Abstract
Type 1 fimbriae have been suggested to play a role in the pathogenesis of extraintestinal Escherichia coli infection. Type 1 fimbriation in E. coli is phase variable and known to be dependent upon FimB and FimE, the two recombinases that invert the molecular switch fimS and control the expression of the downstream fim operon. Here we showed that HbiF, a novel site-specific recombinase, inverted fimS independently of FimB and FimE. HbiF-mediated fimS inversion appeared to be predominantly switching from “off” (termed OFF) to “on” (termed ON) orientation. This is different from the fimS inversion mediated by either FimB (bidirectional ON to OFF and OFF to ON) or FimE (unidirectional ON to OFF). Constitutive expression of the hbiF gene in E. coli resulted in a fimS-locked-ON phenotype, which resulted in the pathogenic E. coli K1 strain being incapable of inducing a high degree of bacteremia in neonatal rats. Discovery of HbiF-mediated OFF-to-ON fimS switching provides a new opportunity to develop a strategy for the prevention and therapy of extraintestinal E. coli infection including bacteremia and meningitis.
Type 1 fimbriae are filamentous surface organelles produced by many members of the Enterobacteriaceae including Escherichia coli and mediate mannose-sensitive adhesion to various eukaryotic cells. The importance of type 1 fimbriae was implicated in E. coli colonization of the gastrointestinal tract and also in the pathogenesis of extraintestinal E. coli infections (20, 41, 45). For example, we have demonstrated that more than 80% of E. coli K1 bacteria bound onto the surface of human brain microvascular endothelial cells are type 1 fimbriated, suggesting that type 1 fimbriae are important in the pathogenesis of E. coli meningitis (41).
Type 1 fimbriae are encoded by the fimAICDFGH gene cluster and are primarily composed of the major subunit FimA and a small tip structure containing FimF, FimG, and the adhesin FimH (18). FimH protein has a lectin-like activity with high affinity to α-d-mannose or mannosylated glycoproteins/glycolipids (13, 39, 41). Type 1 fimbriae are assembled via a chaperone-usher pathway, where two specialized accessory proteins are required, the periplasmic chaperone FimC and the outer membrane usher FimD (30).
The expression of the fim operon is phase variable. The phase variation of type 1 fimbriae is mediated by an invertible 414-bp cis element, fimS, that is located upstream of fimA and contains a vegetative promoter for the fim operon when fimS is in the “on” orientation (1). Flipping of this molecular switch alternates E. coli between type 1 fimbriated and nonfimbriated states, termed ON and OFF, respectively (1, 19). The switch of fimS is a site-specific recombination process and dependent on either FimB (ON-to-OFF or OFF-to-ON switching) or FimE (ON-to-OFF switching only) (19, 22).
In this report, we examined a genomic island deletion mutant (designated E. coli YX101 in this study) that failed to induce a high degree of bacteremia in neonatal rats. We found that E. coli YX101 is a natural switch-locked-ON mutant for type 1 fimbria expression and that type 1 fimbriation was responsible for the failure to induce bacteremia. The fimS switch locking is due to the induction of a tyrosine site-specific recombinase, HbiF, whose effect on fimS is independent of FimB or FimE invertase.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
E. coli strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) or brain heart infusion (BHI) broth supplemented with appropriate antibiotics at 37°C overnight statically unless otherwise specified. The antibiotics were used at the indicated final concentrations: streptomycin, 100 μg/ml; ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 40 μg/ml.
TABLE 1.
Strains and plasmids used in the studies
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| RS218str | Spontaneous streptomycin-resistant mutant of E. coliRS218 (O18:K1:H7), isolated from the cerebrospinal fluid of a neonate with E. coli meningitis | (34) |
| 10G | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG λ−tonA | Lucigen |
| fimS-ON | RS218strfimS-locked-ON | (41) |
| fimS-OFF | RS218strfimS-locked-OFF | (41) |
| YX101 | RS218str ΔRDI 21 | (41) |
| YX102 | YX101 ΔfimAICDFGH::Cm | This study |
| YX104 | YX101 ΔhbiF::Cm | This study |
| YX105 | YX104 carrying pSMART-hbiFc | This study |
| YX106 | YX104 carrying pSMART vector | This study |
| YX107 | YX101 ΔhbiF::Cm ΔfimBE::Kan and fimS-locked-OFF | This study |
| YX108 | YX107 carrying pSMART-hbiFc | This study |
| YX109 | YX107 carrying pSMART vector | This study |
| YX110 | YX101 ΔfimBE::Kan | This study |
| Plasmids | ||
| pBAD-Thio | Expression control vector for pBAD promoter carrying ampicillin resistance gene | Invitrogen |
| pBAD-hbiF | hbiF gene from RS218 under the control of pBAD promoter in pBAD/Thio-TOPO vector | This study |
| pBAD-fimB | fimB gene from RS218 under the control of pBAD promoter in pBAD/Thio-TOPO vector | This study |
| pBAD-fimE | fimE gene from RS218 under the control of pBAD promoter in pBAD/Thio-TOPO vector | This study |
| pSMART vector | Low-copy-number transcription and translation-free cloning vector derived from pSMARTLCAmp carrying ampicillin resistance gene | Lucigen |
| pSMART-hbiFc | hbiF gene under the control of its native promoter in pSMART vector | This study |
Expression profiling of E. coli RS218 and its derivatives with DNA microarray.
Expression profiling of E. coli RS218 and its derivatives was conducted using an E. coli DNA microarray (8, 41). This microarray is composed of 8,144 oligonucleotides that covered open reading frames present in E. coli K-12 strain MG1655, E. coli O157:H7 strains EDL933 and Sakai, uropathogenic E. coli strain CFT073, and other uropathogenic or meningitis-related virulence factors and pathogenicity islands (8, 41).
Overnight cultures of E. coli were grown in fresh BHI broth for 30 min at 37°C. Subsequently, bacteria were recovered with centrifugation and bacterial total RNA was extracted with the Ribopure Bacteria kit (Ambion, Austin, TX) followed by an RNeasy minicolumn cleanup (QIAGEN, Valencia, CA). Cy3- and Cy5-labeled cDNA probes were generated with an indirect labeling procedure as previously described (8, 41). The purified Cy3- and Cy5-coupled cDNAs were combined and hybridized to an E. coli DNA microarray at 42°C for 18 h. After stringent washings, hybridized microarray slides were scanned (GenePix 4000B; Axon Instruments, Foster City, CA). DNA microarray images were analyzed with GenePix 4.0 software (Axon). The signal intensities between two channels were normalized with print-pin group loess normalization. The differential expression of genes was also determined with the Limma package from Bioconductor (http://www.bioconductor.org) (37).
Individual gene or gene cluster knockout.
A targeted gene or gene cluster knockout was constructed with a “one-step PCR” gene inactivation method (7). The targeted gene or gene cluster was replaced with a chloramphenicol or kanamycin cassette that is PCR synthesized with long primers based on the antibiotic resistance gene in the template plasmid (Table 2). Both primers have 50-nucleotide extensions that are homologous to boundaries of the targeted gene or gene cluster. After electroporation into the host strain, the targeted gene or gene cluster was replaced with a chloramphenicol or kanamycin cassette via homologous recombination, which was facilitated by the λ Red system (23). Subsequently, chloramphenicol or kanamycin was used to identify mutants and confirmation of correct gene replacement was conducted by PCR with screening primers and sequencing of PCR products (Table 2).
TABLE 2.
Oligonucleotides used in the studies
| Oligonucleotide | Sequencea | Usage |
|---|---|---|
| fimS-F | AGTAATGCTGCTCGTTTTGC | fimS inversion assay |
| fimS-R | GACAGAGCCGACAGAACAAC | fimS inversion assay |
| hbiF-ATG | ATGACGAGAAAATATCTCACAC | hbiF coding region cloning into pBAD/ Thio-TOPO |
| hbiF_stop2 | GGTACTGTTTTAACGAGGCT | hbiF coding region cloning into pBAD/ Thio-TOPO and hbiF knockout checking |
| fimB-ATG | ATGAAGAATAAGGCTGATAACAA | fimB coding region cloning into pBAD/ Thio-TOPO |
| fimB-stop | CTATAAAACAGCGTGACG | fimB coding region cloning into pBAD/ Thio-TOPO |
| fimE-ATG | GTGAGTAAACGTCGTTAT | fimE coding region cloning into pBAD/ Thio-TOPO |
| fimE-stop | ATTATCAATTAGTTAAATCAAGC | fimE coding region cloning into pBAD/ Thio-TOPO and fimBE knockout checking |
| hbiFc_F | TTCTACTATGAGGACCGAATCCTT | hbiF cloning into pSMART |
| hbiFc_R | TTGAATCAGAAGGTACTGTTTTAACG | hbiF cloning into pSMART |
| fim_KOF | TGGCAGTCAAACTCGTTGACAAAACAAAGTGTACAGAACGACTGCCCATGgtgtaggctggagctgcttc | fim knockout |
| fim_KOR | TAGCTTCAGGTAATATTGCGTACCTGCATTAGCAATGCCCTGTGATTTCTcatatgaatatcctcct | fim knockout |
| fim_CKF | TCTTTTGGGGGAAAACTGTG | fim knockout checking |
| fim_CKR | TCTGGCCTACAAAGGGCTAA | fim knockout checking |
| hbiF_KOF | TACATCGATAACGTTCTGATTGCAGGCATACTTATCTGGGTGTGCCTCTCgtgtaggctggagctgcttc | hbiF knockout |
| hbiF_KOR | GAAGGTACTGTTTTAACGAGGCTTTTTTTTCCACACCCCTTTAAAACGAGcatatgaatatcctcctta | hbiF knockout |
| hbiF_CKF | TGACTTGTAGAAAAGCTGCACAA | hbiF knockout checking |
| fimBE_KOF | ATAATCAGGATTAAAATGTTGGATTATTGCTAACCCAGCACAGCTAGTGCgtgtaggctggagctgcttc | fimBE knockout |
| fimBE_KOR | CCCATAATCCGGCAAAACGAGCAGCATTACTGGCGGTATAACGCACAGTAcatatgaatatcctcctta | fimBE knockout |
| fimBE_CKF | TCACTCAGAAGAACTGGTCCAC | fimBE knockout checking |
Lowercase underlined sequences represent sequences matching the template plasmids pKD3 or pKD4 (23).
Construction of plasmids for expression, complementation, and switch evaluation.
Plasmids with inducible hbiF, fimB, and fimE expression were constructed using the pBAD/Thio-TOPO cloning kit (Invitrogen, Carlsbad, CA). The full gene was amplified by PCR from E. coli RS218 genomic DNA using Elongase (Invitrogen) and cloned into the pBAD/Thio-TOPO vector according to the manufacturer's instructions. The colonies with recombinant pBAD/Thio fusion plasmids were selected using ampicillin, and the clones with an intact invertase insert in the correct orientation were screened using combined primers from both insert and vector. The correctness of each invertase cloning was confirmed by plasmid sequencing.
Gene cloning for complementation was conducted using a pSMARTLCAmp-based low-copy-number cloning vector (Lucigen, Middleton, WI). The individual gene or gene cluster including the upstream promoter region from E. coli RS218 was amplified with Phusion high-fidelity DNA polymerase (New England Biolabs, Inc., Beverly, MA), followed by T4 polynucleotide kinase phosphorylation (Table 2). The phosphorylated PCR product was ligated into pSMARTLCAmp using the CloneSMART kit (Lucigen). Successful cloning of the target gene or gene cluster was confirmed by PCR and plasmid sequencing. For complementation, the resultant plasmid was electroporated into the desired gene knockout mutant.
Invertible element fimS orientation assay.
A qualitative assay of fimS orientation was conducted using the asymmetrical digestion of a fimS-containing fragment as previously described (41). The 602-bp fragment containing the invertible element fimS was PCR amplified using a pair of primers, fimS-F and fimS-R (Table 2), directly from broth culture or colonies. Asymmetrical restriction digestion of this PCR product with SnaBI produced 404- and 198-bp fragments when type 1 fimbriation was in phase-ON or 442- and 160-bp fragments when fimbriation was in phase-OFF. The SnaBI-digested PCR products were separated on a 2.0% agarose gel.
Yeast aggregation assay.
The abilities of E. coli strains to aggregate yeast cells were measured as previously described (39). E. coli strains were washed and resuspended in phosphate-buffered saline at an optical density at 530 nm of 0.4. These normalized E. coli suspensions were subjected to twofold serial dilutions in phosphate-buffered saline and subsequently mixed with commercial baker's yeast cells (5 mg/ml) with or without 1% d-mannose. Aggregation was monitored visually, and the titer was recorded as the highest dilution giving a positive aggregation result.
Animal model of E. coli bacteremia.
E. coli bacteremia was induced in 5-day-old rats by subcutaneous injection of E. coli as previously described (17). The outbred, pathogen-free pregnant Sprague-Dawley rats with timed conception were purchased from Charles River Breeding Laboratories (Wilmington, MA). The rats usually deliver in our vivarium 5 to 7 days after arrival. At 5 days of age, all members of each litter were randomly divided into designated experimental groups to receive subcutaneously 106 CFU of E. coli RS218 or its derivatives. At 18 h after bacterial inoculation, blood specimens were obtained for quantitative cultures as previously described (17).
RESULTS
E. coli YX101 is a type 1 fimbrial molecular switch fimS-locked-ON mutant.
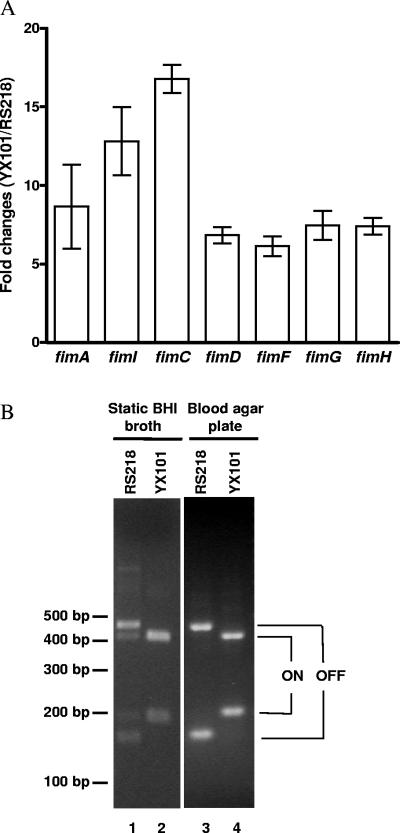
Recently, we showed that E. coli YX101, a genomic island mutant derived from meningitis-causing E. coli K1 strain RS218, failed to induce any detectable bacteremia in neonatal rats after subcutaneous inoculation of 106 CFU of bacteria (44). In contrast, the neonatal rats infected with wild-type E. coli RS218 showed bacteremia of 108 CFU/ml of blood. To understand why E. coli YX101 failed to induce a high degree of bacteremia in the neonatal rats, we compared the expression profiles of this strain with those of E. coli RS218 using DNA microarrays. One of the most drastic changes in E. coli YX101 was the induction of fim operon expression (Fig. 1A). The fimAICDFGH genes in the fim operon were significantly induced 6-to 17-fold in E. coli YX101 compared to the parent strain E. coli RS218, suggesting that E. coli YX101 is highly type 1 fimbriated.
FIG. 1.
E. coli YX101 is a type 1 molecular switch fimS-locked-ON mutant. (A) In E. coli YX101 (an RSI 22 deletion derivative of E. coli RS218), genes in the fim operon were significantly induced compared to E. coli RS218 (values are means ± standard errors of the means). The mRNA levels of the fim gene cluster were profiled using an E. coli DNA microarray. (B) The induction of the fim operon in E. coli YX101 was due to OFF-to-ON orientation switching of fimS. The fimS gene was locked in ON orientation in this strain but phase variable in parental strain RS218.
Since type 1 fimbriation is phase variable, we examined the status of molecular switch fimS in E. coli YX101 with PCR amplification combined with SnaBI asymmetrical restriction digestion (41). The wild-type E. coli RS218 was composed of a mixture of fimbriated and nonfimbriated cells when grown in BHI broth under static conditions as previously shown (Fig. 1B, lane 1). However, it is striking that the fimS switch in E. coli YX101 was completely in switched-ON orientation under similar growth conditions (Fig. 1B, lane 2). We also examined growth conditions favorable for ON-to-OFF switching, such as passage on blood agar plates. Without exception, fimS in E. coli YX101 was orientated in the switched-ON state (Fig. 1B, lane 4). In addition to the fimS orientation, we also examined the type 1 fimbriation using a yeast aggregation assay. E. coli YX101 exhibited abilities to aggregate yeast cells similar to those of highly type 1 fimbriated E. coli strain fimS-ON, a fimS-locked-ON mutant of E. coli RS218 previously constructed through the mutation of inverted repeats in the fimS molecular switch (41) (Table 3). Based on these findings, we conclude that E. coli YX101 is a fimS-locked-ON mutant and highly type 1 fimbriated.
TABLE 3.
Yeast aggregation abilities of E. coli RS218 and its derivatives
| Straina | Yeast aggregation titerb,c
|
|
|---|---|---|
| Without d-mannose | With 1% d-mannose | |
| E. coli RS218 | 1 | − |
| E. coli YX101 | 16 | − |
| E. coli fimS-ON | 16 | − |
| E. coli fimS-OFF | − | − |
The strains were grown overnight in static BHI broth at 37°C.
Agglutination titers were recorded as the highest dilution giving a positive aggregation.
Minus sign shows that no aggregation was observed.
Constitutive induction of hbiF is responsible for the fimS-locked-ON phenotype.
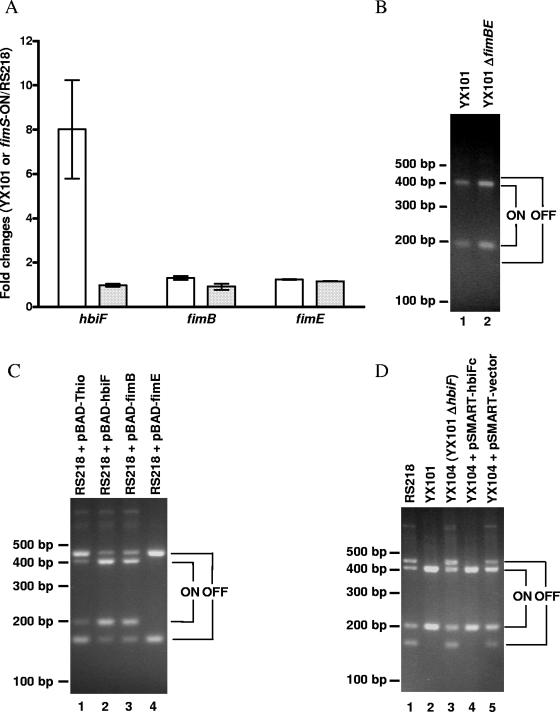
Since E. coli YX101 is a fimS-locked-ON mutant, we examined and compared the expression profiles of E. coli YX101 with those of the genetically constructed fimS-locked-ON strain (fimS-ON) and the parent strain RS218 using DNA microarrays. We showed that the hbiF gene was highly induced in E. coli YX101 compared to E. coli RS218, while it remained relatively unchanged in the genetically constructed fimS-locked-ON mutant (Fig. 2A). In contrast to the strong induction of hbiF, the expression levels of fimB and fimE were not significantly changed in E. coli YX101 compared to E. coli RS218 (Fig. 2A). In addition, deletion of fimBE from YX101 did not affect the fimS inversion and the fimBE deletion mutant remained in the fimS-locked-ON state (Fig. 2B).
FIG. 2.
The fimS-locked-ON phenotype in E. coli YX101 is due to induction of hbiF. (A) DNA microarray results showed that hbiF was significantly induced in E. coli YX101 compared to RS218 (white bars) but not in the engineered fimS-locked-ON mutant (shaded bars) (values are means ± standard errors of the means). Expression of fimB and fimE genes remained unchanged in both YX101 and fimS-ON. (B) Deletion of fimB and fimE in E. coli YX101 did not affect the locked-ON status of fimS. (C) Overexpression of hbiF in E. coli RS218 resulted in fimS switching from OFF to ON. (D) hbiF was responsible for the fimS-locked-ON phenotype in E. coli YX101.
The hbiF gene (GenBank accession number DQ272538) belongs to a gene cluster designated hbi (from HBMEC [human brain microvascular endothelial cell] binding induced) due to their high expression levels in human brain microvascular endothelial cell-associated E. coli (Y. Xie, unpublished data). The hbiF gene was absent in the E. coli K-12 strain MG1655 genome (2) but present in approximately 80% of E. coli K1 isolates derived from blood and cerebrospinal fluid including strain RS218 (data not shown). Although this gene was also present in the genomes of E. coli O157:H7 strains EDL 933 and Sakai, hbiF genes in these two strains were truncated due to a frameshift mutation (14, 27). HbiF protein was homologous to two well-known fimS invertases, FimB and FimE, with percentages of amino acid identity of 55% and 52%, respectively, suggesting that it may belong to the family of tyrosine site-specific recombinases and have an effect on the type 1 molecular switch fimS.
A marked induction of hbiF in E. coli YX101 and similarity between HbiF and FimB as well as FimE prompted us to hypothesize that HbiF protein might be involved in the fimS-locked-ON phenotype in E. coli YX101. We cloned the hbiF gene into the pBAD/Thio-TOPO vector (designated pBAD-hbiF) so that the expression of hbiF was under the tight control of the pBAD promoter. When E. coli RS218 was transformed with pBAD-hbiF, fimS was switched from OFF to ON orientation with 0.1% arabinose overnight induction compared to that in RS218 with control vector pBAD-Thio (Fig. 2C, lanes 1 and 2). When exogenous FimB and FimE were supplied in a similar fashion, FimB favored OFF-to-ON switching while FimE favored ON-to-OFF switching as expected (Fig. 2C, lanes 3 and 4). These findings suggest that HbiF protein is likely to be responsible for the fimS-locked-ON phenotype observed in E. coli YX101.
To further confirm the role of hbiF in type 1 fimbriation, we constructed an hbiF deletion mutant in the background of E. coli YX101. Deletion of hbiF from E. coli YX101 (YX104, the hbiF-negative derivative of YX101) restored the fimS switch from locked-ON orientation to a mixed population of ON/OFF orientations when grown in BHI broth under static conditions (Fig. 2D, lane 3). Complementation of the hbiF gene with its native promoter in the hbiF deletion mutant of E. coli YX101 (YX105, YX104 plus pSMART-hbiFc) changed the fimS switch back to locked-ON orientation (Fig. 2D, lane 4). In contrast, complementation of the hbiF deletion mutant of YX101 with the control vector (YX106, YX104 plus pSMART vector) did not affect the mixed ON/OFF fimS composition as expected (Fig. 2D, lane 5). These findings indicate that the induction of hbiF gene expression is responsible for the fimS-locked-ON phenotype in E. coli YX101.
HbiF invertase switches molecular switch fimS independently of FimB and FimE.
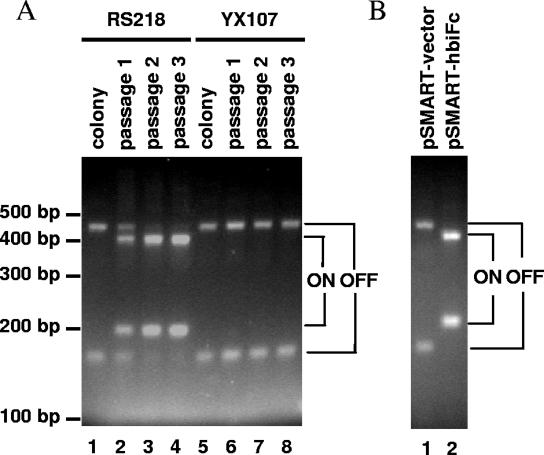
Based on the sequence homology and expression profiling results, we hypothesize that HbiF invertase directly affects fimS inversion, rather than indirectly through FimB or FimE. In order to distinguish the contributions of FimB and FimE from that of HbiF to fimS inversion, we deleted fimBE in the hbiF mutant strain of YX101. Deletion of both fimBE and hbiF in YX101 resulted in strain YX107, whose fimS switch was locked in OFF orientation. For example, the fimS switch in E. coli YX107 (fimBE negative, hbiF negative) was maintained in locked-OFF orientation even after three passages in static BHI broth (Fig. 3A, lanes 5 to 8), while the fimS switch in E. coli RS218 (fimBE+ hbiF+) was switched from OFF to ON orientation during these passages (Fig. 3A, lanes 1 to 4). The fimS locking in fimBE-negative, hbiF-negative strain YX107 suggests that there are no other factors capable of switching fimS in E. coli YX101 except for these three invertases. This is consistent with the findings from genomic sequencing where only three FimB homologues are found in the E. coli RS218 genome (i.e., FimB, FimE, and HbiF; http://www.genome.wisc.edu).
FIG. 3.
HbiF inverts the molecular switch fimS independently of FimB and FimE. (A) The molecular switch fimS was locked OFF in E. coli YX107, where both fimBE and hbiF genes are deleted from the genome of YX101. (B) HbiF was able to switch fimS from OFF to ON and remains in locked-ON orientation in the absence of FimB and FimE, the two well-known fimS invertases.
To further confirm our hypothesis that HbiF activity in fimS inversion is independent of FimB and FimE, we next transformed the invertase-negative mutant YX107 (fimBE negative, hbiF negative and fimS-locked-OFF) with pSMART-hbiFc. Reintroduction of hbiF with its native promoter into this invertase-negative mutant changed the fimS switch from OFF to ON orientation, while the same strain supplemented with the vector control remained in OFF orientation as in the parent strain when grown on solid medium (Fig. 3B). We also examined the fimS status in these transformants growing in static or shaking liquid culture. With no exception, the fimS switches in YX107 carrying pSMART-hbiFc were orientated in locked-ON, while those in the same strain carrying vector control were orientated in locked-OFF (data not shown). These data illustrate that HbiF invertase is working on the molecular switch fimS independently of FimB and FimE recombinases and is responsible for fimS changes from OFF to ON orientation in YX101. FimB- and FimE-independent fimS inversion is also consistent with the observations where deletion of fimBE did not affect the fimS-locked-ON phenotype in YX101 (Fig. 2B). In addition, HbiF-mediated fimS OFF-to-ON switching is consistent with our demonstrations of fimS conversion from locked-OFF in the fimBE-negative, hbiF-negative strain YX107 (YX101 ΔfimBE ΔhbiF, Fig. 3A, lanes 5 to 8) to locked-ON in fimBE-negative, hbiF+ strain YX110 (YX101 ΔfimBE, Fig. 2B, lane 2) with the introduction of hbiF.
HbiF-mediated type 1 fimbriation results in a marked reduction of bacteremia.
We next examined whether type 1 fimbriation in E. coli YX101 is related to this strain's failure to induce bacteremia. To evaluate the role of type 1 fimbriation in induction of bacteremia, we first examined genetically engineered type 1 fimbriation mutants E. coli fimS-ON and fimS-OFF for their abilities to induce bacteremia in neonatal rats. Subcutaneous injection of E. coli fimS-ON into neonatal rats did not induce any detectable bacteremia, which was identical to the result observed with E. coli YX101 (Table 4). In contrast, the E. coli fimS-OFF mutant achieved a magnitude of bacteremia similar to that of wild-type E. coli RS218 (Table 4). These results suggest that type 1 fimbriation in E. coli YX101 is most likely responsible for its failure to induce bacteremia in neonatal rats.
TABLE 4.
Magnitude of bacteremia induced by E. coli RS218 and its derivatives in neonatal rats
| Strain (no. of animals examined) | Bacteremia, log CFU/ml blood (mean ± SD) | Comment(s) |
|---|---|---|
| E. coli RS218 (12) | 8.59 ± 2.70 | Wild type |
| E. coli YX101 (13) | <1.69a,b | Highly type 1 fimbriated, a fimS-locked-ON mutant of RS218 |
| E. coli fimS-ON (6) | <1.69a,b | Highly type 1 fimbriated, a genetically engineered fimS-locked-ON mutant of RS218 |
| E. coli fimS-OFF (6) | 8.93 ± 1.20 | Non-type 1 fimbriated, a genetically engineered fimS-locked-OFF mutant of RS218 |
| E. coli YX102 (5) | 8.15 ± 0.60 | Non-type 1 fimbriated, fim operon deletion mutant of E. coli YX101 |
| E. coli YX104 (5) | 7.30 ± 2.03 | Type 1 fimbriation similar to RS218, hbiF deletion mutant of E. coli YX101 |
Bacterial counts in bloodstream of 5-day-old rats were below the detection limits.
The magnitude of bacteremia induced by the mutant was significantly different from that of wild-type E. coli RS218 (P < 0.01).
Further, we constructed a fim operon deletion mutant of E. coli YX101, where an approximately 7.5-kb chromosomal DNA segment including the molecular switch fimS and its downstream structural gene fimAICDFGH were deleted from E. coli YX101. This type 1 fimbria-negative mutant of E. coli YX101 exhibited an ability to induce a high degree of bacteremia in neonatal rats, similarly to the wild-type E. coli RS218 (Table 4). We also showed that deletion of hbiF from E. coli YX101 (which changed fimS-locked-ON to a mixed population of fimS ON/OFF orientations) restored this bacterial ability to induce a high degree of bacteremia in neonatal rats (Table 4). Taken together, the failure of E. coli YX101 to induce bacteremia in neonatal rats is attributed to HbiF-dependent type 1 fimbriation in this strain.
DISCUSSION
In this study, we identified a novel site-specific recombinase, HbiF, that regulates the inversion of fimS in E. coli. More importantly, HbiF invertase was able to switch fimS in the absence of FimB and FimE. The existence of non-FimB- and non-FimE-dependent fimS inversion was implied for E. coli Nissle 1917. An uncharacterized recombinase was suggested to be responsible for fimS OFF-to-ON switching in a fimBE double mutant of this strain (40). In addition, HbiF-mediated fimS inversion appears to be predominantly unidirectional OFF-to-ON switching. This is different from the fimS switching mode mediated by either FimB (bidirectional ON-to-OFF and OFF-to-ON) or FimE (unidirectional ON-to-OFF). The unique switching mode of HbiF also explains why the induction of HbiF resulted in a fimS-locked-ON phenotype in E. coli YX101. The role of hbiF in the fimS-locked-ON phenotype was genetically confirmed with deletion and complementation of hbiF.
Type 1 fimbrial phase variation is subject to the control of a variety of environmental signals, including physical signals such as temperature, pH, or osmolarity, and nutritional signals such as the availability of amino sugars or branch chain amino acids (10, 11, 26, 32). In addition to environmental signals, alteration of intracellular signals derived from cross-talk among different fimbriae such as P and S fimbriae also affects fimS inversion (15). Despite complex regulation mechanisms and the involvement of multiple regulators such as H-NS, Lrp, leuX, OmpR, PapB, and SfaB, it is invariable that the type 1 fimbrial phase variations due to those external or internal signals are dependent upon the functions of FimB and FimE directly or indirectly (10, 11, 15, 16, 26, 28, 32, 38, 43). Discovery of a novel regulator coordinating FimB- and FimE-independent type 1 fimbriation in E. coli opens a new research opportunity to study the regulation and expression of type 1 fimbriae.
E. coli YX101 is an RDI 21 island deletion derivative of meningitis-causing E. coli K1 strain RS218 (44). This island is the largest genomic island in E. coli RS218 (∼120 kb). Deletion of this island promoted the expression of the hbiF gene, suggesting that the factor(s) inside RDI 21 is likely a negative transcriptional regulator for hbiF expression. There are at least 11 putative proteins in RDI 21 that potentially function as a transcriptional repressor(s) for the hbiF promoter, including PapB, PapI, PapX, a DsdC homologue, and seven other putative transcriptional regulators. It is tempting to speculate that loss of one or more of these transcriptional regulators might be directly or indirectly responsible for hbiF induction in YX101. For example, papB, papI, and papX are located inside the pap gene cluster, which is responsible for biosynthesis of P fimbriae. It is known that there is cross talk between P fimbriae and type 1 fimbriae in uropathogenic E. coli due to the activity of the PapB protein (15, 16, 43). PapB inhibits FimB-mediated fimS OFF-to-ON inversion and activates the expression of FimE. On the other hand, there are two highly homologous copies of DsdC in RS218 (98% amino acid residue identity). Transcriptional activation of dsdXA via the activity of DsdC is required for d-serine detoxification in E. coli (24). Deletion of dsdA in uropathogenic E. coli CFT073 reduced type 1 fimbriation (29). Since CFT073 harbors only a backbone copy of the dsd gene cluster, the effect of deletion of RDI 21-associated dsd on type 1 fimbriation is unclear.
Another important finding of this study is that HbiF-dependent type 1 fimbriation drastically reduced the ability of E. coli K1 to induce bacteremia in neonatal rats (Table 4). Human neonates, especially premature ones, are at a greater risk for bloodstream infection due to enteric gram-negative bacteria (5). E. coli is one of the most common causes of septicemia in infants, which is usually associated with high morbidity and mortality (6, 33). Further, E. coli bacteremia is associated with other complications. For example, a high degree of bacteremia is a prerequisite for the induction of E. coli meningitis (9). The contribution of type 1 fimbriation to E. coli K1 bacteremia has been implied by previous studies (12, 25, 31). For example, type 1 fimbriated E. coli K1 strain IH3080 (O18:K1:H7) was found to be less able to induce bacteremia than nonfimbriated bacteria when examined with both neonatal rat and mouse peritonitis models (25, 31). Conversely, disruption of fimA in E. coli K1 did not affect the ability to induce bacteremia, similar to our results with E. coli fimS-OFF (3, 21). The incapability of type 1 fimbriated E. coli to induce a high degree of bacteremia is probably related to its susceptibility to opsonin-independent phagocytosis by polymorphonuclear leukocytes (35, 36).
Previously we have demonstrated that type 1 fimbriae are critical for E. coli K1 binding to human brain endothelial cells in vitro (41). However, a high level of type 1 fimbriation appears to be detrimental for induction of a high degree of bacteremia as demonstrated with the fimS-locked-ON derivatives of E. coli K1 strains in neonatal rats (Table 4). Since a high degree of bacteremia is prerequisite for E. coli crossing the blood-brain barrier (44), the roles of type 1 fimbriae in E. coli meningitis become an apparent paradox. There are several potential explanations that might reconcile the seemingly conflicting roles of type 1 fimbriation in bacteremia and binding to human brain endothelial cells. For example, it is likely that a high level of type 1 fimbriation, i.e., the fimS-locked-ON phenotype, is required in the efficient blood clearance of type 1 fimbriated E. coli. In contrast, a low level of type 1 fimbriation might be permissible in the host bloodstream, which allows wild-type E. coli K1 strains to cross the blood-brain barrier. Alternatively, it is possible that transient expression of type 1 fimbriation in host blood is sufficient for E. coli K1 to bind to brain endothelial cells and cross the blood-brain barrier. Previous studies have demonstrated that type 1 fimbriated E. coli was present in infant rat blood at 6 h after intraperitoneal administration of E. coli K1 (31). Additional studies are needed to clarify the roles of type 1 fimbriae in E. coli bacteremia and meningitis.
In summary, we identified a novel type 1 fimbriation regulator, HbiF, whose function for the type 1 fimbrial molecular switch fimS is independent of FimB and FimE invertases. Constitutive induction of hbiF in E. coli YX101 results in the fimS-locked-ON phenotype in this strain. HbiF-dependent type 1 fimbriation is responsible for this strain's failure to induce bacteremia in neonatal rats.
ADDENDUM
During the review of the manuscript, a paper on unlinked FimB and FimE homologues in uropathogenic E. coli strain CFT073 was published (4). In CFT073, three non-FimB and non-FimE fimS invertases were identified based on genomic sequences, which were designated IpuA, IpuB, and IpbA. IbpA is identical to HbiF and was demonstrated to be capable of inverting fimS from OFF to ON orientation. However, the expression of the ipbA gene was dormant in wild-type E. coli CFT073, because CFT073 derivatives harboring only ipbA (i.e., CFT073 ΔfimBE ΔipuAB) were locked in either ON or OFF orientation (4). Interestingly, CFT073 lacked a complete RDI 21 genomic island, although it did carry two copies of pap gene clusters (responsible for P fimbria biogenesis) (42).
Acknowledgments
This work was supported by National Institutes of Health grants RO1-NS 26310 and AI 47225.
We are grateful to Frederick R. Blattner at the University of Wisconsin—Madison for allowing us to access the draft sequence of the E. coli RS218 genome.
Editor: F. C. Fang
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, C. A., and P. E. Orndorff. 1990. Impaired colonization by and full invasiveness of Escherichia coli K1 bearing a site-directed mutation in the type 1 pilin gene. Infect. Immun. 58:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordero, L., R. Rau, D. Taylor, and L. W. Ayers. 2004. Enteric gram-negative bacilli bloodstream infections: 17 years' experience in a neonatal intensive care unit. Am. J. Infect. Control 32:189-195. [DOI] [PubMed] [Google Scholar]
- 6.Cross, A. S., P. Gemski, J. C. Sadoff, F. Orskov, and I. Orskov. 1984. The importance of the K1 capsule in invasive infections caused by Escherichia coli. J. Infect. Dis. 149:184-193. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Cello, F., Y. Xie, M. Paul-Satyaseela, and K. S. Kim. 2005. Approaches to bacterial RNA isolation and purification for microarray analysis of Escherichia coli K1 interaction with human brain microvascular endothelial cells. J. Clin. Microbiol. 43:4197-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietzman, D. E., G. W. Fischer, and F. D. Schoenknecht. 1974. Neonatal Escherichia coli septicemia—bacterial counts in blood. J. Pediatr. 85:128-130. [DOI] [PubMed] [Google Scholar]
- 10.El-Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49:1109-1118. [DOI] [PubMed] [Google Scholar]
- 11.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerina, N. G., T. W. Kessler, V. J. Guerina, M. R. Neutra, H. W. Clegg, S. Langermann, F. A. Scannapieco, and D. A. Goldmann. 1983. The role of pili and capsule in the pathogenesis of neonatal infection with Escherichia coli K1. J. Infect. Dis. 148:395-405. [DOI] [PubMed] [Google Scholar]
- 13.Hanson, M. S., and C. C. Brinton, Jr. 1988. Identification and characterization of E. coli type-1 pilus tip adhesion protein. Nature 332:265-268. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 15.Holden, N. J., and D. L. Gally. 2004. Switches, cross-talk and memory in Escherichia coli adherence. J. Med. Microbiol. 53:585-593. [DOI] [PubMed] [Google Scholar]
- 16.Holden, N. J., B. E. Uhlin, and D. L. Gally. 2001. PapB paralogues and their effect on the phase variation of type 1 fimbriae in Escherichia coli. Mol. Microbiol. 42:319-330. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Investig. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm, P. 1984. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur. J. Biochem. 143:395-399. [DOI] [PubMed] [Google Scholar]
- 19.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 21.May, A. K., C. A. Bloch, R. G. Sawyer, M. D. Spengler, and T. L. Pruett. 1993. Enhanced virulence of Escherichia coli bearing a site-targeted mutation in the major structural subunit of type 1 fimbriae. Infect. Immun. 61:1667-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norregaard-Madsen, M., E. McFall, and P. Valentin-Hansen. 1995. Organization and transcriptional regulation of the Escherichia coli K-12 d-serine tolerance locus. J. Bacteriol. 177:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowicki, B., J. Vuopio-Varkila, P. Viljanen, T. K. Korhonen, and P. H. Makela. 1986. Fimbrial phase variation and systemic E. coli infection studied in the mouse peritonitis model. Microb. Pathog. 1:335-347. [DOI] [PubMed] [Google Scholar]
- 26.Olsen, P. B., M. A. Schembri, D. L. Gally, and P. Klemm. 1998. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 162:17-23. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Ritter, A., D. L. Gally, P. B. Olsen, U. Dobrindt, A. Friedrich, P. Klemm, and J. Hacker. 1997. The Pai-associated leuX specific tRNA5(Leu) affects type 1 fimbriation in pathogenic Escherichia coli by control of FimB recombinase expression. Mol. Microbiol. 25:871-882. [DOI] [PubMed] [Google Scholar]
- 29.Roesch, P. L., P. Redford, S. Batchelet, R. L. Moritz, S. Pellett, B. J. Haugen, F. R. Blattner, and R. A. Welch. 2003. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49:55-67. [DOI] [PubMed] [Google Scholar]
- 30.Sauer, F. G., H. Remaut, S. J. Hultgren, and G. Waksman. 2004. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta 1694:259-267. [DOI] [PubMed] [Google Scholar]
- 31.Saukkonen, K. M., B. Nowicki, and M. Leinonen. 1988. Role of type 1 and S fimbriae in the pathogenesis of Escherichia coli O18:K1 bacteremia and meningitis in the infant rat. Infect. Immun. 56:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwan, W. R., J. L. Lee, F. A. Lenard, B. T. Matthews, and M. T. Beck. 2002. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect. Immun. 70:1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siitonen, A., A. Takala, Y. A. Ratiner, A. Pere, and P. H. Makela. 1993. Invasive Escherichia coli infections in children: bacterial characteristics in different age groups and clinical entities. Pediatr. Infect. Dis. J. 12:606-612. [DOI] [PubMed] [Google Scholar]
- 34.Silver, R. P., W. Aaronson, A. Sutton, and R. Schneerson. 1980. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect. Immun. 29:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverblatt, F. J., J. S. Dreyer, and S. Schauer. 1979. Effect of pili on susceptibility of Escherichia coli to phagocytosis. Infect. Immun. 24:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverblatt, F. J., and I. Ofek. 1983. Interaction of bacterial pili and leukocytes. Infection 11:235-238. [DOI] [PubMed] [Google Scholar]
- 37.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Computational biology solutions using R and Bioconductor. Springer, New York, N.Y.
- 38.Sohanpal, B. K., S. El-Labany, M. Lahooti, J. A. Plumbridge, and I. C. Blomfield. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 101:16322-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokurenko, E., H. Courtney, D. Ohman, P. Klemm, and D. Hasty. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stentebjerg-Olesen, B., T. Chakraborty, and P. Klemm. 1999. Type 1 fimbriation and phase switching in a natural Escherichia coli fimB null strain, Nissle 1917. J. Bacteriol. 181:7470-7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng, C. H., M. Cai, S. Shin, Y. Xie, K. J. Kim, N. A. Khan, F. Di Cello, and K. S. Kim. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 73:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia, Y., D. Gally, K. Forsman-Semb, and B. E. Uhlin. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie, Y., K. J. Kim, and K. S. Kim. 2004. Current concepts on Escherichia coli K1 translocation of the blood-brain barrier. FEMS Immunol. Med. Microbiol. 42:271-279. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, G., W. J. Mo, P. Sebbel, G. Min, T. A. Neubert, R. Glockshuber, X. R. Wu, T. T. Sun, and X. P. Kong. 2001. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J. Cell Sci. 114:4095-4103. [DOI] [PubMed] [Google Scholar]