Abstract
Toxoplasma gondii induces the expression of proinflammatory cytokines, reorganizes organelles, scavenges nutrients, and inhibits apoptosis in infected host cells. We used a cDNA microarray of 420 annotated porcine expressed sequence tags to analyze the molecular basis of these changes at eight time points over a 72-hour period in porcine kidney epithelial (PK13) cells infected with T. gondii. A total of 401 genes with Cy3 and Cy5 spot intensities of ≥500 were selected for analysis, of which 263 (65.6%) were induced ≥2-fold (expression ratio, ≥2.0; P ≤ 0.05 [t test]) over at least one time point and 48 (12%) were significantly down-regulated. At least 12 functional categories of genes were modulated (up- or down-regulated) by T. gondii. The majority of induced genes were clustered as transcription, signal transduction, host immune response, nutrient metabolism, and apoptosis related. The expression of selected genes altered by T. gondii was validated by quantitative real-time reverse transcription-PCR. These results suggest that significant changes in gene expression occur in response to T. gondii infection in PK13 cells, facilitating further analysis of host-pathogen interactions in toxoplasmosis in a secondary host.
Toxoplasma gondii is an obligate intracellular Apicomplexan protozoan parasite that replicates within nucleated vertebrate cells (16). The parasite has a ubiquitous distribution and broad secondary host range that includes up to 30% of the human population worldwide (15, 51). Infection with T. gondii is most often benign and asymptomatic in humans due to a rapid, effective immune response that forces encystation of the infectious tachyzoite stage and leads to a lifelong persistence of the parasite in the secondary intermediate host (19). One such host is the pig, where the prevalence of the parasite can be as a high as 93% (13). The process of encystation reportedly includes the induction of proinflammatory cytokines that activate both innate and adaptive immune reactions (4, 10, 33).
Toxoplasma gondii resides and replicates within a parasitophorous vacuole (PV) that is surrounded by host mitochondria and endoplasmic reticulum but free of host membrane proteins (43). The PV does not acidify and is resistant to fusion with host endocytic and lysosomal compartments, providing a safe environment for the parasite to multiply (25). The PV expands in size during the course of infection, and 2 to 3 days after invasion the host cell ruptures, releasing parasites that can actively invade new host cells (16). Once the parasite begins growing and dividing within the PV, it scavenges nutrients such as cholesterol, glucose, and purine nucleosides from the host cell (8, 43).
The parasite uses several strategies to infect and persist intracellularly. These include inducing mitogen-activated protein kinase phosphorylation (18), down-regulation of major histocompatibility complex (MHC) class II molecules (30, 30a), and inhibition of host cell apoptosis, as reported for various T. gondii-infected murine and human cell lines (17, 22, 32, 34, 36). Although systematic analyses of host cell gene expression in response to T. gondii infection have just begun (3, 16), these studies suggest that significant changes in host cell transcription occur in response to a T. gondii infection.
We profiled the transcription of porcine kidney epithelial (PK13) cells in response to a T. gondii infection over a 72-hour period postinfection (p.i.), using a porcine cDNA expressed sequence tag (EST) microarray derived from a variety of porcine tissues (40). The 420 selected ESTs represented genes encoding proteins involved in diverse signaling pathways associated with a T. gondii infection, including host defense strategies as well as parasite growth and pathogenesis. The profiles revealed that a significant number of host genes were induced (65.6% of the total analyzed), while fewer (12%) were down-regulated. Induction occurred early in the infection (1 h to 4 h p.i.), while a significant down-regulation was observed only later in the infection (48 h to 72 h p.i.). At least 12 functional categories of genes were transcriptionally altered by T. gondii during the 72-hour course of the infection, including those encoding proteins involved directly in host cell transcription, signal transduction, immune response, nutrient metabolism, apoptosis, cell cycle, and cell structure (adhesion and cytoskeletal components). These results suggest that significant changes in gene expression occur throughout the course of the host cell's response to T. gondii.
MATERIALS AND METHODS
Host cells and parasites.
Porcine kidney epithelial cell line PK13 (CRL6489; ATTC, Manassas, VA) was cultured in 75-cm2 Greiner tissue culture flasks containing 15 ml of Dulbecco's modified Eagle's medium (DMEM) with 4.0 mM l-glutamine, 4.5g/liter glucose, and 1.0 mM sodium pyruvate (ATCC 30-2002) and supplemented with 10% heat-inactivated fetal bovine serum (HIFBS) (ATCC 30-2020). The PK13 cells were incubated in 5% CO2 at 37°C. Tachyzoites of T. gondii strain TS-4 (ATCC 40050) were propagated in PK13 cells in 75-cm2 Greiner tissue culture flasks containing 15 ml of DMEM (as described above) supplemented with 3% HIFBS and were incubated in 5% CO2 at 35°C.
Infection of PK13 host cells with T. gondii.
T. gondii tachyzoites were harvested from infected PK13 cell monolayers when host cell lysis and parasite egress were almost complete. Briefly, the flasks were scraped and the cells, together with cell debris, harvested and passed twice through a 27-gauge needle to rupture any remaining PK13 cells and release the parasites within. Host cell debris was removed by centrifugation of the cell suspension at 50 × g for 3 min. The supernatant was then centrifuged at 1,300 × g for 10 min, the pellet washed three times in serum-free DMEM, and the tachyzoites counted under light microscopy. The tachyzoites were resuspended in DMEM supplemented with 3% HIFBS and used to infect confluent monolayers of porcine PK13 cells in 75-cm2 tissue culture flasks at a multiplicity of infection of 1:6. Infected cells were cultured in 15 ml of DMEM supplemented with 3% HIFBS and incubated in 5% CO2 at 35°C. Uninfected control cells were maintained under similar conditions. All experiments were carried out in duplicate, with RNA isolation at 0 h, 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h p.i.
RNA extraction.
Total RNA was isolated from T. gondii-infected and uninfected control cells (∼8 × 106 cells per flask) at each time point, using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RNA samples used in cDNA microarray analyses were identical to those used in quantitative real-time reverse transcription-PCR (QRT-PCR) at corresponding time points.
Microarray construction.
Slides were spotted with 420 annotated ESTs sequenced from cDNA clones derived from porcine tissues infected by an intracellular pathogen (40). A list of these ESTs showing porcine cDNA clones identities, GenBank accession numbers for each sequenced EST, the BLASTx protein homologues for each EST, and NCBI accession numbers for the protein homologues, as well as bit scores, E values, and identities (percent similarities), can be found at http://www.ag.unr.edu/ab/Downloads/sup1.xls. Briefly, inserts from these cDNA libraries in the pBSK vector (Stratagene) were amplified using primers T7 (5′CGGGAT ATCACTCAGCATAATG3′) and SK (5′CGCTCTAGAACTAGTGGATC3′), and purified with the QIAquick PCR purification kit (QIAGEN, Valencia, CA), and then printed in triplicate on Telechem Super- Amine aminosilane-coated glass microscope slides (Telechem, Sunnyvale, CA) using the Versarray ChipWriter Pro (Bio-Rad, Hercules, CA) in the Nevada Genomics Center at 23°C and 40% humidity. After being spotted, the slides were rehydrated at 42°C for 10 seconds, snap dried in an 80°C oven for 30 seconds, cross-linked with UV light at 120 mJ/cm2 for 20 seconds in a UVP CL-1000 cross-linker (UVP Inc, Upland, CA), and stored at room temperature.
Indirect aminoallyl labeling of cDNA probes.
Total RNA samples (10 μg) extracted from T. gondii-infected and uninfected control host cells were reverse transcribed into cDNA and labeled with Cytidine 3 (Cy3) and Cytidine 5 (Cy5), respectively (24). Dye swapping experiments were performed, with no significant differences observed between the two labeling regimens (data not shown).
Microarray hybridization.
Microarray slides were prehybridized in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% bovine serum albumin, and 0.1% sodium dodecyl sulfate (SDS) at 42°C for 45 min, dipped five times in distilled water, denatured in boiling water for 2 min followed by 95% ethanol for 1 min, and then centrifuged at 1,000 rpm for 2 min to dry in 50-ml uncapped centrifuge tubes. Blocking agents [1 μl each of 20 μg/μl human Cot1 DNA, 20 μg/μl poly(A) DNA, and 20 μg/μl yeast tRNA] were added to the indirectly aminoallyl-labeled cDNA probes, heated at 95°C for 3 min, snap cooled on ice for 30 s, centrifuged for 1 min, and applied to prehybridized slides in a CMT hybridization chamber (Corning Inc., Corning, NY), and cover slips were added. Humidity was maintained with 10 μl diethyl pyrocarbonate water added to the two reservoir wells of the hybridization chamber. The chamber was tightly sealed and incubated at 42°C in a ProBlot 6 hybridization oven (Labnet, Edison, NJ) overnight for 16 to 20 h. Slides were removed from the chamber; sequentially washed for 10 min in 1× SSC-0.2% SDS, 0.1× SSC-0.2% SDS, and 0.1× SSC buffers; rinsed in distilled water (4 min each wash); and dried by centrifugation at 1,000 rpm for 2 min. The entire experiment, including primary culture of cells, T. gondii infection, and RNA isolation, was performed twice, with probe synthesis and hybridizations performed twice per independent experiment. This experimental design resulted in 12 independent Cy3/Cy5 intensity ratio data points for each spotted cDNA at each time point (representing a total of 96 data points per arrayed sequence).
Fluorescent scanning and image analysis.
Images were acquired by laser confocal scanning at the Nevada Genomics Center, using a ScanArray 4000 microarray analysis system (Perkin-Elmer, Boston, MA) at a resolution of 5 μm. A 16-bit TIFF image was generated for each channel (Cy3 and Cy5). The scanned microarray images for Cy3 and Cy5 were overlaid and quantified to determine the fluorescent intensities of the two dyes for each spot by using the QuantArray microarray analysis version 3.0 software (Perkin-Elmer, Boston, MA). QuantArray extracts Cy3 and Cy5 intensities in a defined pixel area surrounding each spot on every array, which are output along with estimates of local background and measures of local variability in the form of Microsoft Excel-type spreadsheets (Microsoft, Seattle, WA).
Microarray data analysis.
The GeneSpring 6.1 software (Silicon Genetics, Redwood City, CA) was used to normalize and analyze output data imported from the QuantArray software. Briefly, the net signal intensity in each channel (Cy3 or Cy5) was determined by subtracting the local background from signal intensity values. Each experimental result was interpreted as the average of 12 replicates of each gene at each time point (3 technical replicates spotted on each chip and 4 biological replicates in hybridization). The raw data were filtered to eliminate signals with average fluorescent spot intensities (brightnesses) of <500 in both Cy3 and Cy5 channels, which were not included in the final microarray data analysis. Each spot was normalized by averaging replicate intensities, followed by division of each spot's Cy3 signal intensity (T. gondii-infected cells) by the control Cy5 signal intensity (uninfected cells) to determine Cy3/Cy5 ratios. Further normalization was performed by dividing each spot's Cy3/Cy5 ratio by the median fluorescence of the housekeeping GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene. Therefore, the reported differential expression of each gene is relative to that of GAPDH expression. To better visualize and compare expression data, final log2-transformed Cy3/Cy5 ratios were calculated from the normalized values. P values were calculated using Student's t test under the null hypothesis for each experiment (treatments and time points). Genes exhibiting signal-to-control channel (Cy3/Cy5) ratios of ≥2.0 and a P value of ≤0.05 by the t test were defined as significantly up-regulated; those with Cy3/Cy5 ratios of ≤0.5 (P ≤ 0.05) were defined as significantly down-regulated. Differentially expressed genes (displaying a ≥2.0-fold modulation) were clustered by standard correlation by using GeneSpring 6.1 software (Silicon Genetics) and classified into functional subgroups by using Simplified Ontology, a feature of GeneSpring.
QRT-PCR.
QRT-PCR was performed to validate the microarray data for 12 selected genes modulated by T. gondii (Table 1). Primers and probes were designed using the Primer Express 2.0 software (Applied Biosystems [ABI], Foster City, CA). Reverse transcription was performed with 1 μg total RNA isolated from T. gondii-infected and uninfected control cells at experimental time points corresponding to those in the microarray analysis (0 h, 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h p.i.), using 300 ng/μl random primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. QRT-PCR was performed using the 5′ nuclease TaqMan assay (ABI, Foster City, CA) according to the manufacturer's instructions. Briefly, 1 μl of cDNA obtained from the reverse-transcription reaction was amplified in a 25-μl mixture containing 2× TaqMan Universal PCR Mastermix (ABI, Foster City, CA), 900 nmol/liter of forward and reverse primers, and 250 nmol/liter of TaqMan hybridization probe. For each sampled time point, the target gene was amplified concurrently with the GAPDH control gene in separate wells. Nontemplate controls were included in every run. All reactions were performed in triplicate. Thermal cycling was performed in an ABI PRISM 7000 sequence detection system unit (ABI, Foster City, CA).
TABLE 1.
Primer and probe sequences for quantitative real-time RT-PCR
| Accession no.a | Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | TaqMan probeb (5′ to 3′) |
|---|---|---|---|---|
| CAB40144 | C-FOS | CTTCCTGTTCCCAGCATCGT | AAGGAACCAGACAGGTCCATGT | 6FAM-AGCCCGCTCTGTGC-MGBNFQ |
| AAH02844 | NFKB2 | GCGTACGGGAGCCAGTCA | GCCCTCCTGGTGGCTCA | 6FAM-AGAGAAGCTGGGCCC-MGBNFQ |
| P35638 | DDIT3 | GACTGCTTCGTGGAGCTTCTG | GCAATGACTCAGCTGCCATCT | 6FAM-AGCCTAATCCAGCCGCA-MGBNFQ |
| BAD92726 | TTRAP | GACGCGACCGGAGACTTG | TGGTCAGAAACTCGGAAATGTTG | 6FAM-CAACCGCCCTGTCC-MGBNFQ |
| AAQ89327 | APR-3 | CAACACCTTCCGTGGCTTTAC | TCCCGGACAGTTGACACCTT | 6FAM-AGCTCCAGACTCTGATAC-MGBNFQ |
| AAC686533 | BECN1 | AACCCAGTTTAACTCTGAGGAACAGT | CTGTGAGGATACCCAAGCAAGAC | 6FAM-AGCTCTCAAGTTCATGCTG-MGBNFQ |
| AAS91711 | FADD | GGACGCGTGGGCAAGAG | TCCAGGAGCACGGAGAAGAG | 6FAM-AAGCTGGAGCGCGTG-MGBNFQ |
| AAO72308 | KS1/ZNF382 | ATGCCCTCAGTGTGGGAAAG | CCTGTATGAGTTCTTTGATGATGAATG | 6FAM-CCTTTAGCAGGAAATC-MGBNFQ |
| AAH00491 | PCNA | GGTGATGCCAGCACATACTTAATT | GGACTTAAATTGTAGAGAATGGAGGAA | 6FAM-TGTACTCAGAGGTACAAAGA'MGBNFQ |
| AAC39171 | PIAP | TCTAACCTGAGCATGCAGACATATG | TTGCAAGCTGTTCAGGATGAAC | 6FAM-TCTGTAACTGGCCTTCTAGT-MGBNFQ |
| CAI23572 | RIN2 | GCTCTGTGCTGAGAAGTTCAAGGT | CCACGAAGAGGAAGAGGCTGTA | 6FAM-GCACCCCGCGGAA-MGBNFQ |
| P10114 | RAP2A | TCTCGGACAGGCCCTTTG | CGGCCCAGGACGGTTT | 6FAM-CCAAGGCCGTAGCC-MGBNFQ |
| P00355 | GAPDH | GTGACACTCACTCTTCTACCTTTGATG | GGTCCACCACCCTGTTGCT | 6FAM-CAAGCTCATTTCCTGGTAC-MGBNFQ |
From the NCBI protein database.
6FAM, 6-carboxyfluorescein; MGBNFQ, minor groove binding nonfluorescent quencher.
QRT-PCR data were analyzed using the Relative Expression Software Tool (REST) (37) (http://www.gene-quantification.de/rest.html). REST uses a mathematical model based on the PCR efficiencies and the mean threshold value (Ct) deviation between sample and control group, where the ratio (R) = (Etarget)ΔCt Target (control − sample)/(Eref)ΔCt ref (control − sample). The quantitation was done relative to a nonregulated reference housekeeping control gene (GAPDH gene). Differences in expression between control and treated samples for the investigated transcripts were assessed in group means for statistical significance by using the pairwise fixed reallocation randomization test running within REST (23, 37).
Enzyme-linked immunosorbent assay detection of transcription factor DNA-binding activity.
We also characterized the DNA-binding activities of the transcription factors c-Fos and NF-κB2 in whole-cell lysates isolated from T. gondii-infected and uninfected control cells at 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h p.i. to determine whether the induction of gene transcription observed following T. gondii infection was correlated with elevated levels of the respective proteins. Activation assays were performed using Trans-AM c-Fos and NF-κB p52 transcription factor kits (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 25 μg extract was added to 96-well plates coated with oligonucleotides containing the c-Fos and NF-κB2 (p52) consensus sites, and binding of the transcription factors was visualized using anti-c-Fos and anti-p52 antibodies that specifically recognize activated DNA-bound c-Fos and NF-κB2, respectively. Antibody binding was detected with a secondary horseradish peroxidase-conjugated antibody and developed with 3,3′,5,5′-tetramethylbenzidine substrate.
RESULTS
The terminally differentiated PK13 cells used in this study function similarly to epithelial cells, forming the main portal of entry for T. gondii in the intestinal tracts of pigs and humans (12a), and they represent the first line of host defense against infection. The T. gondii strain TS-4 used for the in vitro infections is a slower-growing derivative of the highly virulent RH strain (42). At an MOI of 1:6, parasite egress and host cell lysis occurred at 4 days p.i. (data not shown), allowing us to harvest sufficient intact host cells for RNA extraction at 72 h.
Differential gene expression following T. gondii infection.
PK13 cells were infected with T. gondii, and total RNA was isolated from infected and uninfected control cells at 0 h, 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h p.i. At each experimental time point, microarray analysis compared gene expression between T. gondii-infected and uninfected control cells relative to a GAPDH internal control, which remained constitutively expressed throughout the in vitro T. gondii infection and was not significantly altered, exhibiting nonnormalized expression ratios ranging from 0.75 at 2 h p.i. (equivalent to a −1.33-fold change; P = 0.496) to 1.51 at 72 h p.i. (1.51-fold change; P = 0.062). The application of the GAPDH gene as a housekeeping gene for normalizing gene expression in kidney epithelial cells has been previously reported (14, 35).
Of the original 420 EST sequences, 401 (95.5%) proved to be sufficiently reliable and were selected for further analysis. The expression of 263 genes (65.6% of 401 analyzed) was induced ≥2.00-fold (expression ratio, ≥2.00) at at least one of the eight sampled time points; 48 (12%) were significantly down-regulated ≥2.00-fold (expression ratio, ≤0.50). Of the significantly up-regulated genes, 95 (22.6% of the total analyzed) were induced ≥3-fold, while 8 (1.9%) were induced ≥4-fold. Cluster analysis of time course data for 401 genes (Fig. 1A) suggested two discrete gene modulation patterns, i.e., induced genes and down-regulated genes, over 72 h p.i. The enlarged gene tree (Fig. 1B) revealed the correlated expression of genes involved in diverse functions and cellular pathways.
FIG. 1.
Transcriptional profile of porcine kidney epithelial (PK13) cells infected with T. gondii for 0 to 72 h. (A) Hierarchical clustering of the expression pattern of cDNAs representing 401 known genes, analyzed by microarray. Each row represents an individual cDNA element spotted on the array, and each column represents the expression states of cDNAs at a particular time point postinfection. Each expression data point represents the ratio of the fluorescence intensity of the cDNA from T. gondii-infected cells to the fluorescence intensity of the cDNA from noninfected control cells and is the average value of 12 data points for each time point of the experiment. The cluster is subdivided into two major groups, indicated by roman numerals at the left, consisting of genes that were repressed (i) (green) and genes that were induced (ii) (red). Genes whose expression did not change are colored black on the gene tree. (B) Enlarged view of the cluster from panel A, showing the genes that are modulated at 0 to 72 h postinfection.
T. gondii modulates diverse functional categories of host genes.
Transcripts representing 12 functional categories were significantly modulated by T. gondii (Table 2). The two largest classes of genes were those involved in host cell transcription and signaling, followed by genes involved in metabolism, immune response, cell cycle, and apoptosis. This suggests that numerous cellular processes were transcriptionally altered during the course of the infection. The majority of up-regulated genes were induced early in the infection (1 h to 4 h p.i.), while most down-regulation occurred later in the infection (48 h to 72 h p.i.). Table 3 shows a partial list of genes significantly modulated by T. gondii over the course of infection. A complete list of functionally classified genes regulated by T. gondii can be found in the supplemental information at http://www.ag.unr.edu/ab/Downloads/sup2.xls.
TABLE 2.
Functional categories of host genes modulated by T. gondii over 72 ha
| Functional class | No. of genes modulated at the indicated period postinfection
|
|||||
|---|---|---|---|---|---|---|
| Induced genesb
|
Repressed genesc
|
|||||
| 1-4 h | 6-24 h | 48-72 h | 1-4 h | 6-24 h | 48-72 h | |
| Transcription | 50 | 36 | 10 | 0 | 4 | 6 |
| Signal transduction | 49 | 33 | 7 | 0 | 0 | 7 |
| Nutrient metabolism | 38 | 30 | 4 | 1 | 3 | 13 |
| Immune response | 35 | 30 | 12 | 1 | 1 | 5 |
| Cell cycle | 22 | 16 | 2 | 0 | 0 | 1 |
| Apoptosis | 19 | 17 | 5 | 1 | 1 | 6 |
| Cell structure | 13 | 7 | 2 | 0 | 0 | 5 |
| Protein degradation | 11 | 6 | 1 | 1 | 1 | 2 |
| Protein synthesis | 10 | 7 | 2 | 0 | 0 | 2 |
| Protein trafficking | 9 | 5 | 0 | 0 | 0 | 0 |
| Cell growth and proliferation | 5 | 3 | 0 | 0 | 0 | 0 |
| Heat shock/stress response | 2 | 1 | 0 | 0 | 0 | 0 |
| Total | 263 | 191 | 45 | 4 | 10 | 48 |
Distribution of 401 functionally classified genes with reliable gene expression data.
Significantly up-regulated ≥2-fold (gene expression ratio, ≥2.0; P ≤ 0.05 by t test).
Significantly down-regulated ≥2-fold (gene expression ratio, ≤0.5; P ≤ 0.05 by t test).
TABLE 3.
Host gene expression modulated by T. gondii infectiona
| Functional class | Accession no.b | Protein (gene) | Fold change in transcription at h postinfectionc:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 6 | 24 | 48 | 72 | |||
| Transcription | CAA58780 | Transcription initiation factor TFIID (TAF11) | 2.09 | 3.96 | 2.78 | 3.05 | 2.24 | —d | 1.91 |
| AAH93797 | DNA-directed RNA polymerase II, 16 kDa (POLR2D) | — | 2.34 | — | 2.02 | — | — | — | |
| AAH00889 | DNA-directed RNA polymerase I, 16 kDa (POLR1D) | 2.13 | 3.05 | 2.44 | 2.78 | — | — | — | |
| BAD06318 | Signal transducer and activator of transcription 1 (STAT1) | — | 2.26 | — | — | — | — | — | |
| BAD89432 | Signal transducer and activator of transcription 6 (STAT6) | — | 2.63 | 2.44 | 2.52 | — | — | — | |
| CAA65753 | c-myc proto-oncogene product (MYC) | — | 2.44 | 2.29 | 2.07 | — | — | — | |
| AAH27924 | Myc-binding factor Max (MAX) | — | 3.91 | 3.28 | 3.15 | 2.18 | 1.83 | — | |
| CAB40144 | c-Fos protein (C-FOS)e | — | 2.65 | 2.43 | 2.65 | — | — | — | |
| AAH02844 | Nuclear factor κB p100/p49 (NFKB2)e | — | — | 1.89 | 1.82 | — | 1.65 | — | |
| Signal transduction | AAH00471 | Dual-specificity mitogen-activated protein kinase kinase 2 (MAP2K2) | 2.22 | 4.05 | 2.35 | 4.43 | 2.17 | — | 1.86 |
| AAM01193 | Protein kinase C beta 1 (PKC) | 2.04 | 3.53 | 2.69 | 3.14 | 2.11 | — | — | |
| BAA06171 | Tec protein-tyrosine kinase (TEC) | — | 2.28 | — | — | — | — | −3.10 | |
| CAI23831 | Proto-oncogene tyrosine-protein kinase LCK (LCK) | — | — | — | -2.33 | — | −2.11 | −2.79 | |
| P80310 | Calgranulin C (S100A12) | — | 3.15 | 2.34 | 2.55 | 2.03 | — | 1.90 | |
| AAH00846 | Calcium- and integrin-binding protein 1 (CIB1) | — | 3.09 | 2.45 | 2.53 | — | — | — | |
| AAH25372 | Calponin 3, acidic isoform (CNN3) | — | 2.73 | — | 2.22 | — | — | — | |
| AAS75333 | Rho family small GTP-binding protein RhoG (RHOG) | 2.32 | 4.13 | 3.21 | 3.27 | 2.37 | 2.05 | — | |
| AAV68380 | GTP-binding protein SAR1b (SARA2) | 2.22 | 3.68 | 2.73 | 3.10 | 2.14 | 1.95 | 1.94 | |
| CAB57359 | Rab5 GDP/GTP exchange factor homologue (RABGEF1) | — | — | — | — | — | −2.89 | −2.95 | |
| Immune response | AAH07091 | Interferon-inducible protein with tetratricopeptide repeats 1 (IFIT1) | — | 3.57 | 2.60 | 3.53 | 2.09 | 1.84 | — |
| AAP35948 | Interferon-inducible protein with tetratricopeptide repeats 4 (IFIT4) | 2.15 | 3.80 | 2.66 | 2.76 | 2.26 | 1.91 | — | |
| AAP35538 | Interferon-induced transmembrane protein 3 (IFITM3) | 2.02 | 3.66 | 2.59 | 3.39 | 2.19 | 1.87 | — | |
| AAP30878 | Proteasome activator 28 alpha subunit (PSME1) | — | 2.97 | 2.53 | 2.72 | — | — | — | |
| AAH21136 | Interferon gamma-inducible protein 30 (IFI30) | — | 2.24 | 2.14 | 2.02 | — | — | — | |
| AAD38418 | Immunoglobulin G heavy chain (IGHG) | — | 2.98 | 2.56 | 2.89 | 2.11 | — | — | |
| AAC33488 | Human cytokine receptor/Epstein-Barr virus-induced gene 3 (EBI3) | 2.04 | 3.93 | 2.49 | 2.83 | 2.28 | — | 1.98 | |
| BAC98339 | Interleukin-2 receptor gamma (IL2RG) | 2.12 | 3.86 | 2.59 | 2.73 | 2.18 | — | — | |
| AAH28221 | Interleukin-8 receptor alpha (IL8RA) | — | 2.23 | 2.14 | — | — | — | — | |
| AAQ84094 | CXCL12 chemokine (CXCL12) | — | 2.56 | 2.12 | 2.07 | — | — | — | |
| AAZ32767 | C-X-C chemokine receptor type 4 (CXCR4) | 2.03 | 3.07 | 2.68 | 2.82 | 2.06 | — | — | |
| AAH63122 | Small inducible cytokine B9 precursor (CXCL9) | — | — | — | −2.80 | — | −2.23 | −2.75 | |
| CAH72304 | MHC class I-related protein (MR1) | — | — | — | — | — | — | −2.27 | |
| BAB39859 | MHC class II antigen, SLA-DM alpha chain (SLA-DMA) | 2.04 | 3.73 | 2.65 | 2.89 | 2.11 | — | — | |
| BAD92814 | MHC class II antigen, HLA-DM beta chain (HLA-DMB) | 2.33 | 3.22 | 2.67 | 3.08 | 2.13 | 1.82 | 1.84 | |
| BAD99115 | MHC class II antigen, SLA-DQ alpha chain (SLA-DQA) | 2.21 | 3.66 | 2.60 | 2.93 | 2.08 | — | — | |
| BAD99117 | MHC class II antigen, SLA-DQ beta 1 domain (SLA-DQB1) | 2.28 | 4.12 | 3.11 | 3.37 | 2.36 | 1.94 | 1.80 | |
| BAD99121 | MHC class II antigen, SLA-DR alpha chain (SLA-DRA) | — | — | — | — | — | — | −2.00 | |
| AAR20889 | Complement C1r component (C1R) | 2.26 | 3.91 | 2.94 | 3.80 | 2.29 | 2.00 | 1.87 | |
| AAG40565 | Complement component C3 (C3) | — | 2.29 | 2.11 | 2.21 | — | 2.58 | 2.44 | |
| AAR20891 | Complement component C4 (C4) | 2.25 | 4.26 | 2.48 | 2.86 | 2.29 | 1.83 | 1.85 | |
| Apoptosis | AAC68653 | BCL-2-interacting protein Beclin (BECN1)e | — | 2.41 | 2.17 | 2.19 | — | — | — |
| BAA13115 | Defender against cell death 1 (DAD1) | — | 3.66 | 2.43 | 2.59 | 2.23 | — | 1.82 | |
| AAC39171 | Putative inhibitor of apoptosis (PIAP)e | — | 2.04 | — | — | — | — | — | |
| BC001602 | CASP8- and FADD-like apoptosis regulator (CFLAR) | 2.12 | 3.33 | 2.63 | 2.62 | 2.20 | — | — | |
| AAC98530 | p53-binding protein (TOPORS) | 2.13 | 4.04 | 2.87 | 3.33 | 2.23 | 1.96 | 1.90 | |
| AAC69439 | Inhibitor of p53-induced apoptosis-alpha (NME6) | — | 2.87 | 2.13 | 2.39 | 2.00 | — | — | |
| AAH16040 | Serine-threonine kinase 17b/DAP kinase-related apoptosis-inducing protein kinase 2 (STK17B) | −2.61 | — | — | −3.75 | — | −2.32 | −3.48 | |
| P35638 | Growth arrest- and DNA damage-inducible protein GADD153 (DDIT3) | — | — | — | — | — | −2.75 | −2.87 | |
| CAA61857 | Death-associated protein 5 (EIF4G2) | — | — | — | — | — | −2.16 | — | |
| BAD92726 | Transformation/transcription domain-associated protein (TTRAP)e | — | — | — | — | — | −2.62 | −2.40 | |
| Nutrient metabolism | |||||||||
| Lipid metabolism | BAA22372 | Squalene epoxidase (SQLE) | — | 2.70 | 2.20 | 2.33 | — | — | — |
| AAH08466 | Dolichol-phosphate mannosyltransferase (DPM1) | — | 2.46 | 2.24 | 2.00 | — | — | — | |
| AAH33841 | Peroxisomal D3,D2-enoyl-coenzyme A isomerase, isoform 1 (PECI) | — | 2.98 | 2.75 | 2.80 | — | — | — | |
| BAA13965 | Acyl-coenzyme A dehydrogenase (ACADL) | 2.06 | 2.94 | 2.49 | 2.82 | 2.01 | — | — | |
| P12026 | Acyl-coenzyme A binding protein (DBI) | 2.13 | 2.85 | 2.62 | 2.78 | 2.01 | — | — | |
| AAB09475 | Apolipoprotein AI regulatory protein1 (NR2F2) | — | 2.88 | 2.50 | 2.60 | — | — | — | |
| AAA30994 | Apolipoprotein R (C4BPA) | — | 2.21 | 2.05 | 2.00 | — | — | — | |
| AAF33208 | Glycolipid transfer protein (GLTP) | — | 2.64 | 2.15 | 2.14 | — | — | — | |
| Carbohydrate metabolism | AAH73741 | Phosphoglycerate mutase 2 (PGAM2) | — | 2.50 | — | 2.17 | — | — | — |
| AAQ15274 | Pyruvate kinase, M1 isozyme (PKM1) | — | — | — | — | — | −2.27 | −2.15 | |
| AAH93020 | Isocitrate dehydrogenase 1, NADP+, cytosolic (IDH1) | 2.06 | 3.62 | 2.40 | 2.75 | — | — | 1.89 | |
| P16276 | Aconitate hydratase, mitochondrial (ACO2) | 2.04 | 2.94 | 2.16 | 2.47 | — | — | — | |
| AAA31017 | Citrate synthase, mitochondrial (CS) | — | 2.76 | 2.31 | — | — | — | — | |
| P33198 | Isocitrate dehydrogenase (NADP), mitochondrial (IDH2) | 2.03 | 2.69 | 2.58 | 2.26 | — | — | — | |
| AAC48610 | Malate dehydrogenase, cytosolic (MDH1) | — | 2.34 | 2.05 | 2.06 | — | — | — | |
| AAB94003 | Succinyl-coenzyme A synthetase (SUCLG1) | — | 2.09 | — | — | — | — | — | |
| Oxidative phosphorylation | AAD34190 | ATP synthase F0 subunit 6 (ATP6) | — | 2.93 | — | — | — | −2.72 | −2.51 |
| AAF62364 | Cytochrome b (CYTB) | — | — | — | — | — | −2.98 | −2.70 | |
| AAQ06129 | Cytochrome c oxidase subunit III (COX3) | — | — | — | — | — | −3.37 | −2.70 | |
| AAQ06019 | NADH dehydrogenase subunit 1 (ND1) | — | — | — | — | — | −2.35 | — | |
| AAR01789 | NADH dehydrogenase subunit 4 (ND4) | — | — | — | — | — | −2.93 | −2.96 | |
| Nucleoside metabolism | AAH26106 | 5′-Methylthioadenosine phosphorylase (MTAP) | — | 2.67 | — | — | — | — | — |
| AAH07511 | UMP synthase (UMPS) | 2.32 | 3.73 | 2.77 | 3.22 | 2.48 | 2.00 | 2.04 | |
| AAA57475 | Dihydropyrimidine dehydrogenase NADP+ (DPYD) | — | 3.07 | 2.34 | 2.42 | — | — | — | |
| AAH00293 | Nucleoside-diphosphate kinase 1, isoform 2 (NME1) | — | 2.17 | — | 2.02 | — | — | — | |
| AAH02476 | Nucleoside-diphosphate kinase 2, isoform 2 (NME2) | — | 2.09 | — | — | — | — | — | |
| Cell cycle | AAC39263 | DNase II (DNASE2) | 2.36 | 4.15 | 2.66 | 3.40 | 2.22 | — | 1.99 |
| AAH10022 | CDC45-related protein (CDC45L) | 2.29 | 4.06 | 2.54 | 2.98 | 2.22 | — | 1.92 | |
| AAH93642 | Septin 7/CDC10 protein homologue (SEPT7) | 2.36 | 3.43 | 2.61 | 2.90 | 2.10 | — | — | |
| AAH11616 | G1/S-specific Cyclin D3 (CCND3) | 2.11 | 3.26 | 2.31 | 2.15 | 2.05 | — | — | |
| AAH13919 | Thymidylate synthetase (TYMS) | — | 2.86 | 2.47 | 2.64 | 2.00 | — | — | |
| AAL91594 | DNA polymerase beta (POLB) | — | 2.92 | 2.26 | 2.37 | — | −2.10 | — | |
| AAH14847 | Origin recognition complex 4 (ORC4L) | — | 2.70 | 2.04 | — | — | — | — | |
| Cell structure (cytoskeleton/ membranes) | AAH15516 | Proteoglycan 1, secretory granule (PRG1) | 2.38 | 3.45 | 2.90 | 3.23 | 2.11 | 1.85 | 1.92 |
| O02697 | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform (PIK3CG) | — | 3.26 | 2.89 | 2.86 | — | — | — | |
| AAH18034 | Phosphatidylinositol-4-phosphate 5-kinase type II alpha (PIP5K2A) | 2.08 | 3.05 | 2.61 | 2.50 | 2.08 | — | — | |
| AAH36916 | Phosphatidylinositol glycan, class O, isoform 2 (PIGO) | — | 2.88 | — | 2.19 | — | — | ||
| P50127 | N-Acetyllactosaminide alpha-1,3-galactosyltransferase (GGTA1) | 2.22 | 2.85 | 2.42 | 2.35 | 2.00 | — | — | |
| AAH00590 | Actin-related protein 2/3 complex subunit 2 (ARPC2) | — | 2.33 | — | — | — | — | — | |
| P60982 | Destrin (actin-depolymerizing factor) (DSTN) | — | — | — | — | — | −3.10 | −2.86 | |
| AAH10435 | H1 histone family, member X (H1FX) | — | 2.62 | 2.26 | 2.25 | — | — | — | |
| Protein degradation | AAH66336 | 26S proteasome-associated pad1 homolog (PSMD14) | — | — | — | — | — | −2.57 | −2.18 |
| CAI18623 | Proteasome subunit beta type 8 (PSMB8) | 2.03 | 3.02 | 2.49 | 2.87 | — | — | — | |
| AAH51289 | Ubiquitin-conjugating enzyme E2I (UBE2I) | 2.11 | 3.25 | 2.72 | 2.73 | 2.15 | — | — | |
| AAH33349 | Ubiquitin-conjugating enzyme E2D 2, isoform 1 (UBE2D2) | — | 3.28 | 2.42 | 2.66 | 2.05 | — | — | |
| AAD28788 | Ubiquitin fusion-degradation 1 protein (UFD1L) | — | 2.55 | 2.37 | 2.19 | — | — | — | |
| AAL32103 | Ubiquitin ligase E3 alpha-I (UBR1) | 2.16 | 3.43 | 2.41 | 2.78 | — | — | 1.83 | |
| Protein synthesis | AAG30114 | Lysyl-tRNA synthetase (KARS) | 2.04 | 3.07 | 2.72 | 3.07 | 2.11 | 1.97 | — |
| AAD02220 | Phenylalanyl-tRNA synthetase beta chain (FARSLB) | — | 3.04 | 2.49 | 2.60 | 2.08 | — | — | |
| AAH01870 | Eukaryotic translation initiation factor 2B, subunit 4 delta (EIF2B4) | 2.20 | 3.94 | 2.74 | 3.07 | 2.24 | — | 1.91 | |
| AAH64908 | Ribosomal protein S15 (RPS15) | 2.01 | 3.08 | 2.43 | 2.65 | 2.00 | — | — | |
| Protein trafficking | AAH03382 | Sorting nexin 2 (SNX2) | −2.18 | — | — | −3.29 | — | −2.29 | −3.15 |
| AAA51551 | Alpha-2-macroglobulin precursor (A2M) | — | 3.25 | 2.32 | 2.37 | — | — | — | |
| AAK95565 | Peptide/histidine transporter (SLC15A4) | — | 2.01 | — | — | — | — | — | |
| Cell growth and proliferation | AAH96185 | Proteinase 3/myeloblastin precursor (PRTN3) | 2.16 | 3.36 | 2.41 | 2.50 | — | — | — |
| AAP35490 | Granulin (GRN) | 2.17 | 3.06 | 2.39 | 2.64 | — | — | — | |
| AAH51321 | Heparanase (HPSE) | 2.05 | 2.90 | 2.25 | 2.31 | 2.00 | — | — | |
| AAH00491 | Proliferating cell nuclear antigen (PCNA)e | — | 2.11 | — | — | — | −2.05 | — | |
| Heat shock and stress response | AAH16660 | Heat shock 70-kDa protein 8, isoform 1 (HSPA8) | — | 2.55 | 2.54 | 2.43 | — | — | — |
| AAH12807 | Heat shock 90-kDa protein 1, beta (HSPCB) | — | 2.41 | — | — | — | — | — | |
The T. gondii infection time course experiment was repeated twice, and hybridization of cDNA microarrays with fluorescently labeled cDNA probes was repeated twice per experiment. Modulated transcripts are classified according to functional groups. Only genes with ≥2-fold changes in transcription in T. gondii-infected cells versus uninfected control cells, relative to a GAPDH internal control, are shown.
From the NCBI protein database.
Values are average Cy3/Cy5 ratios from four replicate hybridization experiments (for P values determined by the t test, see the supplemental material at http://www.ag.unr.edu/ab/Downloads/sup2.xls for transcripts showing ≥2-fold changes in expression.
—, no significant change in gene expression.
Microarray data were confirmed by quantitative real-time RT-PCR.
Host cell transcription.
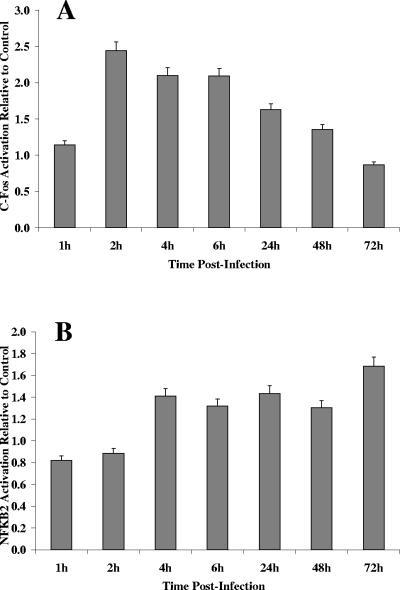
Genes encoding proteins associated directly with host cell transcription (Table 2) were among the majority of genes induced early in the infection (1 h to 4 h p.i.), including those encoding transcription initiation factor TFIID (TAFII), which coordinates the activity of more than 70 polypeptides involved in transcription initiation (28), and those encoding RNA polymerases RPA16 and POLR2D (Table 3). Signal transducer and activator of transcription 1 (STAT1), which is activated by gamma interferon (IFN-γ) to form a critical component of the interferon-induced GTPase-dependent pathway of host resistance to T. gondii (7), and STAT6, which is involved in interleukin-4 signaling (52), were among the transcription factors significantly induced by T. gondii. The proto-oncogene encoding p55 c-Fos (C-FOS), an immediate-early gene expressed transiently in the G1 phase of the cell cycle after antigen stimulation, which plays a role in cell proliferation (2, 49), was significantly induced at 2 h to 6 h p.i., as was the gene encoding nuclear factor κB (NFKB2), a transcription factor whose activation is strongly linked to the inhibition of apoptosis (26, 32, 36). QRT-PCR analysis (Table 4) confirmed the induced levels of C-FOS at 2 h and 6 h p.i. and those of NFKB2 at 4 h and 6 h p.i. In addition, enzyme-linked immunosorbent assay-based DNA-binding assays of c-Fos and NF-κB2 proteins showed increased binding activity of both proteins in T. gondii-infected PK13 cells relative to uninfected control cells for most of the infection period (Fig. 2A and 2B). Peak activation of c-Fos protein was observed 2 h p.i., with an ∼2.4-fold increase in DNA-binding activity (Fig. 2A). At a similar time point (2 h), the C-FOS gene was induced ∼2.7-fold following microarray analysis (Table 3) and ∼2.8-fold following QRT-PCR (Table 4).
TABLE 4.
Confirmation of microarray data by quantitative real-time RT-PCRa
| Accession no.b | Protein (gene) | Fold change in gene expression determined byc:
|
Time (h) p.i. | |
|---|---|---|---|---|
| Microarrays | QRT-PCR | |||
| CAB40144 | C-Fos (C-FOS) | 2.65* | 2.82** | 2 |
| 2.65* | 2.61** | 6 | ||
| AAH02844 | NF-κB p100/p49 (NFKB2) | 1.82* | 2.56** | 6 |
| NSC | NSC | 24 | ||
| P35638 | Growth arrest- and DNA damage-inducible protein (DDIT3) | −2.75* | −4.11** | 48 |
| −2.86* | −5.08** | 72 | ||
| BAD92726 | Transformation transcription domain-associated protein (TTRAP) | −2.62* | −10.86** | 48 |
| −2.40* | −3.14** | 72 | ||
| AAQ89327 | Apoptosis-related protein 3 (APR-3) | 2.04** | 2.43** | 24 |
| NSC | NSC | 48 | ||
| AAC686533 | BCL-2-interacting protein (BECN1) | 2.19* | 2.27** | 6 |
| NSC | NSC | 24 | ||
| AAS91711 | Fas-associating protein (FADD) | NSC | NSC | 1 |
| NSC | NSC | 24 | ||
| AAO72308 | Multiple-zinc-finger-Kruppel-associated box protein KS1 (ZNF382) | 2.23* | 2.84** | 2 |
| NSC | NSC | 6 | ||
| AAH00491 | Proliferating cell nuclear antigen (PCNA) | NSC | NSC | 24 |
| −2.05* | −11.77** | 48 | ||
| AAC39171 | Putative inhibitor of apoptosis (PIAP) | NSC | NSC | 6 |
| NSC | NSC | 24 | ||
| CAI23572 | Ras and Rab interactor 2 (RIN2) | 2.31* | 2.45** | 6 |
| NSC | NSC | 24 | ||
| P10114 | Ras-related protein Rap-2a (RAP2A) | 2.61* | 2.82** | 2 |
| 2.23* | 2.50** | 4 | ||
Quantitative real-time RT-PCR validation of cDNA microarray data for genes that are up- or down-regulated in T. gondii-infected versus uninfected control host cells at various times postinfection. Genes validated by RT-PCR are also marked and listed in Table 3. Total RNAs isolated from T. gondii-infected and uninfected control cells at 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h postinfection were used as templates for TaqMan-based RT-PCR that was run on an ABI 7700 instrument. Quantification of gene expression in T. gondii-infected versus uninfected control cells was done relative to the GAPDH internal control gene.
From the NCBI protein database.
*, P ≤ 0.05; **, P ≤ 0.001 (see the supplemental material). NSC, no significant change in gene expression.
FIG. 2.
Effect of T. gondii infection on c-Fos activity (A) and NF-κB2 activity (B). Activation assays were performed as described in the text, using whole-cell lysates isolated from T. gondii-infected PK13 cells and uninfected PK13 control cells at 1 h, 2 h, 4 h, 6 h, 24 h, 48 h, and 72 h postinfection. Transcription factor activation in infected cells is reported relative to that observed in uninfected control cells. Error bars indicate standard deviations.
Immune response genes.
T. gondii significantly altered the expression of several immune response genes, including interferon-stimulated genes (ISGs) as well as those encoding proinflammatory cytokine receptors, proinflammatory chemoattractant molecules, MHC antigens, complement activation proteins, scavenger receptors involved in phagocytosis, and immunoglobulins (Table 3). IFN-γ constitutes a first line of innate defense against T. gondii infection by inducing the expression of several ISGs (48). As early as 1 h p.i., several ISGs were significantly up-regulated, including those for interferon-induced transmembrane proteins 3 and 4 (IFITM3 and IFIT4), which control cell growth through transduction of antiproliferative and homotypic adhesion signals (12). Additional ISGs induced early in the infection were gamma interferon-up-regulated proteasome activator 28 alpha protein (PSME1), which is involved in assembly of the immunoproteasome required for processing of MHC class I peptides (39), and gamma interferon-inducible protein 30 (IFI30), which plays an important role in MHC class II-restricted antigen processing. We also observed significant induction of several genes encoding swine MHC class II antigens (Table 3), suggesting that T. gondii peptides were presented for antigenic processing via the MHC class II pathway. Although this pathway is thought to be the main mode of presentation of antigenic peptides from T. gondii and other intracellular parasites residing within a PV (30), as the PV might prevent parasite antigens from entering the endogenous MHC class I presentation pathway, recent studies indicate that proteins secreted by T. gondii can escape from the PV and be presented by the endogenous MHC class I pathway, thus allowing direct recognition of infected cells by CD8 T cells (21).
Apoptosis.
T. gondii inhibits host cell apoptosis as an evasion strategy that facilitates its intracellular survival and establishment of persistent infections (17, 22, 32, 36). We observed increased transcription of several antiapoptotic genes (Table 3), namely, those encoding BCL-2-interacting protein Beclin (BECN1), defender against apoptotic cell death (DAD1), putative inhibitor of apoptosis (PIAP), inhibitor of p53-induced apoptosis-alpha (NME6), and CASP8/FADD-like apoptosis regulator (CFLAR). BECN1 interacts with BCL-2, an integral outer mitochondrial membrane protein that blocks cell death by controlling mitochondrial membrane permeability and prevents translocation of proapoptotic caspases (6), while DAD1 is a mammalian cell death suppressor downstream of the BCL-2 protein (47). CFLAR, an inhibitor of tumor necrosis factor (TNF) receptor superfamily 6-mediated apoptosis (45), was significantly induced for most of the infection period, suggesting that blockade of caspase activation and activity may contribute to the inhibition of apoptosis in T. gondii-infected cells. PIAP belongs to a family of antiapoptotic genes, conserved across several species, that are key intrinsic inhibitors of the caspase cascade (50), and it is regulated by NF-κB (46), whose activation is important in T. gondii's inhibition of host cell apoptosis (32, 36). A member of the NF-κB family, NF-κB2, was also overexpressed (Table 3).
In contrast, genes encoding known proapoptotic proteins (Table 3), including DAP kinase-related apoptosis-inducing protein kinase 2 (STK17B), death-associated protein 5 (EIF4G2), growth arrest- and DNA damage-inducible protein GADD153 (DDIT3) and, transformation/transcription domain-associated protein (TTRAP), were down-regulated following exposure to T. gondii. STK17B was down-regulated as early as 1 h p.i. The gene encodes a serine/threonine protein kinase 17β that acts as a positive regulator of apoptosis (41), while EIF4G2, which is down-regulated late in the infection (48 h), modulates IFN-γ-induced apoptosis (29). DDIT3, encoding a stress-inducible transcription factor that induces cell cycle arrest and apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms (31), was unchanged early postexposure but significantly down-regulated at 48 h and 72 h p.i., as was TTRAP, whose product associates with CD40, TNF receptor-75, and TNF receptor-associated factors (TRAFs) and inhibits NF-κB activation (38). The down-regulation of DDIT3 and TTRAP at 48 h and 72 h p.i. was confirmed by QRT-PCR (Table 4).
Nutrient metabolic pathways.
Genes encoding proteins involved in various host nutrient metabolic pathways, including lipid, carbohydrate, nucleoside, protein, and amino acid pathways, were significantly induced by T. gondii (Table 3). T. gondii scavenges host cholesterol, as it is unable to synthesize sterols de novo via the melavonate pathway (8, 43). Squalene epoxidase (SQLE), which catalyzes the first oxygenation step in this pathway and is thought to be one of the rate-limiting enzymes, was significantly induced 2 h to 6 h p.i. (Table 3). The squalene precursor farnesyl pyrophosphate is also a substrate for dolichol biosynthesis. Dolichol-phosphate mannosyltransferase (DPM1), a key enzyme in dolichol metabolism, was significantly induced. The induction of mevalonate biosynthetic enzymes may be necessary to increase cellular levels of squalene for T. gondii to scavenge. Several of the ESTs spotted on the microarray encoded enzymes associated with the glycolytic pathway, of which only phosphoglycerate mutase 2 (PGAM2) was significantly induced, while pyruvate kinase (PKM1) was significantly down-regulated at 48 h and 72 h (Table 3). Six of the eight genes encoding enzymes associated with the tricarboxylic acid cycle were significantly induced (Table 3). In contrast, genes encoding proteins and enzymes associated with the oxidative phosphorylation pathway (electron transport chain) were mostly unchanged earlier in the infection but were significantly down-regulated at 48 h and 72 h p.i. (Table 3).
Differential cellular gene expression in PK13 cells compared to HFFs.
Changes in cellular gene expression induced by T. gondii infection in human foreskin fibroblasts (HFFs) have been previously reported (3). We compared expression data for 20 genes in our study to those for similar genes analyzed by Blader et al. (3) which were modulated ≥2-fold or were unchanged following T. gondii infection (Table 5). Between 2 h and 24 h p.i., genes associated with the host immune response, including CCL2, IFIT1, IFI30, and CXCR4, were significantly induced in both cell lines. Selected carbohydrate metabolism genes (ALDOA, PKM1, PKM2, and GPI) remained unchanged at 24 h p.i. in both cell lines. Despite divergent modulation of certain genes in the two cell lines (including CTSL and PCNA, which at 2 h p.i. were significantly induced in PK13 cells but unchanged in HFFs, and PGK1, GAPDH, and SQLE, which at 24 h p.i. remained unchanged in PK13 cells but were significantly up-regulated in HFFs), these findings suggest some general similarities in the host transcriptional response to T. gondii infection in PK13 cells and HFFs.
TABLE 5.
Comparison of host transcriptional responses to T. gondii in PK13 cells and HFFsa
| Accession no.b | Protein (gene) | Fold change in gene expression in:
|
Time (h) p.i. | |
|---|---|---|---|---|
| PK13 cells | HFFs | |||
| P42831 | Small inducible cytokine A2 (MCP-1) | 3.27 | 10.00 | 2 |
| AAH07091 | Interferon-inducible protein with tetratricopeptide repeats 1 (IFIT1) | 2.09 | 3.34 | 6 |
| AAH21136 | Interferon gamma-inducible protein 30 (IFI30) | 2.02 | 2.19 | 6 |
| AAZ32767 | C-X-C chemokine receptor type 4 (CXCR4) | 2.06 | 9.17 | 24 |
| AAS22336 | CASP8 and FADD-like apoptosis regulator (CFLAR) | 2.20 | 4.05 | 24 |
| Q08353 | NF-κB inhibitor alpha (NFKBIA) | 2.27 | 2.46 | 2 |
| CAC95165 | Fatty acid-binding protein 3 (FABP3) | NSC | NSC | 6 |
| AAH08466 | Dolichol-phosphate mannosyltransferase (DPM1) | NSCc | NSC | 24 |
| AAH16800 | Fructose-bisphosphate aldolase A (ALDOA) | NSC | NSC | 24 |
| S64635 | Pyruvate kinase, M1 isozyme (PKM1) | NSC | NSC | 24 |
| AAQ15274 | Pyruvate kinase, M2 isozyme (PKM2) | NSC | NSC | 24 |
| CAA82246 | Glucose-6-phosphate isomerase (GPI) | NSC | NSC | 24 |
| AAH21901 | Ras suppressor protein 1 (RSU1) | 2.77 | NSC | 2 |
| AAA58623 | Glutathione S-transferase (GSTM) | 3.42 | NSC | 2 |
| AAH12612 | Cathepsin L (CTSL) | 2.67 | NSC | 2 |
| AAH00491 | Proliferating cell nuclear antigen (PCNA) | 2.11 | NSC | 2 |
| AAT77773 | Phosphoglycerate kinase 1 (PGK1) | NSC | 4.00 | 24 |
| AAF71751 | Urokinase plasminogen activator surface receptor (PLAUR) | NSC | 4.25 | 24 |
| P00355 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | NSC | 3.70 | 24 |
| BAA22372 | Squalene epoxidase (SQLE) | NSC | 5.00 | 24 |
Porcine kidney epithelial (PK13) cells were infected with T. gondii for 72 h (this study). HFFs were infected with T. gondii for 24 h (3).
From the NCBI protein database.
NSC, no significant change in gene expression.
DISCUSSION
We used a cDNA microarray to study the transcriptional response of porcine kidney epithelial cells following exposure to T. gondii. Diverse functional classes of genes were altered by T. gondii (Table 2), suggesting that numerous cellular processes were transcriptionally modulated during the course of the infection. The response of the PK13 cells to T. gondii infection was rapid, with induction of gene expression occurring at 1 h to 4 h p.i., while down-regulation occurred later in the infection (48 h to 72 h p.i.). In addition, the number of host genes induced by T. gondii over the course of infection was significantly higher than the number down-regulated (263 versus 48), suggesting that induction of gene expression may be the epithelial cell's predominant response to the infection.
Previous reports of the induction of proinflammatory cytokine genes in pigs infected in vivo with T. gondii (9) and in human epithelial cell lines infected with T. gondii (10, 27), as well as the induction of proinflammatory immune mediators in this study (Table 3), reinforce the significance of cell-mediated immunity in the host's defense mechanism against T. gondii. The induction of several MHC class II antigens, as well as that of genes encoding components of the complement activation pathway (Table 3), suggests possible roles of these pathways in a host's immune defense against T. gondii.
The apoptotic pathways, through which T. gondii inhibits host cell apoptosis (17, 22, 32, 36), were of particular interest. Our results revealed that T. gondii induced the expression of several genes encoding antiapoptotic proteins but down-regulated proapoptotic genes (Table 3). The induced antiapoptotic genes inhibit apoptosis via three diverse pathways, including mitochondrial (DAD1 and BECN1), TNF-α-induced (PIAP, CFLAR, and NFKB2) and p53-induced (NME6) pathways, while the down-regulated proapoptotic genes are involved in TNF-α-induced (STK17B and TTRAP), Fas-induced (EIF4G2), and p38 kinase (DDIT3) apoptosis signaling pathways.
The induction of NFKB2, including activation of its corresponding protein (Fig. 2B) supports the significance of NFKB activation in T. gondii's inhibition of host cell apoptosis (32, 36) through its regulation of antiapoptotic genes in the TNF-α pathway. These genes include TRAF1, TRAF2, c-IAP1, cIAP2, IEX-1L, BCL-XL, and Bfl-1/A1 (20, 26, 53). However, mitochondrial mediation of apoptosis remains the most interesting with regard to T. gondii, given the intimate association of host cell mitochondria with the parasitophorous vacuole membrane (44). At least two known antiapoptotic genes, BECN1 and DAD1, which contribute to apoptosis inhibition via the mitochondrial pathway, were significantly induced (Table 3). Apoptogenic stimuli signaling through this pathway trigger the release of cytochrome c into the cytoplasm, which in turn triggers the oligomerization of the “apoptosome,” a complex of the cytoplasmic protein APAF1, cytochrome c, and caspase 9 (5). Within this complex, caspase 9 is activated by trans-cleavage and proceeds to activate caspase 3, leading to apoptosis. The antiapoptotic activity of T. gondii is accompanied by interference with mitochondrial cytochrome c release and subsequent down-regulation of caspase activation, as well as down-regulation of the protein level of poly-ADP-ribose polymerase (17).
T. gondii modulated host genes involved in diverse nutrient metabolic pathways (Table 3). This suggests that transcriptional alteration of the host's basic metabolism may be necessary to the parasite's intracellular growth. The induction of genes associated with the mevalonate pathway (SQLE and DPM1) supports reports of T. gondii's scavenging of host cholesterol by subverting low-density lipoprotein trafficking (8), as infected cells may respond to the loss of cellular low-density lipoprotein by up-regulating genes encoding these enzymes (3). In contrast to the induction of several genes associated with the tricarboxylic acid cycle, only one gene directly associated with glycolysis (PGAM2) was up-regulated, suggesting that T. gondii may not necessarily scavenge glucose from the host. Moreover, glycolytic enzymes are abundantly expressed in T. gondii, suggesting that parasites can independently utilize glucose as a carbon source (11). The induction of these genes may therefore be nonspecific, possibly an incidental consequence of a response required for other purposes.
The genes identified as being up- or down-regulated following exposure of PK13 cells to T. gondii were solely host derived; hence, any parasite factors involved in host-pathogen interactions were not evident from this study. We have developed cDNA libraries of PK13 cells infected with T. gondii tachyzoites and bradyzoites (M. Okomo-Adhiambo et al., unpublished data) and an intensive annotation scheme, to develop microarrays spotted with parasite-derived genes. By combining expression analysis data from host and parasite microarrays, a full repertoire of genes involved in host-pathogen interactions can be elucidated.
Supplementary Material
Acknowledgments
This research is supported by grant DOA-ARS-BRDC 26-7571 from the Biotechnology Research and Development Corporation (BRDC).
We acknowledge Craig Osborne and Joan Rowe of the Nevada Genomics Center for their contribution to microarray construction. We also thank Ben Roelofs for computer technical support and Katie Eyer for laboratory technical assistance.
Editor: J. F. Urban, Jr.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Reference deleted.
- 2.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 3.Blader, I. J., I. D. Manger, and J. C. Boothroyd. 2001. Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J. Biol. Chem. 276:24223-24231. [DOI] [PubMed] [Google Scholar]
- 4.Bliss, S. K., A. J. Marshall, Y. Zhang, and E. Y. Denkers. 1999. Human polymorphonuclear leukocytes produce IL-12, TNF-alpha, and the chemokines macrophage-inflammatory protein-1 alpha and -1 beta in response to Toxoplasma gondii antigens. J. Immunol. 16:7369-7375. [PubMed] [Google Scholar]
- 5.Cain, K., S. B. Bratton, and G. M. Cohen. 2002. The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie 84:203-214. [DOI] [PubMed] [Google Scholar]
- 6.Chandra, D., J. W. Liu, and D. G. Tang. 2002. Early mitochondrial activation and cytochrome c upregulation during apoptosis. J. Biol. Chem. 277:50842-50854. [DOI] [PubMed] [Google Scholar]
- 7.Collazo, C. M., G. S. Yap, S. Hieny, P. Caspar, C. G. Feng, G. A. Taylor, and A. Sher. 2002. The function of gamma interferon-inducible GTP-binding protein IGTP in host resistance to Toxoplasma gondii is Stat1 dependent and requires expression in both hematopoietic and nonhematopoietic cellular compartments. Infect. Immun. 70:6933-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppens, I., A. P. Sinai, and K. A. Joiner. 2000. Toxoplasma gondii exploits host low-density lipoprotein receptor-mediated endocytosis for cholesterol acquisition. J. Cell Biol. 149:167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson, H. D., E. Beshah, S. Nishi, G. Solano-Aguilar, M. Morimoto, A. Zhao, K. B. Madden, T. K. Ledbetter, J. P. Dubey, T. Shea-Donohue, J. K. Lunney, and J. F. Urban, Jr. 2005. Localized multigene expression patterns support an evolving Th1/Th2-like paradigm in response to infections with Toxoplasma gondii and Ascaris suum. Infect. Immun. 73:1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denney, C. F., L. Eckmann, and S. L. Reed. 1999. Chemokine secretion of human cells in response to Toxoplasma gondii infection. Infect. Immun. 67:1547-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton, H., C. W. Roberts, J. Alexander, K. W. Thong, and G. H. Coombs. 1996. Enzymes of energy metabolism in the bradyzoites and tachyzoites of Toxoplasma gondii. FEMS Microbiol. Lett. 137:103-108. [DOI] [PubMed] [Google Scholar]
- 12.de Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54:267-277. [DOI] [PubMed] [Google Scholar]
- 12a.Dimier, I. H., and D. T. Bout. 1998. Interferon-gamma-activated primary enterocytes inhibit Toxoplasma gondii replication: a role for intracellular iron. Immunology 94:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey, J. P., H. R. Gamble, D. Hill, C. Sreekumar, S. Romand, and P. Thuilliez. 2002. High prevalence of viable Toxoplasma gondii infection in market weight pigs from a farm in Massachusetts. J. Parasitol. 88:1234-1238. [DOI] [PubMed] [Google Scholar]
- 14.Fischereder, M., B. Schroppel, P. Wiese, M. Fink, B. Banas, S. Schmidbauer, and D. Schlondorff. 2003. Regulation of glucose transporters in human peritoneal mesothelial cells. J. Nephrol. 16:103-109. [PubMed] [Google Scholar]
- 15.Flegr, J., J. Havlicek, P. Kodym, M. Maly, and Z. Smahel. 2002. Increased risk of traffic accidents in subjects with latent toxoplasmosis: a retrospective case-control study. BMC Infect. Dis. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gail, M., U. Gross, and W. Bohne. 2001. Transcriptional profile of Toxoplasma gondii-infected human fibroblasts as revealed by gene-array hybridization. Mol. Genet. Genomics 265:905-912. [DOI] [PubMed] [Google Scholar]
- 17.Goebel, S., U. Gross, and C. G. Luder. 2001. Inhibition of host cell apoptosis by Toxoplasma gondii is accompanied by reduced activation of the caspase cascade and alterations of poly(ADP-ribose) polymerase expression. J. Cell Sci. 114:3495-3505. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Marin, J. E., A. Valere, A. Bonhomme, H. el'Btaouri, F. Antonicelli, H. Burlet, D. Aubert, I. Villena, M. Guenounou, B. Haye, and J. M. Pinon. 1998. Interferon-gamma signal transduction during parasite infection: modulation of MAP kinases in the infection of human monocyte cells (THP1) by Toxoplasma gondii. Parasite Immunol. 20:631-635. [DOI] [PubMed] [Google Scholar]
- 19.Gross, U., and F. Pohl. 1996. Influence of antimicrobial agents on replication and stage conversion of Toxoplasma gondii. Curr. Top. Microbiol. Immunol. 219:235-245. [DOI] [PubMed] [Google Scholar]
- 20.Grumont, R. J., I. J. Rourke, and S. Gerondakis. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 13:400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gubbels, M., B. Striepen, N. Shastri, M. Turkoz, and E. A. Robey. 2005. Class I major histocompatibility complex presentation of antigens that escape from the parasitophorous vacuole of Toxoplasma gondii. Infect. Immun. 73:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heussler, V. T., P. Kuenzi, and S. Rottenberg. 2001. Inhibition of apoptosis by intracellular protozoan parasites. Int. J. Parasitol. 31:1166-1176. [DOI] [PubMed] [Google Scholar]
- 23.Horgan, G. W., and J. Rouault. 2000. Introduction to randomisation tests. Biomathematics and Statistics Scotland, Aberdeen, Scotland.
- 24.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 25.Joiner, K. A., S. A. Fuhrman, H. M. Miettinen, L. H. Kasper, and I. Mellman. 1990. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science 249:641-646. [DOI] [PubMed] [Google Scholar]
- 26.Khoshnan, A., C. Tindell, I. Laux, D. Bae, B. Bennett, and A. E. Nel. 2000. The NF-κB cascade is important in Bcl-xL expression and for the anti-apoptotic effects of the CD28 receptor in primary human CD4+ lymphocytes. J. Immunol. 165:1743-1754. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. M., Y. K. Oh, Y. J. Kim, S. J. Cho, M. H. Ahn, and Y. J. Cho. 2001. Nuclear factor-kappa B plays a major role in the regulation of chemokine expression of HeLa cells in response to Toxoplasma gondii infection. Parasitol. Res. 87:758-763. [DOI] [PubMed] [Google Scholar]
- 28.Kuzuhara, T., and M. Horikoshi. 1996. Isolation and characterization of a cDNA encoding a human TFIID subunit containing a variety of putative structural motifs including direct repeats. Biol. Pharm. Bull. 19:122-126. [DOI] [PubMed] [Google Scholar]
- 29.Levy-Strumpf, N., L. P. Deiss, H. Berissi, and A. Kimchi. 1997. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol. Cell. Biol. 17:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luder, C. G., and F. Seeber. 2001. Toxoplasma gondii and MHC-restricted antigen presentation: on degradation, transport and modulation. Int. J. Parasitol. 31:1355-1369. [DOI] [PubMed] [Google Scholar]
- 30a.Luder, C. G., T. Lang, B. Beuerle, and U. Gross. 1998. Down-regulation of MHC class II molecules and inability to up-regulate class I molecules in murine macrophages after infection with Toxoplasma gondii. Clin Exp. Immunol. 112:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maytin, E. V., M. Ubeda, J. C. Lin, and J. F. Habener. 2001. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp. Cell Res. 267:193-204. [DOI] [PubMed] [Google Scholar]
- 32.Molestina, R. E., T. M. Payne, I. Coppens, and A. P. Sinai. 2003. Activation of NF-kappaB by Toxoplasma gondii correlates with increased expression of antiapoptotic genes and localization of phosphorylated IkappaB to the parasitophorous vacuole membrane. J. Cell Sci. 116:4359-4371. [DOI] [PubMed] [Google Scholar]
- 33.Nagineni, C. N., B. Detrick, and J. J. Hooks. 2000. Toxoplasma gondii infection induces gene expression and secretion of interleukin 1 (IL-1), IL-6, granulocyte-macrophage colony-stimulating factor, and intercellular adhesion molecule 1 by human retinal pigment epithelial cells. Infect. Immun. 68:407-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nash, P. B., M. B. Purner, R. P. Leon, P. Clarke, R. C. Duke, and T. J. Curiel. 1998. T. gondii-infected cells are resistant to multiple inducers of apoptosis. J. Immunol. 160:1824-1830. [PubMed] [Google Scholar]
- 35.Nogae, S., M. Michimata, T. Araki, M. Suzuki, I. Kazama, S. Ito, Y. Imai, and M. Matsubara. 2002. Detection of mRNA for alpha-3 chain of type IV collagen in the glomerular epithelium, and the effect of perfused elastase on its expression. Nephron 92:853-859. [DOI] [PubMed] [Google Scholar]
- 36.Payne, T. M., R. E. Molestina, and A. P. Sinai. 2003. Inhibition of caspase activation and a requirement for NF-kappaB function in the Toxoplasma gondii-mediated blockade of host apoptosis. J. Cell Sci. 116:4345-4358. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative Expression Software Tool (REST©) for group wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pype, S., W. Declercq, A. Ibrahimi, C. Michiels, A. G. I. Van Rietschoten, N. Dewulf, M. de Boer, P. Vandenabeele, D. Huylebroeck, and J. E. Remacle. 2000. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 275:18586-18593. [DOI] [PubMed] [Google Scholar]
- 39.Realini, C., W. Dubiel, G. Pratt, K. Ferrell, and M. Rechsteiner. 1994. Molecular cloning and expression of a gamma-interferon-inducible activator of the multicatalytic protease. J. Biol. Chem. 269:20727-20732. [PubMed] [Google Scholar]
- 40.Rink, A., E. M. Santschi, and C. W. Beattie. 2002. Normalized cDNA libraries from a porcine model of orthopedic implant associated infection. Mamm. Genome 13:198-205. [DOI] [PubMed] [Google Scholar]
- 41.Sanjo, H., T. Kawai, and S. Akira. 1998. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J. Biol. Chem. 273:29066-29071. [DOI] [PubMed] [Google Scholar]
- 42.Sayles, P. C., and L. L. Johnson. 1996. Intact immune defenses are required for mice to resist the ts-4 vaccine strain of Toxoplasma gondii. Infect. Immun. 64:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinai, A. P., and K. A. Joiner. 1997. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol. 51:415-462. [DOI] [PubMed] [Google Scholar]
- 44.Sinai, A. P., and K. A. Joiner. 2001. The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J. Cell Biol. 154:95-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivasula, S. M., M. Ahmad, S. Ottilie, F. Bullrich, S. Banks, Y. Wang, T. Fernandes-Alnemri, C. M. Croce, G. Litwack, K. J. Tomaselli, R. C. Armstrong, and E. S. Alnemri. 1997. FLAME-1, a novel FADD-like anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. J. Biol. Chem. 272:18542-18545. [DOI] [PubMed] [Google Scholar]
- 46.Stehlik, C., R. de Martin, B. R. Binder, and J. Lipp. 1998. Cytokine induced expression of porcine inhibitor of apoptosis protein (iap) family member is regulated by NF-kappa B. Biochem. Biophys. Res. Commun. 243:827-832. [DOI] [PubMed] [Google Scholar]
- 47.Sugimoto, A., R. R. Hozak, T. Nakashima, T. Nishimoto, and J. H. Rothman. 1995. Dad-1, an endogenous programmed cell death suppressor in Caenorhabditis elegans and vertebrates. EMBO J. 14:4434-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, Y., M. A. Orellana, R. D. Schreiber, and J. S. Remington. 1988. Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science 240:516-518. [DOI] [PubMed] [Google Scholar]
- 49.Ullman, K. S., J. P. Northrop, C. L. Verweij, and G. R. Crabtree. 1990. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Annu. Rev. Immunol. 8:421-452. [DOI] [PubMed] [Google Scholar]
- 50.Vaux, D. L., and S. J. Korsmeyer. 1999. Cell death in development. Cell 96:245-254. [DOI] [PubMed] [Google Scholar]
- 51.Wong, S. Y., and J. S. Remington. 1993. Biology of Toxoplasma gondii. AIDS 7:299-316. [DOI] [PubMed] [Google Scholar]
- 52.Zamorano, J., H. Y. Wang, L. M. Wang, J. H. Pierce, and A. D. Keegan. 1996. IL-4 protects cells from apoptosis via the insulin receptor substrate pathway and a second independent signaling pathway. J. Immunol. 157:4926-4934. [PubMed] [Google Scholar]
- 53.Zong, W. X., L. C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvial Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNF-α-induced apoptosis. Genes Dev. 13:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.