Abstract
The var gene family of Plasmodium falciparum encodes the variant surface antigen Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1). PfEMP1 is considered an important pathogenicity factor in P. falciparum infection because it mediates cytoadherence to host cell endothelial receptors. var genes can be grouped into three major groups, A, B, and C, and the conserved var genes, var1-4, according to sequence similarities in coding and noncoding upstream regions. Using real-time quantitative PCR in a study conducted in Tanzania, the var transcript abundances of the different var gene groups were compared among patients with severe, uncomplicated, and asymptomatic malaria. Transcripts of var group A and B genes were more abundant in patients with severe malaria than in patients with uncomplicated malaria. In general, the transcript abundances of var group A and B genes were higher for children with clinical malaria than for children with asymptomatic infections. The var group C and var1-like transcript abundances were similar between the three sample groups. A transcript abundance pattern similar to that for var group A was observed for var2csa and var3-like genes. These results suggest that substantial and systematic differences in var gene expression exist between different clinical presentations.
The particular virulence of Plasmodium falciparum is linked to the cytoadhesion properties of infected erythrocytes in deep vascular beds leading to multiple complications and symptoms (19). This process of sequestration is thought to be an immune evasion strategy to avoid splenic clearance (2, 8, 14). Infected erythrocytes also form rosettes with uninfected erythrocytes (27) or form larger groups involving platelets, called clumps (25). P. falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the var gene family and expressed on the surfaces of infected erythrocytes, mediates binding to host endothelial receptors and is an important target for protective immunity (1, 7, 9, 13, 31, 34). Each parasite possesses 50 to 60 var gene copies, and switching between surface expression of the various var gene products results in antigenic variation while maintaining or changing adhesion properties (12, 31). Immunity preventing severe malaria and death develops naturally in exposed populations. In areas of intense transmission, the main burden of malaria morbidity and mortality is among children between 6 months and 5 years of age (32). Adults are often infected asymptomatically, and severe disease is rare.
It has been shown that parasites from patients with severe malaria express a different subset of surface antigens that are more frequently recognized by sera from malaria-exposed individuals, including young children, than parasite antigens from older children with mild malaria (5, 23). It has also been shown that this subset of surface antigens is serologically conserved among different geographical regions (24), and it is therefore crucial to identify the molecular phenotype of such a subset to develop a disease-ameliorating vaccine.
Linking PfEMP1 expression to disease outcome is inherently difficult due to the extensive inter- and intragenomic variation in var genes. Previous studies predominantly relied on reverse transcription-PCR using degenerate primers, with subsequent cloning and sequencing (4, 17). While these studies have shown that the transcription of certain DBL1α domains is associated with either severe malaria or rosette formation, they have been unable to identify a clear correlation between var gene groups and disease outcomes.
Sequencing of the 3D7 genome revealed that P. falciparum parasites contain 50 to 60 var genes that can be grouped into three major groups, A, B, and C, and the single-copy intergenomic conserved var genes, var1 and var2csa, according to sequence similarities in both noncoding and coding sequences (12, 18, 22, 37). Evidence is emerging for the existence of subgroups of var group A, namely, type 3 var (18) and type 4 var (12, 15) genes, referred to here as var3 and var4, respectively. The functional relevance of this genetic structuring is indicated by the fact that CIDR domains of group B and C PfEMP1 variants bind to CD36, in contrast to CIDR domains of group A variants (26). Parasites selected for chondroitin sulfate A and human bone marrow endothelial cell binding in vitro dominantly express var2csa and var4, respectively (15, 29), also supporting the notion of functional genetic substructuring.
The genetic organization of var genes was exploited to design primers targeting the conserved regions defining var1-3 genes and group A, B, and C var genes. Using these primers in quantitative reverse transcription-PCR, the transcript abundances of var genes were measured in parasites collected from P. falciparum-infected Tanzanian children with asymptomatic malaria (AM), uncomplicated malaria (UM), and severe malaria (SM). Our data demonstrate an increase in transcript abundance for group A and B var genes in parasites causing severe malaria compared to that in parasites causing uncomplicated malaria.
MATERIALS AND METHODS
Study design and population.
The study was conducted in Ifakara, a semirural area in southern Tanzania, from June to September 2003. Ifakara is an area of moderate perennial P. falciparum transmission surrounded by areas of more intense transmission.
Of all children seeking medical treatment, 40% were admitted to the hospital due to infection with P. falciparum. Malaria is reported to account for a case fatality rate of 2.4% in this hospital (30). Samples were collected from children (aged 4 to 59 months) presenting with malaria at the hospital. Severe malaria cases were defined according to the World Health Organization criteria for severe malaria (38). Uncomplicated malaria was defined as the presence of asexual P. falciparum, an axillary temperature of >37.5°C, or symptoms of headache or myalgia but no other signs of severe malaria. Exclusion criteria were confirmed coinfection, malnutrition (mid-upper-arm circumference [MUAC] of <12 cm), or antimalarial treatment during the last 14 days. Asymptomatic patients (presence of P. falciparum, axillary temperature of <37.5°C, and no other symptoms) were age-matched as closely as possible to the patients with severe cases by convenience sampling in the same area in January 2005. P. falciparum infection was determined by Giemsa-stained blood films, and parasitemia was counted as parasites per 200 white blood cells. Ethical clearance for this study was obtained from the Ifakara Health Research and Development Centre's scientific review board and the Medical Research Coordinating Committee of the National Institute for Medical Research in Tanzania.
Blood samples.
After obtaining written informed consent from parents, 1 to 2 ml of venous blood from children was collected in EDTA tubes. Erythrocytes were separated from serum by centrifugation and washed with 40 ml phosphate-buffered saline, 5 volumes of TRIzol reagent (Invitrogen) was added to the erythrocyte pellet, and the sample was frozen at −70°C. Samples were collected from 52 patients with SM, 56 patients with UM, and 19 AM children. 3D7 parasite lines were generated as described elsewhere (33).
DNA, RNA, and cDNA.
Genomic DNAs were isolated from infected red blood cells (100 μl blood was frozen after adding 2 volumes of 6 M guanidine HCl, 50 mM Tris, pH 8.0, 20 mM EDTA) with a QIAamp blood kit (QIAGEN), and total RNAs were extracted by using TRIzol reagent (Invitrogen) twice, as recommended by the manufacturer, and treated with DNase I (Invitrogen) for 30 min at 37°C. The absence of DNA in RNA samples was confirmed by stable base fluorescence after 40 cycles of real-time PCR with seryl-tRNA synthetase primers (29). Reverse transcription was performed using Superscript II (Invitrogen) and random hexamer primers in a total volume of 40 μl according to the manufacturer's recommendations. Hereafter, two different real-time PCR methods were used to quantify var transcript abundance, namely, a SYBR green-based assay and a minor groove binder (MGB) probe-based assay.
Validation of SYBR green method and quantification of var gene transcript abundance.
Quantitative real-time PCR using Quantitect SYBR green PCR master mix (QIAGEN) was performed on a Rotorgene thermal cycler system (Corbett Research) as previously described, using the seryl-tRNA synthetase (primer pair p90) and fructose-bisphosphate aldolase (primer pair p61) genes as endogenous controls (23). Primers targeting var gene groups A, B, and C were designed based on sequence similarities in the 3D7 var repertoire (22). The majority of var genes are flanked by conserved upstream region upsA, upsB, or upsC. Primer pairs B1 and B2 target the conserved upstream region of var B genes, whereas C1 and C2 target the upstream region of var C genes. Attempts to design primer pairs targeting upsA with sufficient amplification in the SYBR green assay were unsuccessful. In strain 3D7, grouping into groups A, B, and C is also maintained in the coding sequences for DBLα and ATS. This was exploited to design primer pairs A1, targeting DBLα of group A var genes; A2 and A3, targeting exon 2 of group A var genes; and BC1 and BC2, targeting exon 2 of both B and C var genes. Additional primers were designed to target conserved regions of var1 and var2csa 5′ untranslated regions (UTRs) and var3 coding regions (pvar1utr, pvar2utr, and pvar3). Primers designed to target var4 genes in quantitative reverse transcriptase PCR amplified fragments with the expected melting temperature (Tm) for 3/20 genomic DNAs from field samples only, and this analysis was therefore left out subsequently. Seven annotated genes in 3D7 were predicted not to be targeted by any of these primer pairs. Primers are shown in Table 1, and the 3D7 genes expected to be amplified by the respective primers are listed in Table 2. The primers were validated as follows. Initially, all primers were tested on 10-fold dilutions of 3D7 genomic DNA (gDNA). All primers amplified fragments of the expected size and Tm, and sequencing of one PCR clone from each amplification reaction revealed an expected target sequence. All primers had amplification efficiencies (E) between 1.85 and 2 [E = 10(−1/slope of 10-fold-dilution gDNA standard curve)] (data not shown). Cycle threshold (CT) values for primers targeting multiple genes were compared to those obtained with the control primer p90, targeting a single-copy gene. The observed 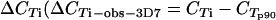 ) was compared to the expected value estimated from the number of predicted genes targeted in 3D7. A ΔCT value reduction of 1 represents a duplication of targeted gene copies, and the estimated
) was compared to the expected value estimated from the number of predicted genes targeted in 3D7. A ΔCT value reduction of 1 represents a duplication of targeted gene copies, and the estimated 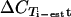 = −log(no. of targeted genes)/log(2) (Table 2). Most primer pairs amplified as expected; the primers were then tested on gDNAs from 20 field isolates, and the amplification results were compared to the p90 CT values (
= −log(no. of targeted genes)/log(2) (Table 2). Most primer pairs amplified as expected; the primers were then tested on gDNAs from 20 field isolates, and the amplification results were compared to the p90 CT values (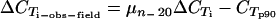 ) (Table 2). All primer pairs yielded fragments with the sizes and Tms expected from the amplifications of 3D7 gDNA, and all primers amplified with similar efficiencies (95% confidence interval for μ(n = 20)[
) (Table 2). All primer pairs yielded fragments with the sizes and Tms expected from the amplifications of 3D7 gDNA, and all primers amplified with similar efficiencies (95% confidence interval for μ(n = 20)[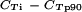 ], <0.8) from all field isolates, with no significant difference (t test) between DNAs from severe and uncomplicated malaria cases. The amplification efficiency for field isolate gDNA was similar to that for 3D7 gDNA for most primer pairs, except C1, C2, and BC2, indicating that these primers targeted fewer genes in field isolates than in 3D7 and might reflect that a larger variation in var group C gene copy numbers exists among parasites from naturally infected individuals. However, none of the primer pairs showed any significant difference in amplification efficiencies between DNAs from severe and uncomplicated malaria cases.
], <0.8) from all field isolates, with no significant difference (t test) between DNAs from severe and uncomplicated malaria cases. The amplification efficiency for field isolate gDNA was similar to that for 3D7 gDNA for most primer pairs, except C1, C2, and BC2, indicating that these primers targeted fewer genes in field isolates than in 3D7 and might reflect that a larger variation in var group C gene copy numbers exists among parasites from naturally infected individuals. However, none of the primer pairs showed any significant difference in amplification efficiencies between DNAs from severe and uncomplicated malaria cases.
TABLE 1.
SYBR green primers
| Primer pair | Sequence (5′-3′)
|
|
|---|---|---|
| Forward | Reverse | |
| A1 | TTGGGRAATBTGTTAGTTAYRGCAA | CTGCAAAACTKCGWGCAAG |
| A2 | AACCCATCTGTRRATGATATACCTATGGA | GTTCCAASGATCCATTRGATGTATTA |
| A3 | AGGTAATGTTTTAGATGATGGTAT | ACCAGAATATACATTATTTGATACATA |
| B1 | CATCCGCCATGCAAGTATAA | CGTGCACGATTTCGATTTTT |
| B2 | ATCAAGGTAATTTCATACATATGTGATA | GTCCGTGCACGATTTCGATTTT |
| C1 | CACATCGATTACATTTTAGCGTTT | TGTGGTAATATCATGTAATGG |
| C2 | GTAGCGACAACCACGRYATCATGG | CATTGTTAACATAGTCTACCATTA |
| BC1 | GACAAAACTTTCACCCAATAGA | AATGATCGGTGTAACCACTATC |
| BC2 | CATCTGTTGCAAATTTATTCCAAATAC | TCAGTAGTATCAGACATAAATGCATA |
| pvar1utr | TGGCACATCTTTGGTATAAAA | AAACCTTTATATTCCTGTAAAATTCA |
| pvar2utr | CACGACATTAACAATACATGCAGA | CATTGCATTCACAGACATTGG |
| pvar3coding | CGTAAAACATGGTGGGATGA | GGCCCATTCAGTTAACCATC |
TABLE 2.
Technical characteristics of var group primers for SYBR green real-time PCRa
| Characteristic | Value or description for indicated primer pair
|
Value or description for indicated primer pair
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | C1 | C2 | BC1 | BC2 | pvar1utr | pvar2utr | pvar3 | |
| Target region | Grp A DBL1α | Group A exon 2 | Group A exon 2 | upsB | upsB | upsC subtype | upsC subtype | Group B and C exon 2 | Group B and C exon 2 | var1 5′ UTR | var2 5′ UTR | var3 coding |
| Targeted genes in 3D7 predicted by alignments | PFD1235w, MAL7P1.1, PF08_0141, PF11_0008, PFD0020c, PF11_0521, PF13_0003, PFA0015c, MAL6P1.314, PFI1820w | PFD1235w, MAL7P1.1, PF08_0141, PF11_0008, PFD0020c, PF11_0521, PF13_0003, PFA0015c, MAL6P1.314, PFI1820w | PF11_0521, PFD1235w, MAL7P1.1, PF13_0003, PFD0020c, PF11_0008 | MAL6P1.1, PF07_0139, PF08_0142, PF10_0001, PF10_0406, PF11_0007, PF13_0001, PF13_0364, PFA0005w, PFA0765c, PFB0010w, PFB1055c, PFC0005w, PFC1120c, PFD0005w, PFD1245c, PFE0005w, PFI0005w, PFI1830c, PFL0005w, PFL0935c, PFL2665c | MAL6P1.1, PF07_0139, PF08_0142, PF10_0001, PF10_0406, PF11_0007, PF13_0001, PF13_0364, PFA0005w, PFA0765c, PFB0010w, PFB1055c, PFC0005w, PFC1120c, PFD0005w, PFD1245c, PFE0005w, PFI0005w, PFI1830c, PFL0005w, PFL0935c, PFL2665c | PFD0625c, PFL1960w, PF07_0048, PFD0630c, PF07_0049, PF07_0051, PFD0615c, PFD1015c | PFD0615c, PF07_0051, PFD0995c, PFD1000c, PF07_0049, PFD1015c, MAL6P1.252 | PF08_0142, MAL6P1.1, PFA0765c, PFC1120c, PFC0005w, PFE0005w, PF10_0406, PFB1055c, MAL7P1.56, PF08_0140, PF13_0364, PF13_0001, PFL2665c, PFI0005w, PFB0010w, PF11_0007, PF07_0139, PFD1005c, PFD1000c, PF08_0107, PFD0005w, PFL0005w, PFD1015c, PFI1830c, PF07_0050, PF10_0001 | MAL7P1.50, PF07_0048, PF08_0103, PF08_0106, PFA0005w, PFD1245c, PFL0935c, PFL1950w | PFE1640w | PFL0030c | PFA0015c, MAL6P1.314, PFI1820w |
| Coverage in 3D7 (group) | All A genes, including var3 types | All A genes, including var3 types | All A genes, excluding var3 types | All B genes | All B genes | 8/13 C | 7/13 C | 17/22 B, 4/13 C, 1/4 BA, 2/9 BC | 4/9 BC, 3/22 B, 1/13 C | var1 | var2 | var3 |
| Fragment Tm (°C) in 3D7 | 77.2 | 73.6 | 72.1 | 75.4 | 75.6 | 74.1 | 73.5 | 75.9 | 75.5 | 69.6 | 74 | 75.5 |
| Fragment length (bp) | 110-120 | 100 | 160 | 260 | 190 | 106 | 120 | 110 | 170 | 87 | 184 | 155 |
| Fragment Tm (°C)b in field isolates (SD) [no. of PCR-negative genomes/total] | 77.2 (0.4) [0/20] | 73.7 (0.2) [0/20] | 72.2 (0.4) [0/20] | 75.1 (0.4) [0/20] | 75.3 (0.4) [0/20] | 74.2 (0.4) [0/20] | 73.5 (0.3) [0/20] | 75.9 (0.3) [0/20] | 75.6 (0.3) [0/20] | 69.6 (0.2) [0/20] | 74.3 (0.4) [0/20] | 75.6 (0.2) [0/20] |
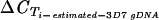 [−log(no. of targeted genes)/log(2)] [−log(no. of targeted genes)/log(2)] |
−3.3 | −3.3 | −2.6 | −4.5 | −4.5 | −3 | −2.8 | −4.7 | −3 | 0 | 0 | −1.6 |
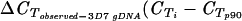 ) ) |
−2.0 | −2.5 | −1.9 | −3.7 | −3.9 | −2.2 | −0.7 | −4.5 | −1.67 | 2.8 | −0.8 | −1.8 |
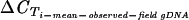 [μ( [μ(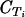 − − 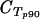 ) (95% CI)]c ) (95% CI)]c
|
−1.2 (0.4) | −1.9 (0.3) | −1.9 (0.3) | −4 (0.2) | −3.6 (0.2) | 0.3 (0.8) | 0.9 (0.5) | −4.55 (0.2) | 0.7 (0.3) | 2.6 (0.2) | 0.3 (0.3) | ND |
The following 3D7 var genes were not targeted by any primer pair: PFD0635c, PF07_0050, PFL1955w, MAL7P1.55, MAL6P1.4, MAL6P1.316, and PFL0020w (three BA and four BC genes).
Mean Tm based on real-time PCR on 20 gDNA and 108 cDNA field isolates.
95% confidence intervals based on real-time PCR on gDNAs from 20 field isolates. The design and validation of primers and probes for the MGB real-time PCR assay were described by Kaestli et al. (16).
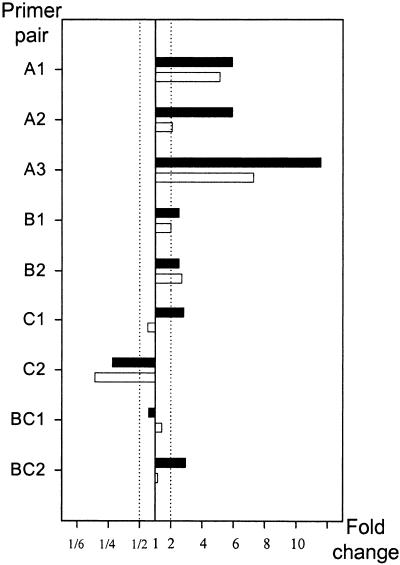
Next, primers targeting var groups were tested on cDNAs from isogenic but phenotypically distinct 3D7 parasites with known differential var transcript abundance patterns. These were nonmanipulated 3D7UM and 3D7SM selected on hyperimmune serum (15). The transcription measured by group-specific primers was compared to predicted changes calculated from absolute quantifications using gene-specific primers. There was a clear association (R = 0.904; P = 0.0008 [Pearson correlation]) between results obtained with gene-specific and group-specific primers (Fig. 1).
FIG. 1.
Differences in var transcript abundance (x-fold changes) between 3D7UM and antibody-selected 3D7SM parasites (15). Transcript abundance was measured by using primers targeting var gene groups (white) or primers targeting single var genes (black) and are summarized corresponding to var gene groups. A twofold change in var transcript abundance (dashed lines) was arbitrarily defined as the cutoff for biologically significant changes in var transcript abundance.
Assessment of Tms of fragments generated from cDNAs from collected field samples showed that all primers amplified fragments with the expected Tms in 90 to 99% of all included samples (not shown), indicating that the primers targeted var sequences conserved in the parasite isolates. The CT values for the two internal control genes showed with cDNAs from field samples that reliable quantification could be performed from the collected samples 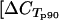 (μ, σ) = 21.2 and 2.6;
(μ, σ) = 21.2 and 2.6; 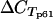 (μ, σ) = 19.0 and 2.5]. As expected, there was a negative association between parasitemia and control gene p90 CT values (representing the overall amount of cDNA) (regression coefficient[log2(parasitemia/blood sample volume):
(μ, σ) = 19.0 and 2.5]. As expected, there was a negative association between parasitemia and control gene p90 CT values (representing the overall amount of cDNA) (regression coefficient[log2(parasitemia/blood sample volume): 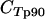 ], −0.58; P = 0.005).
], −0.58; P = 0.005).
Validation of MGB method and quantification of var transcript abundance.
Quantitative real-time PCR using MGB probes was performed using an ABI PRISM 7200 sequence detection system (Applied Biosystems) as described by Kaestli et al. (16), with few modifications. Briefly, cDNAs were synthesized from total RNA, and a primary PCR with 16 cycles over the var 5′ UTR-DBL1α target sequence was performed prior to real-time PCR for var groups A, B, and C. Primers and probes targeting upsA, -B, and -C for the MGB probe assay were described by Kaestli et al. (16) (upsA-probe, upsB-probe, and upsC-probe). The seryl-tRNA synthetase internal control gene was used for relative quantification without prior amplification. The primers and probe were designed using Primer Express software 2.0 (Applied Biosystems) and had the following sequences: primer p90Probe_for, 5′-ACCTCAGAACAACCATTATGTGCTT-3′; primer p90Probe_rev, 5′-TGTGCCCCTGCTTCTTTTCTAA-3′; and p90Probe, 5′-6-carboxyfluorescein-AGGTTACCACTCAAATACGCTGGATTCTCATCTTG- 6-carboxytetramethylrhodamine-6-carboxyfluorescein-3′.
Data analysis.
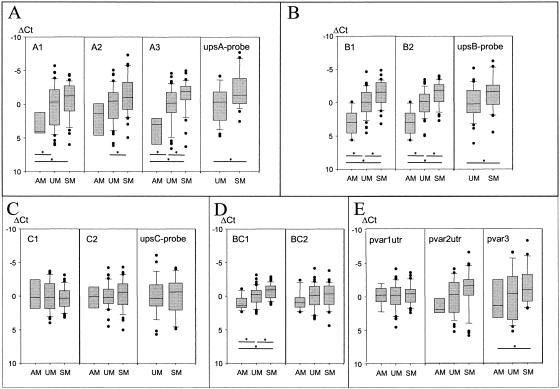
After all samples had been subjected to real-time PCR, the data set was cleaned for subsequent statistical analysis. Data points were not considered if the Tm diverged more than 1°C from the expected value or if the CT value was above 30. Transcript abundances were compared between clinical groups after normalization to internal controls (yielding ΔCT values) (Fig. 2). Based on these, x-fold changes were calculated by the ΔΔCT method (see Table 4). Comparisons between groups were made with one-way analysis of variance and Intercooled Stata 8.0 analysis software.
FIG. 2.
Transcript abundances of var gene groups in parasites from children with AM and from children suffering from UM or SM. Transcript abundances are shown relative to the average abundances in uncomplicated cases (ΔCT values). Panel A shows var group A transcript abundances measured with primers A1-3 in quantitative PCR and with upsA-probe in the MGB assay. Similarly, panels B and C show the transcript abundances of group B and C var genes, respectively. Panel D shows the transcript abundances measured with primers BC1 and BC2, targeting group B and C genes. Panel E shows transcript abundances measured with primers targeting the conserved var genes var1-3. Boxes outline 25th to 75th percentiles, with medians indicated as a line inside each box and whiskers illustrating the 5th and 95th percentiles. Horizontal lines with asterisks below the plots indicate statistically significant differences in transcript abundance between groups (one-way analysis of variance; P < 0.05 after Bonferroni correction).
TABLE 4.
Comparison of var group transcript abundances in parasites from patients with SM or UM
| Primer pair | Fold changea (by ΔΔCT method) | Confidence interval | P value (t test) |
|---|---|---|---|
| A1 | 1.6 | 0.6, 3.9 | 0.3067 |
| A2 | 2.6 | 1.2, 5.6 | 0.0175 |
| A3 | 2.6 | 1.2, 5.6 | 0.0148 |
| UpsA-probe | 4.3 | 1.6, 11.5 | 0.0051 |
| B1 | 2.7 | 1.5, 4.9 | 0.0014 |
| B2 | 2.5 | 1.4, 4.5 | 0.0020 |
| UpsB-probe | 2.8 | 1.2, 6.5 | 0.0208 |
| C1 | 0.8 | 0.4, 1.5 | 0.4779 |
| C2 | 1.1 | 0.6, 2.1 | 0.6836 |
| UpsC-probe | 1.1 | 0.4, 3.1 | 0.8494 |
| BC1 | 1.7 | 1.2, 2.4 | 0.0050 |
| BC2 | 1.1 | 0.6, 1.8 | 0.7928 |
| Pvar1utr | 1.1 | 0.5, 2.7 | 0.4375 |
| Pvar2utr | 2.0 | 0.8, 4.8 | 0.1383 |
| Pvar3coding | 2.6 | 0.7, 10.2 | 0.1611 |
Transcript abundance in SM parasites/transcript abundance in UM parasites.
RESULTS
Sample collection and clinical data.
Samples were collected from 52 children admitted to hospital with SM and 56 children with UM. var gene transcription analysis was performed on cDNAs from 42 SM and 52 UM cases. Twelve samples from 19 children with asymptomatic P. falciparum infections collected during a village survey could be analyzed for var transcript abundance. Clinical characteristics of all children from whom cDNAs were available are presented in Table 3.
TABLE 3.
Clinical and parasitological details of subjects
| Parameter | Value for indicated groupa
|
Significant relationship(s) between groups (P)b | ||
|---|---|---|---|---|
| AM (n = 12) | UM (n = 52) | SM (n = 42) | ||
| Mean age (mo) | 46 (38, 54) | 30 (26, 34) | 28 (24, 32) | AM > UM/SM (<0.001)* |
| Sex (no. of males/no. of females) | 6/6 | 22/30 | 18/24 | |
| Mean parasitemia (parasites/200 leukocytes) | 336 (144, 527) | 867 (1,067, 2,206) | 2,105 (1,883, 3,230) | AM < UM < SM (<0.001)* |
| Mean PCV (%) | 29.28 (25.60, 32.95) | 27.98 (26.32, 29.64) | 25.37 (23.34, 27.4) | AM > UM/SM (<0.001)* |
| Mean lactate (mmol/liter) | 3.82 (2.79, 4.84) | 2.67 (2.4, 2.9) | 2.9 (2.6, 2.3) | AM > UM (0.02)* |
| Mean glucose level (mmol/liter) (95% CI) | 4.97 (4.22, 5.71) | 5.1 (4.62, 5.58) | 5.24 (4.68, 5.81) | 0.491* |
| No. of days of illness | Not applicable | 2.6 (2.3, 2.9) | 2.8 (2.5, 3.1) | 0.526** |
| Mean MUAC (cm) | Not determined | 15.96 (15.02, 15.71) | 15.36 (15.56, 16.36) | 0.01** |
| No. of patients with prostration/total no. of patients | 0/12 | 40/42 | ||
| No. of patients with impaired consciousness, coma, or neurological alterations (Blantyre score of ≤3)/total no. of patients | 0/12 | 0/52 | 7/42 | |
Values in parentheses are 95% confidence intervals.
*, analysis of variance/Kruskal-Wallis test; **, t test.
Comparison of var transcript abundance profiles.
The var group A transcript abundance was lowest in AM cases, higher in children with UM, and highest in children suffering from SM (Fig. 2; Table 4). Similar findings were obtained with primers targeting the upsB upstream region, whereas data obtained with primers targeting group C var genes indicated that group C var genes were transcribed at the same level in the three groups of children.
The BC1 primer pair was predicted to predominantly amplify fragments of B var genes, whereas BC2 primers were expected to amplify a smaller subset of genes consisting of group B, BC, and C var genes (Table 2). The BC1 primer results showed that the targeted genes were transcribed at higher levels by SM parasites than by UM or AM parasites. No significant changes were observed in transcription of genes targeted by the BC2 primer pair.
The primer pairs targeting the var1 and var2csa (Table 1) gene family showed no significant difference in transcript abundance between the cohorts, although a trend of increased var2csa transcript abundance with increased severity of disease was observed. In contrast, primers targeting the var3 family showed significantly higher transcript abundance in SM than in AM samples and a trend of higher transcript abundance with increased severity of disease (Fig. 2). var3 belongs to the group A var genes, and a correlation between the transcript abundances of var3 and group A var genes would be expected. The strongest correlation of var3 transcript abundance was found with transcripts measured by A2 (Rpvar3:A2 = 0.482; P = 0.0012 [Spearman rank]).
High transcript abundances of group A and B var genes are associated with severe disease.
The association between transcript abundance and clinical presentation of malaria was tested in logistic regression models in which the dependent variable was the clinical presentation (SM or UM) and the independent variables were age, MUAC, parasitemia, sex, and transcript abundance measured by the respective primer pair. The logistic regression models were built for primer sets exhibiting statistically significant differences in transcript abundance (Table 4) and showed that young age and increased MUAC significantly increased the risk of severe disease. No significant association was found for parasitemia and sex (data not shown). According to the model, the risk of severe malaria is increased 20 to 61% with a twofold increased var group A or var group B transcript abundance (Table 5).
TABLE 5.
Logistic regression model showing the risk of severe malaria for a twofold increase in transcript abundance of specific var gene groups after correcting for the effects of age, MUAC, parasitemia, and sex
| Primer pair | Odds ratio | 95% Confidence interval | P |
|---|---|---|---|
| A2 | 1.28 | 1.06, 1.56 | 0.012 |
| A3 | 1.37 | 1.08, 1.75 | 0.010 |
| upsA | 1.35 | 1.02, 1.79 | 0.037 |
| B1 | 1.47 | 1.08, 2.00 | 0.014 |
| B2 | 1.52 | 1.14, 2.04 | 0.006 |
| BC1 | 1.61 | 1.06, 2.44 | 0.023 |
| upsB | 1.20 | 0.93, 1.56 | 0.170 |
Seven severe cases were classified as cerebral malaria due to Blantyre scores of ≤3 (data not shown). Parasites from these children showed a trend towards a larger abundance of var group A transcripts than those for all other clinical cases (for upsA-probe, P = 0.1660; for A2 primer, P = 0.0077). Primer pairs A1 and A3 showed no difference in transcript abundance between the groups (data not shown).
Associations of var transcript abundance with other clinical features.
Linear regression models showed that increased var group B transcript abundances measured by both primer pairs B1 and B2 were positively associated with parasitemia. This was also the case when corrected for age [R(parasites/200 leukocytes):ΔCT(B1/B2) = −226/−223; P = 0.045/0.019; an increase of parasitemia by 226 parasites/200 leukocytes resulted in a twofold increase in var group B transcript abundance (1 CT value decrease)]. In contrast, there was a nonsignificant trend for var group C transcript abundance, as measured by C1 or C2 primers, to be negatively correlated with parasitemia [R(parasites/200 leukocytes):ΔCT(C1/C2) = 224/207; P = 0.091/0.074). There was also no association between var group A transcript abundance and parasitemia, but var group A transcript abundance tended to decrease with age among the UM cases (Rage (months):ΔCT(A1/A2/A3) = 1.58/1.67/1.72; P = 0.050/0.058/0.081). No such trends were found with var group B or C.
DISCUSSION
Studies on the development of natural acquired immunity have suggested a genetic structuring of the PfEMP1 protein family leading to niche characteristics with regards to host receptor binding (6, 7, 9, 13). This is supported by var expression analyses of in vitro manipulated parasites (15, 31). The present study aimed to analyze the expression of var genes in naturally infected individuals presenting with different forms of malaria. Based on the sequence of the P. falciparum clone 3D7, it appears that most var genes fall into one of three main groups, A, B, and C, according to both coding and noncoding regions. Four interclonally conserved var genes (var1-4) have also been identified. This genetic structuring might reflect functional differences of the encoded PfEMP1 proteins, a notion that is supported by observations with regards to CD36 binding properties of CIDR domains (26), differences in survival rates in vivo (21), and variant surface antigen serotypes of genotypically identical parasites with diverse PfEMP1 expression profiles (15, 29). However, only a few studies have aimed to directly correlate var/PfEMP1 expression in naturally infected individuals with different presentations of malaria. One study showed that parasites from Chinese children suffering from cerebral malaria expressed larger PfEMP1s than did parasites from other malaria patients (3). This may indicate an involvement of group A genes in severe malaria, as large size is characteristic of but not unique to group A PfEMP1s (22).
PfEMP1 variants of group A were also associated with severe malaria in a study with Brazilian children. The dominant DBL1α transcripts were determined by their amplification frequencies upon reverse transcription-PCR using degenerate primers (17). However, it has been shown to be difficult to reliably reproduce results using this approach (10). Bull et al. (4) used a similar approach to study var gene expression in 12 clinical isolates from Tanzania. Although they were able to identify unique short DBLα sequence markers for var group A that correlated with the formation of rosettes, no association between var group expression and disease outcome was found. Recently, Kaestli et al. (16) analyzed parasites from Papua New Guinean children with asymptomatic, uncomplicated, or severe malaria infections by using quantitative real-time PCR to investigate changes in the proportion of var A, B, or C transcripts, using primers targeting the corresponding upstream regions. This study showed a significant increase in proportions of var group B transcripts in clinical cases, whereas var group C transcript levels were increased in asymptomatic cases. No particular involvement of var group A was reported.
The above-mentioned primers and probes were also applied in the present study, but to allow for relative comparisons to internal control genes, a new set of primers with specificity for the three main groups (A, B, and C) and var1-3 was designed. By using these primers, var transcript levels were measured in cDNAs from 106 Tanzanian children with asymptomatic, uncomplicated, or severe P. falciparum infections.
Similar to the results of the study by Kaestli et al. (16), a larger transcript abundance of var group B genes was observed with an increasing degree of disease severity. Importantly, a similar pattern of transcript abundance was found for the var genes of group A (including var3). Conversely, var genes of group C were found to be transcribed at the same level in all sample groups. The differential transcript abundance patterns determined with the BC1 and BC2 primers corresponded to the transcript abundance patterns measured by B and C group-specific primers, respectively. These conclusions were supported by data generated by two quantitative PCR methods with primers targeting both the 5′ and 3′ ends of the genes. Although we cannot exclude the possibility that the difference in PfEMP1 expression patterns in asymptomatic children and younger symptomatic children is due to the marked age difference or the severity of infection, the data suggest that var group C genes are not involved in severe childhood malaria. The fact that no var gene group was detected at higher transcription levels in AM and UM than in SM samples is puzzling. This could be explained in several ways. Firstly, the categorization of patients into AM, UM, or SM is operational, and other host and parasite factors, including other variant surface antigens, might play equally important roles in disease outcome. Secondly, since the current knowledge of the global var sequence repertoire is limited, unknown transcripts not targeted by our primers may be excluded from the analysis. Finally, there is a possibility that the present primers result in biased amplification of a subset of predicted target genes. In particular, the last case might be true for the var group C primer pairs C2 and BC2, as indicated by the relatively large differences between estimated and observed CT values for the 3D7 genomic DNA amplifications. Alternatively, group A or B var genes might be expressed in relatively larger abundance than var group C genes. This explanation would assume that not only the type of adhesion ligand, but also the amount of ligand, determines the adhesion phenotype. Future studies will test this hypothesis.
In logistic regression models, there was a statistically significant association between the risk of developing severe malaria and the transcript abundance of group A or B var genes. Thus, a twofold increase in var group A or B transcript abundance was associated with an increase of 20 to 61% in the risk of developing severe malaria. This observation does not indicate any difference between var group A and B transcript abundances in relation to disease severity. However, some indications of functional differences in group A and B var genes may be found in the linear regression models of var transcript abundance and clinical features. The association of increased parasitemia with var group B transcript abundance suggests that the parasites expressing these genes caused the severe infections in these children. In contrast, the lack of association between var group C transcription and parasitemia or clinical presentation supports a previous finding which suggested that group C var genes may be involved in establishing chronic infections (16). Similar to the case for var group B transcripts, a positive correlation between var group A transcript abundance and parasitemia would be expected in these data and in previous findings indicating the involvement of group A var genes in severe malaria (3, 15, 17). The lack of such an association might be explained if group A PfEMP1 variants confer the strongest cytoadhesion in naïve individuals only and if group A variants only dominate in first malaria infections. Since the average age of the children enrolled in this study was 29 months, most children would have undergone several, sometimes severe, infections and would have developed some immunity against PfEMP1 variants of group A. This is supported by a trend towards a lower var group A transcript abundance with increasing age in UM cases.
The observed trend of more abundant var group A transcripts in cerebral malaria cases than in all other cases might indicate that these var genes play a specific role in cerebral malaria. var2csa has been identified as the main chondroitin sulfate A binding ligand in pregnancy-associated malaria (28). The difference seen in var2csa transcript abundance between the disease groups was therefore unexpected. However, for all samples, the var2csa transcript abundance was >100-fold lower (data not shown) than that reported for placental parasites (35) or parasites selected in vitro on CSA (29). In addition, while var2csa transcription appears to be controlled by similar mechanisms to those controlling group A var genes (11), the translation of var2csa transcripts, unlike that of other var transcripts, seems to be controlled by translation of an upstream open reading frame (22; K. W. Deitsch, personal communication). Thus, var2csa is most likely not responsible for the disease outcomes of these children.
var1-like genes are unique among var genes because they are highly conserved between parasite genomes and appear to be controlled by a unique 5′ region (36), which might indicate a specialized function for var1 products similar to the var2csa gene in pregnancy-associated malaria. However, the similar abundances of var1 transcripts in all three groups gave no indications of the function of var1 products. This, together with the observed constitutive var1 transcription throughout the intraerythrocytic stages in isogenic but phenotypically different parasite lines (20, 21, 29), leaves the function of var1 molecules enigmatized.
In conclusion, the data presented here show an association between disease outcomes and the transcription of var subtypes in African areas where malaria is endemic. Of specific importance, the association between severe malaria in young children and var group A and B transcription is demonstrated and supports the notion that a vaccine based on selected PfEMP1 molecules might be feasible.
Acknowledgments
We express our thanks to Boniface Idindili and Wilbert Manyilizu for the recruitment and assessment of patients, Irene Kasiga for clinical supervision, and the staffs of the Saint Francis Designated District Hospital Ifakara and the Ifakara Health Research and Development Centre for their field assistance. We highly appreciate the cooperation of the parents and children who were willing to participate in this study. The PlasmoDB database (http://www.plasmodb.org) has been a valuable resource for this work, and the database developers and researchers who made their data available are thanked.
This study was supported by the Swiss National Science Foundation (grant number 3100 AO 104043/1) and by grants from the Danish Medical Research Council (no. 22-02-0220) and the Commission of the European Communities (grant no. QLK2-CT-1999-01293) (EUROMALVAC).
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Baruch, D. I., B. L. Pasloske, H. B. Singh, X. Bi, X. C. Ma, M. Feldman, T. F. Taraschi, and R. J. Howard. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77-87. [DOI] [PubMed] [Google Scholar]
- 2.Berendt, A. R., D. J. Ferguson, and C. I. Newbold. 1990. Sequestration in Plasmodium falciparum malaria: sticky cells and sticky problems. Parasitol. Today 6:247-254. [DOI] [PubMed] [Google Scholar]
- 3.Bian, Z., and G. Wang. 2000. Antigenic variation and cytoadherence of PfEMP1 of Plasmodium falciparum-infected erythrocyte from malaria patients. Chin. Med. J. 113:981-984. [PubMed] [Google Scholar]
- 4.Bull, P. C., M. Berriman, S. Kyes, M. A. Quail, N. Hall, M. M. Kortok, K. Marsh, and C. I. Newbold. 2005. Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS Pathog. 18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, P. C., M. Kortok, O. Kai, F. Ndungu, A. Ross, B. S. Lowe, C. I. Newbold, and K. Marsh. 2000. Plasmodium falciparum-infected erythrocytes: agglutination by diverse Kenyan plasma is associated with severe disease and young host age. J. Infect. Dis. 182:252-259. [DOI] [PubMed] [Google Scholar]
- 6.Bull, P. C., B. S. Lowe, M. Kortok, and K. Marsh. 1999. Antibody recognition of Plasmodium falciparum erythrocyte surface antigens in Kenya: evidence for rare and prevalent variants. Infect. Immun. 67:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull, P. C., B. S. Lowe, M. Kortok, C. S. Molyneux, C. I. Newbold, and K. Marsh. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat. Med. 4:358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David, P. H., M. Hommel, L. H. Miller, I. J. Udeinya, and L. D. Oligino. 1983. Parasite sequestration in Plasmodium falciparum malaria: spleen and antibody modulation of cytoadherence of infected erythrocytes. Proc. Natl. Acad. Sci. USA 80:5075-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodoo, D., T. Staalsoe, H. Giha, J. A. Kurtzhals, B. D. Akanmori, K. Koram, S. Dunyo, F. K. Nkrumah, L. Hviid, and T. G. Theander. 2001. Antibodies to variant antigens on the surfaces of infected erythrocytes are associated with protection from malaria in Ghanaian children. Infect. Immun. 69:3713-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy, M. F., J. C. Reeder, and G. V. Brown. 2003. Regulation of antigenic variation in Plasmodium falciparum: censoring freedom of expression? Trends Parasitol. 19:121-124. [DOI] [PubMed] [Google Scholar]
- 11.Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel-Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25-36. [DOI] [PubMed] [Google Scholar]
- 12.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giha, H. A., T. Staalsoe, D. Dodoo, C. Roper, G. M. Satti, D. E. Arnot, L. Hviid, and T. G. Theander. 2000. Antibodies to variable Plasmodium falciparum-infected erythrocyte surface antigens are associated with protection from novel malaria infections. Immunol. Lett. 71:117-126. [DOI] [PubMed] [Google Scholar]
- 14.Howard, R. J., and J. W. Barnwell. 1984. Roles of surface antigens on malaria-infected red blood cells in evasion of immunity. Contemp. Top. Immunobiol. 12:127-200. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, A. T., P. Magistrado, S. Sharp, L. Joergensen, T. Lavstsen, A. Chiucchiuini, A. Salanti, L. S. Vestergaard, J. P. Lusingu, R. Hermsen, R. Sauerwein, J. Christensen, M. A. Nielsen, L. Hviid, C. Sutherland, T. Staalsoe, and T. G. Theander. 2004. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 199:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaestli, K., I. A. Cockburn, A. Cortés, K. Baea, J. A. Rowe, and H.-P. Beck. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 17.Kirchgatter, K., and H. A. Portillo. 2002. Association of severe noncerebral Plasmodium falciparum malaria in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine residues. Mol. Med. 8:16-23. [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer, S. M., and J. D. Smith. 2003. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 50:1527-1538. [DOI] [PubMed] [Google Scholar]
- 19.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55:673-707. [DOI] [PubMed] [Google Scholar]
- 20.Kyes, S. A., Z. Christodoulou, A. Raza, P. Horrocks, R. Pinches, J. A. Rowe, and C. I. Newbold. 2003. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 48:1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavstsen, T., P. Magistrado, C. C. Hermsen, A. Salanti, A. T. Jensen, R. Sauerwein, L. Hviid, T. G. Theander, and T. Staalsoe. 2005. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar. J. 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavstsen, T., A. Salanti, A. T. Jensen, D. E. Arnot, and T. G. Theander. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, M. A., T. Staalsoe, J. A. Kurtzhals, B. Q. Goka, D. Dodoo, M. Alifrangis, T. G. Theander, B. D. Akanmori, and L. Hviid. 2002. Plasmodium falciparum variant surface antigen expression varies between isolates causing severe and nonsevere malaria and is modified by acquired immunity. J. Immunol. 168:3444-3450. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, M. A., L. S. Vestergaard, J. Lusingu, J. A. Kurtzhals, H. A. Giha, B. Grevstad, B. Q. Goka, M. M. Lemnge, J. B. Jensen, B. D. Akanmori, T. G. Theander, T. Staalsoe, and L. Hviid. 2004. Geographical and temporal conservation of antibody recognition of Plasmodium falciparum variant surface antigens. Infect. Immun. 72:3531-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pain, A., D. J. Ferguson, O. Kai, B. C. Urban, B. Lowe, K. Marsh, and D. J. Roberts. 2001. Platelet-mediated clumping of Plasmodium falciparum-infected erythrocytes is a common adhesive phenotype and is associated with severe malaria. Proc. Natl. Acad. Sci. USA 98:1805-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson, B. A., T. L. Welch, and J. D. Smith. 2003. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol. Microbiol. 47:1265-1278. [DOI] [PubMed] [Google Scholar]
- 27.Rowe, A., J. Obeiro, C. I. Newbold, and K. Marsh. 1995. Plasmodium falciparum rosetting is associated with malaria severity in Kenya. Infect. Immun. 63:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salanti, A., M. Dahlback, L. Turner, M. A. Nielsen, L. Barfod, P. Magistrado, A. T. Jensen, T. Lavstsen, M. F. Ofori, K. Marsh, L. Hviid, and T. G. Theander. 2004. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 200:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salanti, A., T. Staalsoe, T. Lavstsen, A. T. Jensen, M. P. Sowa, D. E. Arnot, L. Hviid, and T. G. Theander. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49:179-191. [DOI] [PubMed] [Google Scholar]
- 30.Schellenberg, D., C. Menendez, J. Aponte, C. Guinovart, H. Mshinda, M. Tanner, and P. Alonso. 2004. The changing epidemiology of malaria in Ifakara Town, southern Tanzania. Trop. Med. Int. Health 9:68-76. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snow, R. W., J. A. Omumbo, B. Lowe, C. S. Molyneux, J. O. Obiero, A. Palmer, M. W. Weber, M. Pinder, B. Nahlen, C. Obonyo, C. Newbold, S. Gupta, and K. Marsh. 1997. Relation between severe malaria morbidity in children and level of Plasmodium falciparum transmission in Africa. Lancet 349:1650-1654. [DOI] [PubMed] [Google Scholar]
- 33.Staalsoe, T., M. A. Nielsen, L. S. Vestergaard, A. T. Jensen, T. G. Theander, and L. Hviid. 2003. In vitro selection of Plasmodium falciparum 3D7 for expression of variant surface antigens associated with severe malaria in African children. Parasite Immunol. 25:421-427. [DOI] [PubMed] [Google Scholar]
- 34.Su, X. Z., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. A. Herrfeldt, D. S. Peterson, J. A. Ravetch, and T. E. Wellems. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89-100. [DOI] [PubMed] [Google Scholar]
- 35.Tuikue Ndam, N. G., A. Salanti, G. Bertin, M. Dahlback, N. Fievet, L. Turner, A. Gaye, T. Theander, and P. Deloron. 2005. High level of var2csa transcription by Plasmodium falciparum isolated from the placenta. J. Infect. Dis. 192:331-335. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez-Macias, A., P. Martinez-Cruz, M. C. Castaneda-Patlan, C. Scheidig, J. Gysin, A. Scherf, and R. Hernandez-Rivas. 2002. A distinct 5′ flanking var gene region regulates Plasmodium falciparum variant erythrocyte surface antigen expression in placental malaria. Mol. Microbiol. 45:155-167. [DOI] [PubMed] [Google Scholar]
- 37.Voss, T. S., J. K. Thompson, J. Waterkeyn, I. Felger, N. Weiss, A. F. Cowman, and H. P. Beck. 2000. Genomic distribution and functional characterisation of two distinct and conserved Plasmodium falciparum var gene 5′ flanking sequences. Mol. Biochem. Parasitol. 107:103-115. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization, Communicable Diseases Cluster. 2000. Severe falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 94(Suppl. 1):S1-S90. [PubMed] [Google Scholar]