Abstract
It had been previously determined that the presence of FoF1 ATP synthase was required for microcin H47 antibiotic action. In this work, microcin-resistant atp mutants were genetically analyzed. Their mutations, originated by Tn5 insertion, in all cases were found to affect determinants for the Fo portion of ATP synthase. To discern if microcin action required the presence of the entire complex or if the Fo proton channel would suffice, recombinant plasmids carrying different segments of the atp operon were constructed and introduced into an atp deletion strain. The phenotypic analysis of the strains thus obtained clearly indicated that the presence of the Fo proton channel was absolutely required for microcin H47 action, while the F1 catalytic portion was found to be dispensable. Furthermore, when any of the three components of the proton channel was missing, total resistance to the antibiotic ensued. Complementation analysis between atp::Tn5 chromosomal mutations and recombinant atp plasmid constructions further supported the idea that the proton channel would be the minimal structure of the ATP synthase complex needed for microcin H47 antibiotic action.
The production of peptides with antibiotic action is a resource frequently employed in nature and, as judged by its widespread occurrence among bacteria, can be considered a successful strategy in mediating bacterial antagonistic relationships. Microcins are a group of antibiotic peptides produced by strains of Gram-negative bacteria. At present, only a few microcins have been thoroughly studied. Nevertheless, it is already clear that they exhibit a noteworthy diversity in their mechanism of action; e.g., colicin V and microcin E492 act as ionophores, microcin C7 inhibits protein synthesis, microcin B17 inhibits DNA replication due to its interaction with DNA gyrase, and microcin J25 acts upon RNA polymerase (6, 8, 11, 21, 24). Therefore, the study of microcins' mode of action presents a particularly interesting aspect: the identification of new antagonistic activities against basic cellular functions.
In the present work, we deal with microcin H47 (MccH47), a gene-encoded peptide antibiotic produced by a naturally occurring strain of Escherichia coli isolated in Uruguay. The MccH47 genetic system is clustered in a 10-kb DNA segment located in the chromosome (13). The information encoded by this system determines the three basic functions related to antibiosis: synthesis of the antibiotic, its secretion to the extracellular medium, and immunity. In brief, the MccH47 genetic system comprises four genes involved in microcin synthesis (the smallest being the structural gene), two further genes devoted to microcin secretion, and an immunity gene whose product protects the cell against its own antibiotic production (2, 7, 16, 17).
MccH47 is ribosomally synthesized as a peptide precursor which already possesses antibiotic activity of the same specificity as that of mature microcin. This toxic effect was detected in cells carrying the MccH47 structural gene and lacking the remaining genes from the microcin system. Although cells with such a genetic construction were nonviable, some mutant clones grew which exhibited Atp− and MccH47-resistant (MccH47r) phenotypes. This fact, together with the isolation and characterization of MccH47r mutants affected in the atp locus, led us to consider ATP synthase as the possible target of MccH47 antibiotic action (17, 20).
ATP synthase of E. coli consists of a membrane-bound Fo sector to which a cytoplasmic F1 sector is bound. This complex is made up of eight different polypeptides: three subunits form the Fo proton channel in a ratio of a1, b2, c9-12; and five subunits compose the catalytic F1 portion in a stoichiometry of α3, β3, γ1, δ1, ɛ1. The atp operon, which encodes these subunits, is arranged as follows: atpIBEFHAGDC, corresponding, respectively, to i (a protein of unknown function that does not form part of ATP synthase) and subunits a, c, b, δ, α, γ, β, and ɛ (23).
To enhance our understanding of the mechanism of action of MccH47, we concentrated on two aspects: (i) the genetic characterization of previously isolated MccH47r atp mutants and (ii) the identification of the minimal structure of the ATP synthase complex capable of conferring sensitivity to MccH47.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophages.
The strains, plasmids and bacteriophages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| BZB1011 | gyrA | Laboratory collection |
| FGB64 | BZB1011 atpB::Tn5 64 | 20 |
| MC4100 | araD139 lacU169 relA rpsL thiA | Laboratory collection |
| RYC816 | gyrA (λ+) recA56 srl::Tn10 | Laboratory collection |
| MC1000 Δ(uncI-uncC) | araD139 Δ(araABC-leu)7679 Δ(atpI-atpC) galK galU Δ(lac)X74 recA rpsL thi | 19 |
| FGB084 | MC4100 atpF::Tn5 84 | This work |
| FGB089 | MC4100 atpF::Tn5 89 | This work |
| FGB091 | MC4100 atpE::Tn5 91 | This work |
| FGB099 | MC4100 atpB::Tn5 99 | This work |
| Bacteriophages | ||
| MuctsApr | Mucts carrying the bla gene of Tn3 | 14 |
| MudII4042 | Mucts A+ B+ repP15A lac(′ZYA)931 (Cmr) | 9 |
| Plasmids | ||
| pEG109 | MudII4042::phoA proC (Cmr) | 9 |
| pBJC917 | pBR322 carrying atpBEFHAGDC (Apr) | 22 |
| pER900 | BamHI deletion of pBJC917 | This work |
| pER20 | pBR322 with an EcoRI-SphI fragment carrying atpBEFH (Apr) | This work |
| pER90 | BamHI deletion of pER20 | This work |
| pER1 | pBR322 with an HindIII-AvaI fragment carrying atpB (Apr) | This work |
| pER12 | HpaI deletion of pBJC917 | This work |
| pER2 | BamHI deletion of pER12 | This work |
| pER13 | NruI deletion of pER20 | This work |
| pER3 | HincII partial deletion of pER13 | This work |
| pMVD31 | pACYC184 with an EcoNI-EcoNI fragment carrying atpHAGDC (Cmr) | This work |
Media and culture conditions.
Luria-Bertani (LB) rich medium, R medium, and M63 minimal medium supplemented either with glucose or succinate were used (15). The following antibiotics were added to media at the indicated final concentrations: ampicillin, 50 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 30 μg/ml; tetracycline, 12 μg/ml. When necessary, l-amino acids were added at a final concentration of 40 μg/ml. All strains were grown at 37°C, except for those carrying Mucts, which were grown at 30°C.
Phenotype assays.
Microcin sensitivity was assayed by patch test as described previously (20), the level of sensitivity being estimated by the diameter of the zone of inhibition. The Atp phenotype was analyzed through the ability of clones to grow on minimal medium supplemented with succinate. Mutants affected in the atp locus are impaired for oxidative phosphorylation and, consequently, are unable to grow on minimal medium supplemented with a nonfermentable carbon source, such as succinate (4). Strains to be assayed were streaked on minimal medium supplemented with succinate and, in parallel, on the same medium supplemented with glucose instead. The plates were incubated 24 h at 37°C. Strains able to grow on either carbon source were considered Atp+, while those only capable of growing on glucose were scored as Atp−.
Genetic techniques.
P1 transduction was performed as described (15). MccH47r atp strains were made deficient for homologous recombination by introducing the recA56 allele from RYC816 by cotransduction with the neighboring srl::Tn10 marker, selecting transductants on LB plates supplemented with tetracycline. The RecA phenotype of the transductants was detected by UV sensitivity. Tn5 insertions of MccH47r atp mutants were cloned in vivo from the chromosome by means of phage MudII4042 lytic propagation as described previously (9, 20). The transducing phage lysates were used to infect MC4100 Mucts Apr, and transductant clones were selected on LB plates supplemented with chloramphenicol and kanamycin.
Manipulation and sequencing of DNA.
Routine DNA manipulations were carried out as described previously (18). DNA sequencing was performed by the Centro Técnico de Análisis Genéticos (Facultad de Ciencias, Montevideo, Uruguay). A specific Tn5 sequencing primer was used: 5′ CGA TGA AGA GCA GAA GTT AT 3′.
RESULTS
Localization of the Tn5 insertion sites in MccH47-resistant atp mutants.
As was previously described, a group of chromosomal Tn5 insertion mutants was preliminarily characterized as presenting a high level of resistance to MccH47; phenotypic and complementation analyses revealed they were affected in the atp locus. The transposon insertion in one of these, FGB64, was located after nucleotide 1038 of the reported atp sequences (accession number J01594), interrupting the expression of the atpB gene, which encodes subunit a of the proton channel (20). P1 transducing phage was grown on four additional MccH47r atp mutants, and the lysates were used to infect MC4100, selecting clones on LB plates supplemented with kanamycin. All the transductants were MccH47r, thus confirming that the Tn5 insertion was responsible for the MccH47r phenotype in each case. The insertions were now called Tn5 84, 89, 91, and 99. One transductant from each group was chosen (FGB084, FGB089, FGB091, and FGB099) and used in further studies.
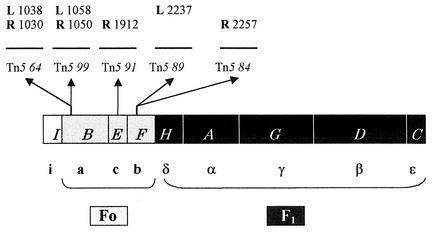
To identify the gene affected by each of the Tn5 insertions mentioned above, as well as to precisely identify, at the nucleotide level, the sites of insertion, these mutations were first cloned in vivo using the chimerical phage MudII4042, as described in Materials and Methods. In each of the four cloning experiments, transductants of strain MC4100 Mucts Apr should carry recombinant plasmids containing MudII4042 DNA, part or all of the Tn5 sequence, and, presumably, chromosomal DNA adjacent to the transposon. These plasmid constructions were used as start points for cloning DNA segments containing the chromosomal DNA-Tn5 junction sites into multicopy vectors. Nucleotide sequencing was performed upon these latter constructions. In this way, the mutations of strains FGB084, FGB089, FGB091, and FGB099 were cloned; this attempt was extended to the analysis of insertion Tn5 64, whose left end had already been precisely located (20). The following junction sites were cloned and sequenced: the right end of Tn5 64, 84, and 91; the left end of Tn5 89; and both ends of Tn5 99 (Fig. 1). It should be noted that in those cases where information was available from both junction sites (Tn5 64 and 99) a distance of 9 nucleotides was found between them, corresponding to the duplication of the Tn5 target (3). The Tn5 insertions were all found to interrupt the atp operon: Tn5 64 and 99 were located to atpB, Tn5 91 was located to atpE, and Tn5 84 and 89 were located to atpF. These three genes are precisely those that encode the three subunits that constitute the Fo proton channel. To discern if the presence of the entire ATP synthase complex was required for antibiotic action or if the proton channel by itself would suffice, the following experimental series was performed.
FIG. 1.
Tn5 insertions in the atp operon conferring resistance to MccH47. The atp operon is represented as follows: white box, atpI; grey boxes, atp genes encoding Fo polypeptides; black boxes, atp genes encoding F1 polypeptides. The corresponding Atp products are designated below. The sites of Tn5 insertions are shown by arrows, and the left (L) and right (R) nucleotides immediately adjacent to the transposon sequences are indicated, numbered according to the sequence of Walker et al. (23).
Identification of the minimal atp genetic information capable of conferring sensitivity to MccH47.
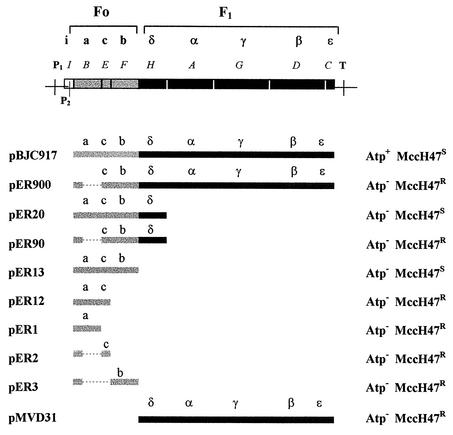
In order to obtain strains carrying different genetic information for ATP synthase, plasmid pBJC917 was used as a start point to construct a series of derivative plasmids with different DNA segments of the atp operon (Table 1 and Fig. 2). These plasmids were used in transformation experiments, and in all cases, a completely deficient atp chromosomal mutant, MC1000 Δ(uncI-uncC), was used as the receptor strain. In the resulting transformant clones, two phenotypes were systematically analyzed: (i) the Atp phenotype, related to the capacity to carry out oxidative phosphorylation, as evidenced by growth on succinate as the sole carbon source, and (ii) the MccH47 sensitivity phenotype, detected by the appearance of growth inhibition halos in patch tests performed with a MccH47-producing strain, as described in Materials and Methods (Fig. 2). It should be mentioned that, as expected, MC1000 Δ(uncI-uncC) exhibited Atp− and MccH47r phenotypes.
FIG. 2.
Recombinant atp constructions and conferred phenotypes. The atp operon is represented at top: white box, atpI; grey boxes, atp genes encoding Fo polypeptides; black boxes, atp genes encoding F1 polypeptides. The corresponding Atp products are indicated. P1, main promoter; P2, secondary promoter; T, terminator. Below, pBJC917 and a series of derivative plasmids are depicted, following the same color code as described above; only atp DNA is represented, with the encoded polypeptides indicated in each case. Deletions are shown as dashed lines. pBJC917 and all the plasmids containing Fo genes keep the P2 promoter. A more detailed description of the construction of each plasmid is given in Table 1. To the right, the Atp and MccH47 sensitivity phenotypes conferred by plasmids to strain MC1000 Δ(uncI-uncC) are indicated.
With respect to the Atp phenotype of the transformant clones, all of them were defective for oxidative phosphorylation with the exception of transformants with pBJC917. As to MccH47 sensitivity, whenever a recombinant plasmid carried the genes encoding the three subunits of the Fo proton channel, the presence of the plasmid conferred antibiotic sensitivity. This was the case regardless of the remaining atp context, thus indicating that the minimal structure necessary for MccH47 action to take place could be the proton channel. Accordingly, total resistance to MccH47 was found in those constructions lacking one or more genes for components of this channel. Among the MccH47-sensitive clones, the degree of sensitivity differed depending on the recombinant plasmid. The strain carrying pBJC917 exhibited the highest level of sensitivity: inhibition halos about 12 mm in diameter were observed. When the same assay was carried out with MC1000 Δ(uncI-uncC) with pER20 (encoding subunits a, c, b, and δ) or with pER13 (encoding subunits a, c, and b), the inhibition halos had a diameter of ca. 9 mm (see below).
Complementation assays.
We knew from previous results that when MccH47r atp mutants were transformed with plasmid pBJC917 the resulting strains were MccH47s and Atp+; i.e., the atp operon complemented for both phenotypes (20). The recombinant plasmids mentioned above, encoding different portions of ATP synthase, were now used in complementation assays with the five chromosomal MccH47r atp mutants under study. Prior to these assays, all five strains were made deficient for homologous recombination by means of P1 transduction, as described in Materials and Methods. Once these derivative strains were obtained, they were transformed with all of the recombinant plasmids carrying Fo determinants shown in Fig. 2. The same two phenotypes, oxidative phosphorylation and MccH47 sensitivity, were assayed for in each transformant clone (Table 2).
TABLE 2.
Complementation analysis for oxidative phosphorylation and for MccH47 sensitivitya between atp::Tn5 chromosomal mutationsc and recombinant plasmids carrying atp genes
| Plasmidb | FGB64 and FGB099 (a)
|
FGB091 (c)
|
FGB084 and FGB089 (b)
|
|||
|---|---|---|---|---|---|---|
| Phenotype | Sensitivity | Phenotype | Sensitivity | Phenotype | Sensitivity | |
| Vector alone | Atp− | MccH47r | Atp− | MccH47r | Atp− | MccH47r |
| pBJC917 (acbδαγβɛ) | Atp+ | MccH47s *** | Atp+ | MccH47s *** | Atp+ | MccH47s *** |
| pER900 (cbδαγβɛ) | Atp− | MccH47r | Atp+ | MccH47s *** | Atp+ | MccH47s *** |
| pER20 (acbδ) | Atp− | MccH47s ** | Atp− | MccH47s ** | Atp− | MccH47s ** |
| pER90 (cbδ) | Atp− | MccH47r | Atp− | MccH47s ** | Atp− | MccH47s ** |
| pER13 (acb) | Atp− | MccH47s ** | Atp− | MccH47s ** | Atp− | MccH47s ** |
| pER12 (ac) | Atp− | MccH47s * | Atp− | MccH47s * | Atp− | MccH47r |
| pER1 (a) | Atp− | MccH47s * | Atp− | MccH47r | Atp− | MccH47r |
| pER2 (c) | Atp− | MccH47r | Atp− | MccH47s * | Atp− | MccH47r |
| pER3 (b) | Atp− | MccH47r | Atp− | MccH47r | Atp− | MccH47s * |
The level of sensitivity to MccH47 was assigned according to the diameter of the antibiosis halos detected by patch test (***, 12 to 15 mm; **, 6 to 8 mm; *, 3 to 4 mm).
Plasmid-encoded Atp polypeptides are shown.
The Atp subunit affected is indicated in parentheses after each mutant or group of mutants.
As to the capacity to carry out oxidative phosphorylation, only the entire operon (pBJC917) restored the Atp+ phenotype in all five mutants. However, the complementation pattern differed upon transformation with plasmid pER900: those mutants defective for subunit a remained Atp−, whereas mutants defective for subunits c or b turned Atp+. In no other context was the Atp+ phenotype restored (Table 2).
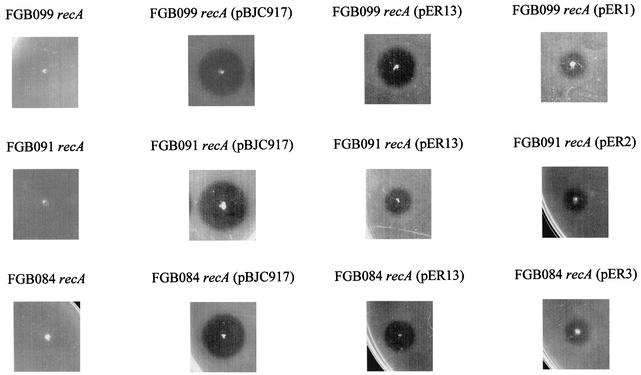
In all five mutants, sensitivity to MccH47 was restored by introduction of plasmids carrying the wild-type atp gene corresponding to the chromosomal gene affected by Tn5 insertion. In the strains where complementation took place, different degrees of microcin sensitivity were observed: in general terms, the more atp information was present in trans, the greater the sensitivity to MccH47 was (Table 2 and Fig. 3). It is noteworthy that the single presence in trans of the wild-type atp gene affected in each of the five mutants gave rise to complementation for the MccH47 sensitivity phenotype.
FIG. 3.
Complementation for MccH47 sensitivity between atp::Tn5 chromosomal mutations and recombinant plasmids carrying atp genes. A patch test was performed on the strains indicated, as described in Materials and Methods.
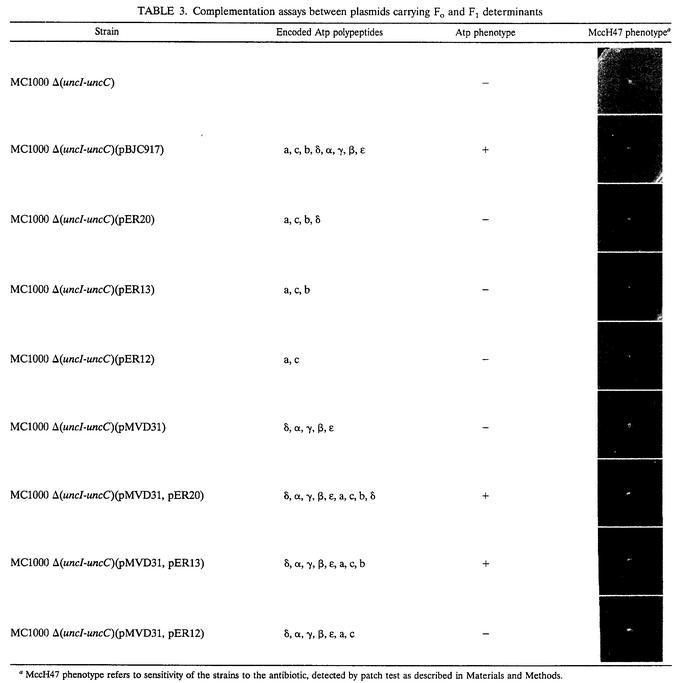
Although the results so far obtained clearly demonstrated that subunits a, c, and b were all three absolutely required for MccH47 action, they did not prove that these subunits were indeed arranged into a proton channel in our constructions. To elucidate this point, the following complementation assay was performed: strain MC1000 Δ(uncI-uncC) carrying pMVD31 (coding for the five F1 subunits) was transformed with plasmid pER13 (coding for the a, c, and b subunits). The resultant strain recovered the wild-type Atp+ and MccH47 sensitivity phenotypes, indicating that a functional ATP synthase complex had indeed been assembled. In a parallel assay, the same result was obtained when pER20 (coding for a, c, b, and δ subunits) was employed instead of pER13; as expected, no such complementation was observed with pER12 (coding for a and c subunits) (Table 3).
TABLE 3.
Complementation assays between plasmids carrying Fo and F1 determinants
MccH47 phenotype refers to sensitivity of the strains to the antibiotic, detected by patch test as described in Materials and Methods.
DISCUSSION
MccH47r atp Tn5 insertion mutants were extensively analyzed, and their mutations were located at the nucleotide level. It was important to discern if their MccH47r phenotype was due to the disruption of one or more atp genes. Complementation assays provided the certainty that only one atp gene had been affected in each mutant analyzed: in every case, when the single wild-type atp gene corresponding to the chromosomal gene affected by Tn5 insertion was introduced, mutants regained sensitivity to the antibiotic. These results showed that there was only one atp gene affected in each case and that its mutation was the sole responsible for the MccH47r phenotype, ruling out the possibility that rearrangements or secondary mutations had occurred. This fact enabled us to analyze our set of results on the basis of a strict correlation between genotype and phenotype.
The Tn5 insertions of the atp mutants analyzed clustered in the genes encoding the three subunits that compose the Fo proton channel; conversely, no Tn5 insertion was found to affect determinants for the F1 catalytic portion of ATP synthase. These results indicated that each of the components of the proton channel would be directly involved in the mode of action of MccH47, but they did not enable us to discard the participation of the catalytic portion. The construction of strains with genetic information for different portions of ATP synthase as well as the results from complementation experiments clearly indicated that the presence of subunits a, c, and b was at the same time necessary and sufficient for conferring MccH47 sensitivity. Whenever the three components of the proton channel were present, the strain was sensitive to the antibiotic. This proved to be the case regardless of whether the Fo polypeptides were encoded by the chromosome, by a recombinant plasmid, or by a combination of both. These results discard the catalytic F1 portion as being directly involved in MccH47 mode of action. On the other hand, the Atp+ phenotype was not always restored, even in the presence of all the information necessary for the ATP synthase complex. The importance of a cis arrangement of the atp genes to produce a correctly assembled ATP synthase has already been reported (5).
It was still necessary to discern if the subunits a, c, and b encoded by our constructions were correctly assembled into a proton channel. The complementation assays carried out in the MC1000 Δ(uncI-uncC) context provided information on this matter. When this strain was transformed with two plasmids, pMVD31 (encoding only F1 polypeptides) and pER13 (encoding only Fo polypeptides), the resulting strain regained full sensitivity to MccH47 as well as the capacity to carry out oxidative phosphorylation. This would imply that each set of atp genes separately determined the adequate formation of Fo and F1, and that both portions would meet to conform the entire ATP synthase complex. Other authors arrived at the same conclusion through in vitro reconstitution experiments in membranes, making use of similar Fo- and F1-encoding constructions. Although the assembly of the ATP synthase complex might involve interactions between the proton channel and the catalytic portion, they found that each moiety would be able to assemble separately in vivo (1, 10). Thus, it could be deduced that cells only bearing the Fo determinants succeed to assemble the proton channel and, therefore, that Fo itself would be the minimal structure necessary for MccH47 action.
We had previously considered that ATP synthase could be the target of MccH47 antibiotic action (17, 20); now we focus this presumption on the Fo proton channel. In light of this hypothesis, and knowing that MccH47 exerts a bactericidal effect on sensitive E. coli K-12 cells carrying the entire ATP synthase or only the proton channel (12; our unpublished results), it could be reasoned that the interaction of MccH47 with the latter structure would be responsible for cell death. If this were the case, antibiotic action would be related to Fo function, i.e., proton translocation. The passage of protons through Fo can conceivably be altered in three ways: blockage of translocation, proton pumping against the gradient, and proton entry downstream of the gradient. The first possibility cannot account for cell death, since cells would survive by means of substrate-level phosphorylation under most of the culture conditions employed. The second possibility has an energetic cost; with the entire ATP synthase complex, the energy required to extrude protons could be provided by the coupled hydrolysis of ATP by F1, but in cells with only Fo, no such energy would be supplied. Therefore, only the third possibility remains clearly feasible, since it is the only one that can account for the bactericidal action of MccH47 in both contexts, with the entire ATP synthase and with only Fo: the membrane potential would be dissipated by the action of MccH47, which would provoke an unregulated entry of protons. It should also be taken into account that the presence of the entire ATP synthase confers higher levels of sensitivity to MccH47 than does the presence of the single proton channel, indicating that the F1 portion, although not being strictly required for antibiotic action, enhances microcin's effect in a way that remains to be elucidated.
In summary, results presented in this work support our previous findings on the need of FoF1 ATP synthase for MccH47 antibiotic action (20). Here we demonstrate that the entire enzymatic complex is not required: the Fo proton channel is both necessary and sufficient for conferring sensitivity to MccH47. So far, all the in vivo analyses performed on MccH47 mode of action indicate that the Fo portion of ATP synthase would be the target of this antibiotic. MccH47 thus appears to be a natural product with an unprecedented mode of action on prokaryotes. This hypothesis will need to be tested in in vitro studies.
Acknowledgments
This work was supported by the International Foundation for Science (grant F/3011-1) and also by the Programa de Desarrollo de las Ciencias Básicas and the Comisión Sectorial de Investigación Científica de la Universidad de la República, Uruguay.
We thank W. S. A. Brusilow for kindly providing the strain MC1000 Δ(uncI-uncC). We are also grateful to María Parente for excellent technical assistance.
REFERENCES
- 1.Aris, J. P., D. J. Klionsky, and R. D. Simoni. 1985. The Fo subunits of the Escherichia coli F1Fo-ATP synthase are sufficient to form a functional proton pore. J. Biol. Chem. 260:11207-11215. [PubMed] [Google Scholar]
- 2.Azpiroz, M. F., E. Rodríguez, and M. Laviña. 2001. The structure, function, and origin of the microcin H47 ATP-binding cassette exporter indicate its relatedness to that of colicin V. Antimicrob. Agents Chemother. 45:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, D. E. 1977. Insertion and excision of a transposable kanamycin resistance determinant Tn5, p. 205-212. In A. I. Bukhari, J. A. Shapiro, and S. L. Adhya (ed.),DNA insertion elements, plasmids and episomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Boogerd, F. C., L. Boe, O. Michelsen, and P. R. Jensen. 1998. atp mutants of Escherichia coli fail to grow on succinate due to a transport deficiency. J. Bacteriol. 180:5855-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brusilow, W. S. A. 1993. Assembly of the Escherichia coli F1Fo ATPase, a large multimeric membrane-bound enzyme. Mol. Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 6.Delgado, M. A., M. R. Rintoul, R. N. Farías, and R. A. Salomón. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaggero, C., F. Moreno, and M. Laviña. 1993. Genetic analysis of microcin H47 antibiotic system. J. Bacteriol. 175:5420-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Bustos, J. F., N. Pezzi, and E. Méndez. 1985. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob. Agents Chemother. 27:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groisman, E. A., B. A. Castilho, and M. J. Casadaban. 1984. In vivo DNA cloning and adjacent gene fusing with a mini-Mu-lac bacteriophage containing a plasmid replicon. Proc. Natl. Acad. Sci. USA 81:1480-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky, D. J., and R. D. Simoni. 1985. Assembly of a functional F1 of the proton-translocating ATPase of Escherichia coli. J. Biol. Chem. 260:11200-11206. [PubMed] [Google Scholar]
- 11.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229-243. [DOI] [PubMed] [Google Scholar]
- 12.Laviña, M., and C. Gaggero. 1992. Genetic determinants for microcin H47, an Escherichia coli chromosome-encoded antibiotic, p. 413-416. In R. James, F. Lazdunski, and F. Pattus (ed.), Bacteriocins, microcins and lantibiotics. Springer-Verlag KG, Heidelberg, Germany.
- 13.Laviña, M., C. Gaggero, and F. Moreno. 1990. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J. Bacteriol. 172:6585-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach, D., and N. Symonds. 1979. The isolation and characterization of a plaque-forming derivative of bacteriophage Mu carrying a fragment of Tn3 conferring ampicillin resistance. Mol. Gen. Genet. 172:179-184. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Rodríguez, E., and M. Laviña. 1998. Genetic analysis of microcin H47 immunity. Can. J. Microbiol. 44:692-697. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez, E., C. Gaggero, and M. Laviña. 1999. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 43:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Solomon, K. A., D. K. W. Hsu, and W. S. A. Brusilow. 1989. Use of lacZ fusions to measure in vivo expression of the first three genes of the Escherichia coli unc operon. J. Bacteriol. 171:3039-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trujillo, M., E. Rodríguez, and M. Laviña. 2001. ATP synthase is necessary for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 45:3128-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizán, J. L., C. Hernández-Chico, I. del Castillo, and F. Moreno. 1991. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 10:467-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Meyenburg, K., B. B. Jorgensen, and B. van Deurs. 1984. Physiological and morphological effects of overproduction of membrane-bound ATP synthase of Escherichia coli K-12. EMBO J. 3:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker, J. E., M. Saraste, and N. J. Gay. 1984. The unc operon, nucleotide sequence, regulation, and structure of ATP-synthase. Biochim. Biophys. Acta 768:164-200. [DOI] [PubMed] [Google Scholar]
- 24.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158:757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]