24-26). The functionality of this gene (designated stcE) was recently elucidated only for the latter pathogen, where it is encoded on the pO157 virulence plasmid (18, 24, 26). In our study, we identified, cloned, and functionally characterized the chromosomally encoded tagA gene from a diarrheal isolate, SSU, of A. hydrophila. Further, the role of A. hydrophila TagA in altering bacterial virulence was evaluated using both in vitro and in vivo models. We showed for the first time the role of TagA in a mouse model of lethality as well as visualized colocalization of this metalloprotease to its target C1-INH on the bacterial surface.
TagA is regulated by the ToxR regulon, with the latter linked to the virulence potential of the Vibrio species. Although little is known regarding the host signals that impact the ToxR regulatory cascade, it is clear that these intraintestinal signals play an important role in maximizing the ability of bacteria to survive and multiply within the host (47). Whether TagA of A. hydrophila is regulated by a mechanism similar to that of Vibrio species is not known. TagA's role in bacterial pathogenesis is only now beginning to be understood.
To date, three proteases have been reported for A. hydrophila: (i) a thermostable 38-kDa metalloprotease (27, 40), (ii) a 19-kDa zinc protease (27), and (iii) a 68-kDa temperature-labile serine protease (39). The 38-kDa metalloprotease (AhyB) has elastolytic activity, and its isogenic mutant showed decreased virulence in rainbow trout (8). Colony blot hybridization analysis in our laboratory showed that TagA was widely distributed in Aeromonas species. Of 165 water isolates of Aeromonas screened for the tagA gene, 59% were positive. Further, 21% of 52 clinical isolates, obtained from patients with gastroenteritis, were also found to possess the tagA gene (data now shown). The presence of the tagA gene was noted in isolates obtained from patients with both gastroenteritis and wound infections.
When the metalloprotease StcE was initially identified in E. coli O157:H7, it was believed to contribute to the pathophysiological derangements of hemolytic uremic syndrome (HUS) by degrading the serpin C1-INH (26). The degradation of C1-INH would result in the loss of control of multiple proteolytic cascades, including the classical and alternative complement pathways, intrinsic coagulation, and contact activation (26). Aeromonas spp. are also known to cause HUS (4, 41). Therefore, it is plausible that TagA could play a role in HUS during Aeromonas infections. However, we noted that the cleavage of C1-INH by TagA enhances, rather than inhibits, the serpin's ability to downregulate the classical complement cascade (as demonstrated in E. coli O157:H7), thereby protecting bacterial and host cells from the deadly lytic effects of complement activation (24). The mechanism by which TagA potentiates the ability of C1-INH to increase complement inhibition is not fully understood and requires further investigation.
The pathogenic and virulence characteristics of A. hydrophila have been shown to be associated with the production of type II secretion system-associated exoenzymes, such as proteases and lipases (10, 21). Our results showed that TagA possessed protease activity by acting on the substrates C1-INH and hide azure powder. Earlier studies indicated that StcE from E. coli O157:H7 was not able to degrade casein, although elastase was able to do so, indicating the specificity of the metalloprotease StcE for the substrate C1-INH (26). However, later studies did increase the range of potential substrates for StcE to include mucin 7 and glycoprotein 340 (18). We would like to explore further potential substrates that TagA could act upon on the gut mucosal surface, a major site of inflammation in gastroenteritis caused by A. hydrophila.
Previous studies showed that StcE did not play a role in the general adherence of E. coli O157:H7 to HEp-2 cells but that it contributed significantly to their intimate adherence (18). Likewise, our results indicated that TagA did not contribute to the adherence of A. hydrophila cells to HT-29 colonic epithelial cells (31; data not shown) or in vivo.
Lathem et al. (24) reported that native C1-INH could not bind to erythrocyte surfaces. However, when treated with StcE of E. coli O157:H7, the serpin was able to bind to the cell surface and hence increased the local concentration of the inhibitor at sites of potential lytic complex formation (24). Different serogroups of A. hydrophila are able to evade complement by preventing the formation of this complex on their cell surface. For example, A. hydrophila strains devoid of the S-layer are resistant to complement-mediated killing because C3b is rapidly degraded and, therefore, the lytic membrane attack complex is not formed (34). Other factors can also contribute to the serum resistance of A. hydrophila, such as the long O-polysaccharide chain of lipopolysaccharide and a capsule-like outer layer, which help protect the bacterium from the killing effects of complement (2, 32, 33). Two capsular genes of A. hydrophila (serogroup O:34) were determined to confer serum resistance to the E. coli K-12 serum-sensitive strains (2). Our studies indicated that TagA from the clinical isolate SSU of A. hydrophila plays a similar role, as it could confer serum resistance not only to the homologous strain but also to a serum-sensitive E. coli strain.
It is reported that StcE is expressed on the surface of E. coli and could act as a “bridge” between C1-INH and the cell surface (24). Using confocal fluorescence microscopy, we illustrated colocalization of these two proteins in WT A. hydrophila and in the tagA mutant complemented with the tagA gene on a plasmid. How does the binding of this metalloprotease to the inhibitor of the classical complement cascade relate to virulence of the bacterium within a host? This question was addressed by our animal studies.
We provided evidence for the first time of TagA's role in vivo and demonstrated reduced lethality in mice infected with the tagA mutant. If TagA is contributing to increased complement inhibition, then why would deletion of this gene result in less host killing? One would expect that the absence of this gene would lead to a more striking virulent phenotype of the bacterium, and hence, more pathology, such as inflammation and tissue damage, would be expected in the host, eventually resulting in death. Perhaps TagA's role in serum resistance is quite significant. Without the coordinated effort of TagA and other factors, A. hydrophila could succumb to the bactericidal effects of human serum more readily. Further, TagA's role as a protease could play a significant role in the in vivo situation. It is known that metalloproteases from other bacteria such as P. aeruginosa as well as from A. hydrophila are crucial to the virulence potential of these pathogens (8, 12).
Other explanations can also be offered to describe the phenomenon observed in vivo. TagA may play a dual role in the virulence of A. hydrophila. In addition to potentiating the inhibition of complement, it could cause damage in the host by acting synergistically with other virulence factors. Vollmer et al. (50) uncovered a novel mechanism by which microbial proteases possibly provoke long-range biological effects in the host cell. Specifically, this group discovered that certain membrane-anchored proteins, including several cytokines and cytokine receptors, were released into tissue culture supernatants of tissue fluid in vivo through the action of endogenous membrane-bound metalloproteinases. The shed molecules were then able to perform biological functions; for example, soluble interleukin-6 receptor could bind to bystander cells, rendering these cells sensitive to the action of interleukin-6 (50). Our future studies will focus on elucidating TagA's effect on host immune status, specifically with regard to cytokine production/secretion. The other side of this possible dual nature of TagA is illustrated by another important metalloprotease, the lethal toxin of Bacillus anthracis, which cleaves upstream mitogen-activated protein kinases and promotes immune evasion of the bacterium by suppressing activation of macrophages and dendritic cells (1, 36).
This study highlighted a unique role for the A. hydrophila SSU metalloprotease TagA in potentiating the activity of the serpin C1-INH in inhibiting complement activation. Our data illustrated that this enzyme also contributed to the serum resistance of the bacterium and played a direct role in its virulence. Research conducted on metalloproteases, such as TagA, illuminated a fascinating role for these proteinases in bacterial virulence; in many cases, they are key players in subverting host immune defenses (38). It is obvious that bacterial proteases represent very attractive targets for the generation of a novel class of therapeutics, since these enzymes are ubiquitously found in many bacterial species, and the inhibition of such critical enzymes would presumably lead to the death of the invading pathogen.
Acknowledgments
This work was supported by a grant from the NIH/NIAID (AI41611) and by the American Water Works Association Research Foundation. L. Pillai, a predoctoral fellow, obtained funding from the NIH T32 training grant in Emerging and Tropical Infectious Diseases. A. A. Fadl was supported by the McLaughlin Postdoctoral Fellowship.
We thank R. A. Welch (University of Wisconsin, Madison, WI) for providing the antibodies to TagA (StcE) from E. coli O157:H7 and M. J. Susman for editing the manuscript. All of the DNA sequencing was performed at the Protein Chemistry Core Facility, UTMB, Galveston, TX. We also acknowledge the Optical Imaging Laboratory of the Department of Microbiology and Immunology and the expertise of T. Albrecht and E. Knutson in obtaining the confocal images. We also thank S. F. Wang for help in editing figures.
Editor: J. T. Barbieri
REFERENCES
- 1.Agrawal, A., J. Lingappa, S. H. Leppla, S. Agrawal, A. Jabbar, C. Quinn, and B. Pulendran. 2003. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 424:329-334. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar, A., S. Merino, M. M. Nogueras, M. Regue, and J. M. Tomas. 1999. Two genes from the capsule of Aeromonas hydrophila (serogroup O:34) confer serum resistance to Escherichia coli K12 strains. Res. Microbiol. 150:395-402. [DOI] [PubMed] [Google Scholar]
- 3.Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdanovic, R., M. Cobeljic, M. Markovic, V. Nikolic, M. Ognjanovic, L. Sarjanovic, and D. Makic. 1991. Haemolytic-uraemic syndrome associated with Aeromonas hydrophila enterocolitis. Pediatr. Nephrol. 5:293-295. [DOI] [PubMed] [Google Scholar]
- 5.Burr, S. E., K. Stuber, and J. Frey. 2003. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J. Bacteriol. 185:6583-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliezi, C., W. A. Wuillemin, S. Zeerleder, M. Redondo, B. Eisele, and C. E. Hack. 2000. C1-esterase inhibitor: an anti-inflammatory agent and its potential use in the treatment of diseases other than hereditary angioedema. Pharmacol. Rev. 52:91-112. [PubMed] [Google Scholar]
- 7.Carugati, A., E. Pappalardo, L. C. Zingale, and M. Cicardi. 2001. C1-inhibitor deficiency and angioedema. Mol. Immunol. 38:161-173. [DOI] [PubMed] [Google Scholar]
- 8.Cascon, A., J. Yugueros, A. Temprano, M. Sanchez, C. Hernanz, J. M. Luengo, and G. Naharro. 2000. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 68:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauret, C., C. Volk, R. Creason, J. Jarosh, J. Robinson, and C. Warnes. 2001. Detection of Aeromonas hydrophila in a drinking-water distribution system: a field and pilot study. Can. J. Microbiol. 47:782-786. [PubMed] [Google Scholar]
- 10.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 11.Chopra, A. K., J. W. Peterson, X. J. Xu, D. H. Coppenhaver, and C. W. Houston. 1996. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 21:357-377. [DOI] [PubMed] [Google Scholar]
- 12.Cowell, B. A., S. S. Twining, J. A. Hobden, M. S. Kwong, and S. M. Fleiszig. 2003. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology 149:2291-2299. [DOI] [PubMed] [Google Scholar]
- 13.Davis, A. E., III. 2004. Biological effects of C1 inhibitor. Drug News Perspect. 17:439-446. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 15.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa, J. E., and P. Densen. 1991. Infectious diseases associated with complement deficiencies. Clin. Microbiol. Rev. 4:359-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galindo, C. L., A. A. Fadl, J. Sha, C. Gutierrez, Jr., V. L. Popov, I. Boldogh, B. B. Aggarwal, and A. K. Chopra. 2004. Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J. Biol. Chem. 279:37597-37612. [DOI] [PubMed] [Google Scholar]
- 18.Grys, T. E., M. B. Siegel, W. W. Lathem, and R. A. Welch. 2005. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanninen, M. L., S. Salmi, L. Mattila, R. Taipalinen, and A. Siitonen. 1995. Association of Aeromonas spp. with travellers' diarrhoea in Finland. J. Med. Microbiol. 42:26-31. [DOI] [PubMed] [Google Scholar]
- 20.Harkey, C. W., K. D. Everiss, and K. M. Peterson. 1995. Isolation and characterization of a Vibrio cholerae gene (tagA) that encodes a ToxR-regulated lipoprotein. Gene 153:81-84. [DOI] [PubMed] [Google Scholar]
- 21.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 22.Jung, C. M., O. Matsushita, S. Katayama, J. Minami, J. Sakurai, and A. Okabe. 1999. Identification of metal ligands in the Clostridium histolyticum ColH collagenase. J. Bacteriol. 181:2816-2822.10217773 [Google Scholar]
- 23.Kirov, S. M., K. Sanderson, and T. C. Dickson. 1998. Characterisation of a type IV pilus produced by Aeromonas caviae. J. Med. Microbiol. 47:527-531. [DOI] [PubMed] [Google Scholar]
- 24.Lathem, W. W., T. Bergsbaken, and R. A. Welch. 2004. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J. Exp. Med. 199:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lathem, W. W., T. Bergsbaken, S. E. Witowski, N. T. Perna, and R. A. Welch. 2003. Acquisition of stcE, a C1 esterase inhibitor-specific metalloprotease, during the evolution of Escherichia coli O157:H7. J. Infect. Dis. 187:1907-1914. [DOI] [PubMed] [Google Scholar]
- 26.Lathem, W. W., T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr, and R. A. Welch. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45:277-288. [DOI] [PubMed] [Google Scholar]
- 27.Loewy, A. G., U. V. Santer, M. Wieczorek, J. K. Blodgett, S. W. Jones, and J. C. Cheronis. 1993. Purification and characterization of a novel zinc-proteinase from cultures of Aeromonas hydrophila. J. Biol. Chem. 268:9071-9078. [PubMed] [Google Scholar]
- 28.Maeda, H., and A. Molla. 1989. Pathogenic potentials of bacterial proteases. Clin. Chim. Acta 185:357-367. [DOI] [PubMed] [Google Scholar]
- 29.Masada, C. L., S. E. LaPatra, A. W. Morton, and M. S. Strom. 2002. An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 51:13-25. [DOI] [PubMed] [Google Scholar]
- 30.Merino, S., A. Aguilar, M. M. Nogueras, M. Regue, S. Swift, and J. M. Tomas. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merino, S., A. Aguilar, X. Rubires, N. Abitiu, M. Regue, and J. M. Tomas. 1997. The role of the capsular polysaccharide of Aeromonas hydrophila serogroup O:34 in the adherence to and invasion of fish cell lines. Res. Microbiol. 148:625-631. [DOI] [PubMed] [Google Scholar]
- 32.Merino, S., S. Camprubi, and J. M. Tomas. 1991. The role of lipopolysaccharide in complement-killing of Aeromonas hydrophila strains of serotype O:34. J. Gen. Microbiol. 137:1583-1590. [DOI] [PubMed] [Google Scholar]
- 33.Merino, S., M. M. Nogueras, A. Aguilar, X. Rubires, S. Albertí, V. J. Benedí, and J. M. Tomás. 1998. Activation of the complement classical pathway (C1q binding) by mesophilic Aeromonas hydrophila outer membrane protein. Infect. Immun. 66:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merino, S., X. Rubires, A. Aguilar, S. Alberti, S. Hernandez-Alles, V. J. Benedi, and J. M. Tomas. 1996. Mesophilic Aeromonas sp. serogroup O:11 resistance to complement-mediated killing. Infect. Immun. 64:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino, S., X. Rubires, S. Knochel, and J. M. Tomas. 1995. Emerging pathogens: Aeromonas spp. Int. J. Food Microbiol. 28:157-168. [DOI] [PubMed] [Google Scholar]
- 36.Paccani, S. R., F. Tonello, R. Ghittoni, M. Natale, L. Muraro, M. M. D'Elios, W. J. Tang, C. Montecucco, and C. T. Baldari. 2005. Anthrax toxins suppress T lymphocyte activation by disrupting antigen receptor signaling. J. Exp. Med. 201:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paton, A. W., and J. C. Paton. 2002. Reactivity of convalescent-phase hemolytic-uremic syndrome patient sera with the megaplasmid-encoded TagA protein of Shiga toxigenic Escherichia coli O157. J. Clin. Microbiol. 40:1395-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24:153-192. [DOI] [PubMed] [Google Scholar]
- 39.Rivero, O., J. Anguita, D. Mateos, C. Paniagua, and G. Naharro. 1991. Cloning and characterization of an extracellular temperature-labile serine protease gene from Aeromonas hydrophila. FEMS Microbiol. Lett. 65:1-7. [DOI] [PubMed] [Google Scholar]
- 40.Rivero, O., J. Anguita, C. Paniagua, and G. Naharro. 1990. Molecular cloning and characterization of an extracellular protease gene from Aeromonas hydrophila. J. Bacteriol. 172:3905-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robson, W. L., A. K. Leung, and C. L. Trevenen. 1992. Haemolytic-uraemic syndrome associated with Aeromonas hydrophila enterocolitis. Pediatr. Nephrol. 6:221. [DOI] [PubMed] [Google Scholar]
- 42.Rose, J. M., C. W. Houston, and A. Kurosky. 1989. Bioactivity and immunological characterization of a cholera toxin-cross-reactive cytolytic enterotoxin from Aeromonas hydrophila. Infect. Immun. 57:1170-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34:195-204. [DOI] [PubMed] [Google Scholar]
- 44.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sha, J., M. Lu, and A. K. Chopra. 2001. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 69:6370-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 48.Supuran, C. T., A. Scozzafava, and B. W. Clare. 2002. Bacterial protease inhibitors. Med. Res. Rev. 22:329-372. [DOI] [PubMed] [Google Scholar]
- 49.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 50.Vollmer, P., I. Walev, S. Rose-John, and S. Bhakdi. 1996. Novel pathogenic mechanism of microbial metalloproteinases: liberation of membrane-anchored molecules in biologically active form exemplified by studies with the human interleukin-6 receptor. Infect. Immun. 64:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei, Z., J. F. Kim, and S. V. Beer. 2000. Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two-component system, and HrpS. Mol. Plant-Microbe Interact. 13:1251-1262. [DOI] [PubMed] [Google Scholar]
- 52.Xu, X. J., M. R. Ferguson, V. L. Popov, C. W. Houston, J. W. Peterson, and A. K. Chopra. 1998. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect. Immun. 66:3501-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]