Abstract
Visceral leishmaniasis (VL) is a form of leishmaniasis, which is caused by infection with the protozoan parasite Leishmania, and is often fatal unless it is treated. Rapid and accurate diagnosis of VL is important for effective treatment. Here we report the cloning of previously undescribed tandem repeat (TR) proteins of Leishmania infantum and an evaluation of VL patient antibody responses to the corresponding proteins. By screening an L. infantum expression library with sera from human VL patients or infected hamsters, we identified 43 genes encoding B-cell antigens. Surprisingly, 19 of the 43 genes (44%) were TR proteins, and that percentage was significantly higher than that for genes picked randomly from the database. We then expressed the TR regions of LinJ16.1750, LinJ22.1590, and LinJ33.2870 and the entire LinJ28.2310 protein. These recombinant proteins were all recognized by Sudanese VL patient sera in an enzyme-linked immunosorbent assay. Recombinant LinJ16.1750 (rLinJ16.1750) showed the best performance among these antigens in terms of both sensitivity and specificity. Serological evaluation revealed that 97% (34 of 35) of Sudanese VL patients had significantly elevated antibody levels to rLinJ16.1750. Furthermore, when eight of the patient sera which had low reactivities to rK39 were tested with the novel recombinant antigens, some of the sera showed stronger antibody responses to these antigens than to rK39. Our results suggest that TR regions from the novel L. infantum proteins identified in this study are immunodominant B-cell epitopes and may represent good candidates for serodiagnosis of VL.
The leishmaniases are a spectrum of diseases caused by protozoan parasites of the genus Leishmania. The parasites are transmitted by the bites of sandflies, and the infecting promastigotes differentiate into and replicate as amastigotes within macrophages in the vertebrate host. Clinical manifestations range from self-healing skin lesions to diffuse cutaneous and mucosal manifestations or severe visceral manifestations. Visceral leishmaniasis (VL), also known as kala-azar, is generally caused by Leishmania infantum in the Mediterranean region, by the same parasite, known as L. chagasi, in South America, and by L. donovani in India and Africa. Acute VL includes disorders of hematologic and hepatosplenic functions and is often fatal unless treated. Thus, a rapid and accurate diagnosis is necessary for effective treatment of this lethal disease.
The diagnosis of VL cannot always be made on the basis of only clinical symptoms because VL shares its clinical features with other diseases, such as malaria, typhoid fever, and tuberculosis, occurring commonly in the same areas of endemicity. Thus, the diagnosis of VL largely relies on parasitological and serological methods (16, 22, 26). The former include microscopic detection of amastigotes in aspirates of spleen and bone marrow and detection of promastigotes through cultivation of the aspirates. However, these methods are invasive, time-consuming, and not sufficiently sensitive, rendering them inefficient. Methods for serological diagnosis, including a direct agglutination test (14), immunofluorescent antibody test (3), enzyme-linked immunosorbent assay (ELISA) (17), and immunochromatographic strip test (24), are often more sensitive than invasive parasitological diagnostic methods. Also, serological diagnostic methods are more rapid and user-friendly than parasitological methods. However, the choice of antigens seems to be important because the usage of crude antigens or whole parasites often results in a low specificity in detecting disease-specific antileishmanial antibody (21). Thus, the discovery of antigens is necessary for more accurate diagnosis of VL.
Among defined leishmanial antigens reported previously, rK39 appears to be the best antigen for serodiagnosis of VL in terms of both sensitivity and specificity (9, 19). rK39 is sensitive and reliable, even in a strip format which is feasible for field use, and the rK39 strip test has 100% sensitivity in India, Nepal, and Brazil (6, 10, 11, 24, 25). In Sudan, however, the sensitivity of the strip test falls to 67%, and the negative responses on the strip test appear to correlate with lower reactivities by ELISA (28). Thus, new diagnostic antigens are needed to complement rK39 to contribute to the development of more accurate diagnosis of Sudanese VL.
Tandem repeats (TR), which consist of two or more copies of a pattern of nucleotides, have been found in many protozoan parasites, including Plasmodium. These repetitive domains are often B-cell epitopes (12, 23). In leishmaniasis, rK39, the most reliable antigen for serodiagnosis of VL, is also a TR protein (9). Other TR proteins have also been reported as candidates for serodiagnosis of leishmaniasis (8, 13, 15). However, because only a few TR antigens have been identified previously, there is still no consensus that TR proteins are more antigenic than non-TR proteins in VL. Thus, we hypothesized that TR proteins are more antigenic than non-TR proteins and are preferentially recognized by VL patient sera.
To address this hypothesis, we serologically screened an L. infantum expression library and processed the identified genes by a TR gene analysis program. As a result of the screening, we identified 43 genes encoding B-cell antigens, and surprisingly, 44% of the identified genes (19/43) encoded TR proteins. This percentage is significantly higher than that found for genes picked from the database randomly (0/108). Taken together, the results of this study provide the first evidence that leishmanial TR proteins have a greater potential to be B-cell antigens than do non-TR proteins, indicating that they are a promising resource for novel antigens for serodiagnosis of VL. In addition, the recombinant antigens used in this study were recognized by Sudanese VL patient sera which had low reactivities to rK39, suggesting their usefulness for more accurate diagnosis of Sudanese VL by complementing rK39.
MATERIALS AND METHODS
Parasites and infection of hamsters.
L. infantum, L. donovani, L. major, L. amazonensis, and Trypanosoma cruzi were used in this study. Hamsters were infected intracardially with 1 × 107 L. infantum promastigotes in stationary phase. After 8 to 12 weeks, the infected hamsters were sacrificed, and the sera were collected.
Patient sera.
Sudanese VL patient sera were collected from patients with active disease that was diagnosed clinically and proven parasitologically. Sera from patients with cutaneous leishmaniasis (CL) (Brazil), tuberculosis (United States), and malaria (Brazil) were collected from well-characterized patients, with parasitological diagnosis (cutaneous leishmaniasis and malaria) or culture-positive diagnosis (tuberculosis). Normal sera were obtained from healthy individuals in the United States.
Serological screening of L. infantum expression library.
Construction and screening of the L. infantum library were performed as reported previously (18). In brief, total DNA from L. infantum was randomly sheared by sonication to an average size of ∼2 kb, blunt ended with T4 DNA polymerase, and treated by the addition of EcoRI adaptors. The inserts were subsequently ligated into the ZAP Express vector predigested with EcoRI (Stratagene, La Jolla, CA) and were packaged using Gigapack III Gold packaging extract (Stratagene). The phage library was amplified and then screened according to the manufacturer's instructions with pooled L. infantum-infected hamster sera or the pooled Sudanese VL patient sera described above. Approximately 5 × 105 plaques were screened by using the sera at a dilution of 1:100, and immunoreactive plaques were detected with alkaline phosphatase-conjugated goat anti-hamster immunoglobulin G or goat anti-human immunoglobulin G (KPL, Gaithersburg, MD) and the substrate 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT; KPL). pBK-cytomegalovirus phagemid vectors were excised from the immunoreactive phage clones according to the manufacturer's protocol. The inserts were sequenced and analyzed using the Leishmania infantum gene database (GeneDB [www.genedb.org]; The Wellcome Trust Sanger Institute).
TR gene analysis.
The genes identified by screening were analyzed to determine whether they are TR genes. Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html), a program used to locate and display TR in DNA sequences, was used for this analysis (5). As a control for the screened genes, 108 genes which were randomly picked from the L. infantum gene database were also analyzed for the presence of TR motifs. The program calculates a score according to the nature of TR genes, such as the period size of the repeat, the number of copies aligned with the consensus pattern, and the percentage of matches between adjacent copies overall. In this study, the genes were regarded as TR genes if the scores from the Tandem Repeats Finder analysis were higher than 500, thus excluding genes containing small TR motifs, such as LinJ01.0470 and LinJ13.0810 (Table 1).
TABLE 1.
L. infantum proteins identified by serological screening
| Gene DB identifiera | Gene product | Size (kDa) | TR analysisb
|
||
|---|---|---|---|---|---|
| PS (bp) | CN | Score | |||
| LinJ01.0470 | Hypothetical protein | 151 | 3 | 11 | 66 |
| LinJ03.0120 | Hypothetical protein | 237 | 117 | 31.8 | 7,033 |
| LinJ05.0380 | Microtubule-associated protein | 165 | 114 | 28.5 | 6,336 |
| LinJ05.0590 | Hypothetical protein | 86 | |||
| LinJ08.0860 | Mitochondrial DNA polymerase beta-PAK | 154 | |||
| LinJ08.1010 | Stress-induced protein Sti1 | 62 | |||
| LinJ08.1130 | Hypothetical protein | 50 | |||
| LinJ10.1460 | Hypothetical protein | 56 | |||
| LinJ13.0810 | Hypothetical protein | 76 | 15 | 3.7 | 74 |
| LinJ14.1160 | Kinesin K39 (rK39) | 241 | 117 | 27.9 | 5,237 |
| LinJ14.1190 | Kinesin K39 | 95 | 105 | 6.2 | 1,198 |
| LinJ14.1540 | Hypothetical protein | 112 | 72 | 6.1 | 806 |
| LinJ15.0490 | Tb-292 membrane-associated protein-like protein | 164 | 105 | 31.6 | 6,027 |
| LinJ16.1540 | Kinesin | 230 | 42 | 138.5 | 10,588 |
| LinJ16.1560 | Kinesin | 88 | 42 | 2.5 | 172 |
| LinJ16.1750 | Hypothetical protein | 346 | 219 | 8.7 | 3,691 |
| LinJ17.0100 | Elongation factor 1-alpha | 49 | |||
| LinJ17.0610 | Hypothetical protein | 86 | |||
| LinJ18.0610 | Hypothetical protein | 68 | |||
| LinJ18.1150 | Hypothetical protein | 107 | |||
| LinJ21.0440 | la RNA binding protein | 37 | |||
| LinJ22.0680 | Hypothetical protein | 45 | 30 | 34.2 | 1,137 |
| LinJ22.1590 | Hypothetical protein | 234 | 84 | 29.2 | 3,993 |
| LinJ24.1570 | Basal body component | 164 | |||
| LinJ26.0980 | Dynein heavy chain | 458 | |||
| LinJ26.1200 | Hsp70.4 heat shock protein 70 | 70 | |||
| LinJ27.0290 | Nucleoporin | 159 | 27 | 2.4 | 74 |
| LinJ27.0410 | Calpain-like cysteine peptidase | 702 | 12 | 2.6 | 53 |
| LinJ27.1480 | ;hypothetical protein | 247 | |||
| LinJ28.2310 | Glycoprotein 96-92 | 61 | 315 | 2.2 | 1,398 |
| LinJ28.3170 | Hypothetical protein | 75 | 60 | 23.4 | 2,546 |
| LinJ31.0530 | Amastin | 21 | |||
| LinJ31.1430 | Hypothetical protein | 95 | |||
| LinJ32.2730 | Hypothetical protein | 173 | 150 | 10.3 | 2,916 |
| LinJ32.2780 | Membrane-associated protein | 131 | 30 | 60.9 | 3,125 |
| LinJ33.2870 | Hypothetical protein | 413 | 444 | 7 | 6,041 |
| LinJ34.0710 | Hypothetical protein | 306 | 168 | 16.3 | 4,487 |
| LinJ34.2140 | Hypothetical protein | 296 | 249 | 7.4 | 3,604 |
| LinJ35.0590 | Proteophosphoglycan Ppg4 | 536 | 45 | 246.1 | 10,667 |
| LinJ35.0600 | Proteophosphoglycan Ppg3 | 135 | 37.8 | 8,773 | |
| LinJ35.4250 | Poly(A) binding protein | 65 | 36 | 3.6 | 111 |
| LinJ36.4560 | Chaperonin Hsp60, mitochondrial precursor | 59 | |||
| LinJ36.4930 | Sterol 24-c-methyltransferase | 40 | |||
Identification numbers in GeneDB are temporary and may vary.
Data for TR analysis are from a program analysis using Tandem Repeats Finder. Data for TR genes are shown in bold. Blanks in TR analysis columns indicate that no repeats were found in those genes. PS, period size of the repeat; CN, number of copies aligned with the consensus pattern.
PCR analysis and cloning of TR genes.
The TR sequences of LinJ16.1750, LinJ22.1590, and LinJ33.2870 were amplified by PCR, using primers corresponding to both ends of a single copy of the TR (primer sequences are shown in Table 2). The entire gene sequence of LinJ28.2310 was amplified by PCR, and the primers used for this reaction are also shown in Table 2. To analyze whether these genes are conserved among Leishmania species, total DNAs of L. infantum, L. donovani, L. major, L. amazonensis, and Trypanosoma cruzi were used as templates. As a control, a T. cruzi gene (GenBank accession no. XM_810936) was amplified by PCR, using L. infantum and Trypanosoma cruzi DNAs as templates (primer sequences are given in Table 2). For cloning of the genes to produce recombinant proteins, L. infantum total DNA was used as a template for PCR.
TABLE 2.
Primers used in this study
| Gene | Primer sequence |
|---|---|
| LinJ16.1750 | 5′-CAA TTA CAT ATG TAC CCG TTC CTA CGG CTG |
| 3′-CAA TTA GGA TCC CTA GCG CGA CGC CAG CTC GTC | |
| LinJ22.1590 | 5′-CAA TTA CAT ATG GCT GAC CTG AGG GAG CAG |
| 3′-CAA TTA GGA TCC CTA CAC CTC GGC GTC CCT GTC | |
| LinJ28.2310 | 5′-CAA TTA CAT ATG AGC GCT GCA CCG TCC |
| 3′-CAA TTA GAA TTC CTA CGC AAG TCC GAG GGC | |
| LinJ33.2870 | 5′-CAA TTA CAT ATG CAG CGG CTG GTG CTC |
| 3′-CAA TTA GGA TCC CTA CGA CGT CCG CGG CAG CGC | |
| T. cruzi XM_810936 | 5′-CAA TTA CAT ATG TGC ATT GCT CTT GGC ATCGTC |
| 3′-CAA TTA AAG CTT CTG GGG CGT GAA GCG TAT GTA CTC |
Expression of recombinant proteins.
The PCR product corresponding to two copies of the LinJ16.1750 repeat (LinJ16.1750r2), three copies of the LinJ22.1590 repeat (LinJ22.1590r3), a copy of the LinJ33.2870 repeat (LinJ33.2870r1), or the entire gene of LinJ28.2310 was inserted into the NdeI/BamHI or NdeI/EcoRI sites of pET28 vector. The sequences of the inserts were then analyzed. These pET28 vectors were transformed into Escherichia coli Rosetta for expression of the recombinant proteins. Expression of the recombinant proteins was induced by cultivation with 1 M isopropyl-β-d-thiogalactopyranoside. The recombinant proteins were then purified as six-His-tagged proteins by using Ni-nitrilotriacetic acid agarose (QIAGEN Inc., Valencia, CA). The purity of the proteins was assessed by Coomassie blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
ELISA.
rLinJ16.1750r2, rLinJ22.1590r3, rLinJ28.2310r1, rLinJ33.2870, rK39, and L. infantum promastigote soluble lysate antigen (LiSLA) were diluted in ELISA coating buffer, and 96-well plates were coated with rLinJ16.1750r2, rLinJ22.1590r3, rLinJ28.2310, rLinJ33.2870r1 (200 ng [each]), rK39 (50 ng), or LiSLA (1 μg), followed by blocking with phosphate-buffered saline containing 0.05% Tween 20 and 1% bovine serum albumin. The plates were then incubated with patient sera (diluted 1:100) as well as healthy controls and then with horseradish peroxidase-conjugated protein G (Zymed Laboratories, South San Francisco, CA). The plates were developed with tetramethylbenzidine peroxidase substrate (KPL) and read by a microplate reader at a 450-nm wavelength.
RESULTS
High prevalence of TR genes in serologically screened genes.
Pooled sera from L. infantum-infected hamsters or Sudanese VL patients were used to screen an L. infantum expression library. A half-million plaques covering approximately eight L. infantum genomic equivalents were screened. pBK-CMV phagemids were excised from positive clones, and the inserts were sequenced and analyzed using GeneDB. A total of 43 genes were identified as genes encoding B-cell antigens (Table 1). Some of the genes encoded previously identified antigens, such as HSP70 and K39 (9, 20), but most of them encoded previously unidentified antigens.
The genes identified by screening were then analyzed by Tandem Repeats Finder. In this study, a gene was regarded as a TR gene if the score from the analysis was 500 or higher, thereby excluding genes containing small TR domains. Of the 43 genes identified by the serological screening, 19 genes were identified as TR genes (Table 1). For example, LinJ16.1750 contained 8.7 copies of a 219-bp sequence, and LinJ28.3170 contained 23.4 copies of a 60-bp sequence. With the exception of LinJ14.1160 (known as rK39), these genes were newly identified as genes encoding B-cell antigens. One hundred eight genes which were picked randomly from the database (three genes from each chromosome) were also analyzed with Tandem Repeats Finder to compare the prevalence of TR genes with that among the genes identified by screening. In contrast to the screened genes, no TR genes were found in the 108 randomly picked genes.
PCR analyses of TR genes.
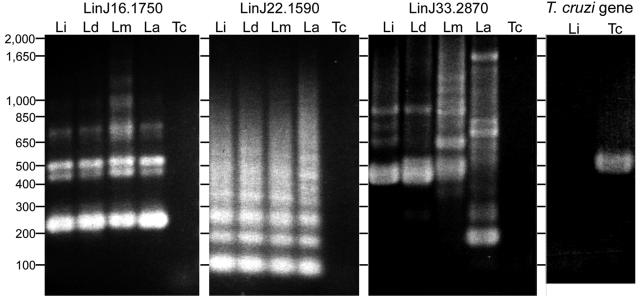
PCR amplifications were performed with total DNAs from Leishmania species and T. cruzi to analyze whether or not TR genes were conserved among these parasites. Using sets of primers specific for the TR domains of LinJ16.1750 and LinJ22.1590, the PCR products showed ladder bands corresponding to one or multiple copies of the repeats (Fig. 1). These genes were conserved among Leishmania species, as similar band patterns were observed among all four Leishmania species tested. When primers for the TR domains of LinJ33.2870 were used, the PCR products also showed bands whose sizes corresponded to one or two copies of the repeat in three Leishmania species but not in L. amazonensis (Fig. 1). When primers for the whole gene of LinJ33.2870 were used, a 1.5-kb band was found for L. infantum and L. donovani, and bands of 2.1 kb and 1.8 kb were found for L. major and L. amazonensis, respectively. In all cases, no bands were found for PCRs using T. cruzi DNA. In contrast, a single band with an expected size was found for T. cruzi, but not L. infantum, when primers for a T. cruzi gene were used for PCR. Thus, while the TR genes are not conserved between Leishmania and T. cruzi, they are well conserved between Leishmania spp.
FIG. 1.
PCR analysis of TR genes. PCRs were performed with primer sets specific for the TR regions of LinJ16.1750, LinJ22.1590, and LinJ33.2870 and for a T. cruzi gene, using total DNAs of L. infantum (Li), L. donovani (Ld), L. major (Lm), L. amazonensis (La), and T. cruzi (Tc) as templates. Sizes are shown in base pairs.
Recognition of recombinant TR proteins by VL patient sera.
To formally test the identified TR proteins as potential diagnostic candidates, LinJ16.1750r2, LinJ22.1590r3, LinJ28.2310, and LinJ33.2870r1 were expressed as recombinant proteins (Fig. 2).
FIG. 2.
Expression and purification of L. infantum recombinant proteins. The images show Coomassie blue-stained sodium dodecyl sulfate-4 to 20% polyacrylamide gradient gels containing uninduced E. coli lysates (lanes 1), induced lysates (lanes 2), and purified proteins (lanes 3). Sizes are shown in kDa. The apparent molecular masses of the proteins were comparable to their predicted molecular masses.
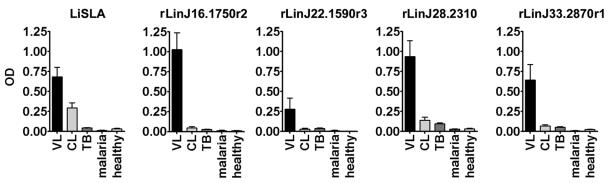
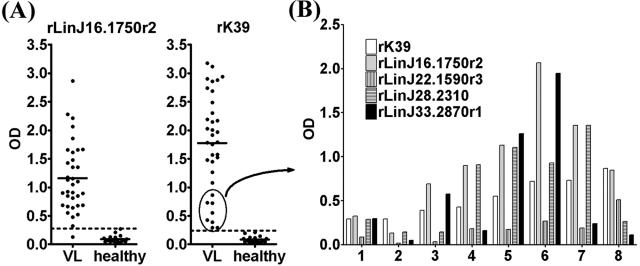
We then examined the prevalence of antibodies to the recombinant proteins in a preliminary screen of 10 Sudanese VL patient sera by ELISA. The VL patient sera showed significantly stronger reactivities to all of the recombinant proteins than did sera from non-VL groups, i.e., tuberculosis, malaria, and cutaneous leishmaniasis patients or healthy individuals, despite the finding that cutaneous leishmaniasis patients showed antibody responses to LiSLA (Fig. 3). Among the tested proteins, rLinJ16.1750r2 was the best antigen, as it was recognized strongly by VL patient sera with a high degree of specificity. We then tested rLinJ16.1750r2 with additional Sudanese VL patient sera. Among 35 sera tested, 34 showed antibody responses to rLinJ16.1750r2 (the cutoff value was the mean + 3 standard deviations [SD] of healthy control values) (Fig. 4A). The sensitivity using rLinJ16.1750r2 was 97%, which is comparable to that of rK39 (35/35 [100%]) (Fig. 4A). Mean optical densities (ODs) for rLinJ16.1750r2 and rK39 were 1.159 and 1.771, respectively.
FIG. 3.
ELISA evaluation of patient seroreactivities to L. infantum recombinant proteins and LiSLA. Sera from patients with visceral leishmaniasis (VL, n = 10), cutaneous leishmaniasis (CL, n = 10), tuberculosis (TB, n = 10), and malaria (n = 6) and sera from healthy controls in the United States (n = 10) were used. The mean and standard error of the mean of the OD values for each group are shown.
FIG. 4.
Reactivities of human VL patient sera to TR proteins. (A) Sera from VL patients (n = 35) and healthy controls (n = 20) were tested for reactivity to rLinJ16.1750r2 or rK39 by ELISA. The mean for each group is shown as a solid line. Dotted lines represent the cutoff value, calculated as the mean plus 3 SD of the OD values for healthy controls. (B) Recognition of recombinant proteins by sera from eight VL patients which showed low reactivities to rK39 (ODs of <1.0 [circled in panel A]).
Although these 35 sera were 100% positive for a response to rK39, 8 sera showed low reactivities to rK39 (OD values of <1.0) (Fig. 4A). These eight samples were tested for reactivity to rLinJ16.1750r2, rLinJ22.1590r3, rLinJ28.2310, and rLinJ33.2870r1 to examine whether or not these antigens could complement rK39 for more sensitive antibody detection. Five of the eight sera showed better responses to the new antigens than to rK39 (no. 3 to 7 in Fig. 4B). Five patients (no. 3 to 7) showed better responses to rLinJ22.1590r3, four (no. 4 to 7) showed better responses to rLinJ28.2310, and three (no. 3, 5, and 6) showed better responses to rLinJ33.2870r1. rLinJ22.1590r3 was recognized weakly by all eight sera compared to the rK39 responses.
DISCUSSION
In this study, we identified L. infantum proteins for serodiagnosis of VL and showed a high prevalence of TR genes in the screened antigens. To date, there have been only a few TR proteins reported as antigens for serodiagnosis of VL. Because those antigens were reported individually, there was still not a consensus that TR proteins are more antigenic than non-TR proteins in VL. In contrast, our approach in this study was directed toward elucidating the immunodominance of TR proteins, using a combination of mass screening and a program analysis. We chose L. infantum as a source of total DNA for library construction because L. infantum is the only species of the L. donovani complex for which genome data are available from the Leishmania genome projects through Gene DB and because this organism, considered to be identical to L. chagasi, is the most widely occurring of the complex. By serological screening of an L. infantum expression library, we identified 43 genes encoding B-cell antigens. It is intriguing that these genes encode many previously unidentified antigens as well as HSP70 and K39 (9, 23). But it is more intriguing that 44% of the screened genes encode TR proteins and that this percentage is significantly higher than that for genes picked randomly from the database. These findings suggest that TR proteins are more potent as immunodominant B-cell antigens recognized by VL patient sera than are non-TR proteins.
The immunodominance of TR antigens may be derived from their multiple identical epitopes. Also, the accessibility of TR domains on the surface of the protein may also be important for detection by antibodies (7). Most of the TR antigens identified in this study are strongly acidic or basic and rich in hydrophilic amino acids. In addition, the repetitive regions generally comprise large portions of the entire proteins. These features of TR proteins may contribute to their strong B-cell antigenicities. At the same time, though, it is still possible that the high prevalence of TR domains in the screened genes is because we chose a randomly sheared L. infantum total DNA library for screening instead of a cDNA library. We used a total DNA library because a cDNA library has a bias toward expression of each gene at the mRNA level. Although a total DNA library should not have such a bias, the existence of multiple copies of TR sequences results in an increased frequency of identical serological epitopes in the library compared to non-TR genes. Thus, it is suggested that large TR genes have a greater tendency to be detected by serological screening of a genomic DNA library than do small, non-TR genes. This may explain why TR antigens have been identified by the use of genomic libraries in this and previous studies (8, 9, 13) and, in contrast, why other groups have picked up highly expressed proteins, such as heat shock proteins, by serological screening using cDNA libraries (1, 2, 20, 27).
Although such biases may influence the high prevalence of TR genes in the screened genes, the results of the screening indicate that repetitive regions of the TR antigens include B-cell epitopes. We found that inserts of the phage clones picked up as TR antigens encoded repetitive regions of the antigens. For example, LinJ22.1590 was identified six times during the screening. Although those clones had different patterns of insertion, all of the inserts contained the repetitive regions, and no clones encoding only the nonrepetitive regions were found. We used randomly sheared L. infantum total DNA to construct the expression library. Thus, there should be phage clones containing not only repetitive regions but also nonrepetitive regions of the TR antigens in the library. Taken together, these findings indicate stronger B-cell antigenicities of the repetitive regions of the TR proteins than of nonrepetitive regions.
The high prevalence of antibodies to the repeat domains of the TR antigens in VL patients was confirmed by ELISAs using the recombinant proteins. We expressed recombinant antigens containing only partial repetitive regions of LinJ16.1750, LinJ22.1590, and LinJ33.2870 and tested these by ELISA, using Sudanese VL patient sera. These recombinants were recognized strongly and specifically by VL patient sera, and no or very low reactivities were observed for malaria and tuberculosis patients, who share similar clinical features with VL patients. Even when tested with more patient sera, 97% of Sudanese VL patient sera (34/35) were significantly reactive to rLinJ16.1750r2. These findings suggest that recombinant proteins that are not entire proteins but only small repeat portions are sufficient as serodiagnostic antigens for recognition by patient sera. This finding has several advantages for antigen expression. Leishmania TR proteins are relatively large proteins compared with nonrepetitive proteins (the average size of the 19 TR antigens in Table 1 is 223 kDa). It is difficult to express and purify the recombinant proteins from TR genes. In contrast, one copy of the repeat protein is often a small polypeptide stretch. Therefore, such partial repetitive regions are suitable for molecular biology manipulations, such as cloning, expression, and purification of the recombinant proteins, and even for making fusion proteins of several candidate antigens. These findings also suggest that a bioinformatic approach may lead to the discovery of new B-cell antigens. Sequence data for 8,173 L. infantum genes are now available in GeneDB. It might be useful to seek TR genes from the database in combination with gene analysis programs such as Tandem Repeats Finder.
PCR analyses demonstrated that the repeat domains of the TR genes are conserved between L. infantum and L. donovani, suggesting that the encoded proteins can be used to diagnose VL in different areas of endemicity. These genes seem to be conserved among other Leishmania species, too, although CL patients did not show strong antibody responses to the antigens. Taken together, the results show that the VL-specific responses to the recombinant TR proteins may be due to different immune responses during VL and CL (16) rather than to L. donovani complex-specific expression of the proteins.
We also showed that these recombinant TR antigens are capable of compensating for low reactivities of Sudanese VL patient sera to rK39. Among defined leishmanial antigens reported previously, K39 seems to be the best antigen for serodiagnosis of VL in terms of both sensitivity and specificity (19). rK39 is sensitive and reliable, even in a strip format adapted for field use, and the strip test has at or near 100% sensitivity in India, Nepal, and Brazil (6, 10, 11, 24, 25). In Sudan, however, the sensitivity of the strip test is lower, and the negative responses on the strip test appear to correlate with low reactivities by ELISA (28). With ELISA, there is a clear-cut difference in reactivities to rK39 between Indian VL patients and healthy controls (4), whereas Sudanese VL patients show a wide range of reactivities. Thus, additional antigens reactive to Sudanese VL patient sera could assist in more accurate diagnosis of VL in both ELISA and strip test formats.
In this study, we demonstrated that novel TR antigens were recognized by patient sera, even those possessing weak reactivities to rK39. Our data suggest that these antigens in combination with rK39 may lead to the production of more accurate diagnostic tests for VL in Sudan and other regions of Africa. We are now evaluating antigen combinations to achieve maximal sensitivity and specificity for serodiagnosis of African VL. Indeed, a mixture of rLinJ16.1750r2 and rK39 seems to be better than rK39 alone. We found that using a mixture of these antigens in ELISA increased the reactivities of Sudanese VL patient sera compared to those using rK39 alone without increasing OD values for healthy controls (data not shown).
In conclusion, we have identified a number of L. infantum TR genes as B-cell antigens. In addition, we have demonstrated that repetitive domains of the TR proteins are strong serological antigens and that these proteins are useful for serodiagnosis of VL. We are currently using the bioinformatic approach to find more TR genes. This effort should lead to the discovery of new antigens for more accurate serodiagnosis of VL.
Acknowledgments
We thank Malcolm Duthie and Stephen Reece for critical comments and helpful suggestions and Jennifer Martino for help with manuscript preparation.
This work was supported by National Institutes of Health grant AI25038 and a grant from the Bill and Melinda Gates Foundation.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Angel, S. O., J. M. Requena, M. Soto, D. Criado, and C. Alonso. 1996. During canine leishmaniasis a protein belonging to the 83-kDa heat-shock protein family elicits a strong humoral response. Acta Trop. 62:45-56. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., P. C. Melby, and S. Sehgal. 1995. Lack of serological specificity of recombinant heat shock protein of Leishmania donovani. Immunol. Cell Biol. 73:446-451. [DOI] [PubMed] [Google Scholar]
- 3.Badaro, R., S. G. Reed, and E. M. Carvalho. 1983. Immunofluorescent antibody test in American visceral leishmaniasis: sensitivity and specificity of different morphological forms of two Leishmania species. Am. J. Trop. Med. Hyg. 32:480-484. [DOI] [PubMed] [Google Scholar]
- 4.Badaro, R., D. Benson, M. C. Eulalio, M. Freire, S. Cunha, E. M. Netto, D. Pedral-Sampaio, C. Madureira, J. M. Burns, R. L. Houghton, J. R. David, and S. G. Reed. 1996. rK39: a cloned antigen of Leishmania chagasi that predicts active visceral leishmaniasis. J. Infect. Dis. 173:758-761. [DOI] [PubMed] [Google Scholar]
- 5.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bern, C., S. N. Jha, A. B. Joshi, G. D. Thakur, and M. B. Bista. 2000. Use of the recombinant K39 dipstick test and the direct agglutination test in a setting endemic for visceral leishmaniasis in Nepal. Am. J. Trop. Med. Hyg. 63:153-157. [DOI] [PubMed] [Google Scholar]
- 7.Berzofsky, J. A. 1985. Intrinsic and extrinsic factors in protein antigenic structure. Science 229:932-940. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia, A., N. S. Daifalla, S. Jen, R. Badaro, S. G. Reed, and Y. A. Skeiky. 1999. Cloning, characterization and serological evaluation of K9 and K26: two related hydrophilic antigens of Leishmania chagasi. Mol. Biochem. Parasitol. 102:249-261. [DOI] [PubMed] [Google Scholar]
- 9.Burns, J. M., Jr., W. G. Shreffler, D. R. Benson, H. W. Ghalib, R. Badaro, and S. G. Reed. 1993. Molecular characterization of a kinesin-related antigen of Leishmania chagasi that detects specific antibody in African and American visceral leishmaniasis. Proc. Natl. Acad. Sci. USA 90:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho, S. F., E. M. Lemos, R. Corey, and R. Dietze. 2003. Performance of recombinant K39 antigen in the diagnosis of Brazilian visceral leishmaniasis. Am. J. Trop. Med. Hyg. 68:321-324. [PubMed] [Google Scholar]
- 11.Chappuis, F., S. Rijal, R. Singh, P. Acharya, B. M. Karki, M. L. Das, P. A. Bovier, P. Desjeux, D. Le Ray, S. Koirala, and L. Loutan. 2003. Prospective evaluation and comparison of the direct agglutination test and an rK39-antigen-based dipstick test for the diagnosis of suspected kala-azar in Nepal. Trop. Med. Int. Health 8:277-285. [DOI] [PubMed] [Google Scholar]
- 12.Cowman, A. F., R. B. Saint, R. L. Coppel, G. V. Brown, R. F. Anders, and D. J. Kemp. 1985. Conserved sequences flank variable tandem repeats in two S-antigen genes of Plasmodium falciparum. Cell 40:775-783. [DOI] [PubMed] [Google Scholar]
- 13.Dillon, D. C., C. H. Day, J. A. Whittle, A. J. Magill, and S. G. Reed. 1995. Characterization of a Leishmania tropica antigen that detects immune responses in Desert Storm viscerotropic leishmaniasis patients. Proc. Natl. Acad. Sci. USA 92:7981-7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Harith, A., A. H. Kolk, J. Leeuwenburg, R. Muigai, E. Huigen, T. Jelsma, and P. A. Kager. 1988. Improvement of a direct agglutination test for field studies of visceral leishmaniasis. J. Clin. Microbiol. 26:1321-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghedin, E., W. W. Zhang, H. Charest, S. Sundar, R. T. Kenney, and G. Matlashewski. 1997. Antibody response against a Leishmania donovani amastigote-stage-specific protein in patients with visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 4:530-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 354:1191-1199. [DOI] [PubMed] [Google Scholar]
- 17.Ho, M., J. Leeuwenburg, G. Mbugua, A. Wamachi, and A. Voller. 1983. An enzyme-linked immunosorbent assay (ELISA) for field diagnosis of visceral leishmaniasis. Am. J. Trop. Med. Hyg. 32:943-946. [DOI] [PubMed] [Google Scholar]
- 18.Lodes, M. J., D. C. Dillon, R. L. Houghton, and Y. A. Skeiky. 2004. Expression cloning. Methods Mol. Med. 94:91-106. [DOI] [PubMed] [Google Scholar]
- 19.Maalej, A. A., M. Chenik, H. Louzir, A. Ben Salah, C. Bahloul, F. Amri, and K. Dellagi. 2003. Comparative evaluation of ELISAs based on ten recombinant or purified Leishmania antigens for the serodiagnosis of Mediterranean visceral leishmaniasis. Am. J. Trop. Med. Hyg. 68:312-320. [PubMed] [Google Scholar]
- 20.MacFarlane, J., M. L. Blaxter, R. P. Bishop, M. A. Miles, and J. M. Kelly. 1990. Identification and characterisation of a Leishmania donovani antigen belonging to the 70-kDa heat-shock protein family. Eur. J. Biochem. 190:377-384. [DOI] [PubMed] [Google Scholar]
- 21.Reed, S. G., R. Badaro, and R. M. Lloyd. 1987. Identification of specific and cross-reactive antigens of Leishmania donovani chagasi by human infection sera. J. Immunol. 138:1596-1601. [PubMed] [Google Scholar]
- 22.Singh, S., and R. Sivakumar. 2003. Recent advances in the diagnosis of leishmaniasis. J. Postgrad. Med. 49:55-60. [DOI] [PubMed] [Google Scholar]
- 23.Stahl, H.-D., P. E. Crewther, R. F. Anders, G. V. Brown, R. L. Coppel, A. E. Bianco, G. F. Mitchell, and D. J. Kemp. 1985. Interspersed blocks of repetitive and charged amino acids in a dominant immunogen of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 82:543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundar, S., S. G. Reed, V. P. Singh, P. C. Kumar, and H. W. Murray. 1998. Rapid accurate field diagnosis of Indian visceral leishmaniasis. Lancet 351:563-565. [DOI] [PubMed] [Google Scholar]
- 25.Sundar, S., K. Pai, M. Sahu, V. Kumar, and H. W. Murray. 2002. Immunochromatographic strip-test detection of anti-K39 antibody in Indian visceral leishmaniasis. Ann. Trop. Med. Parasitol. 96:19-23. [DOI] [PubMed] [Google Scholar]
- 26.Sundar, S., and M. Rai. 2002. Laboratory diagnosis of visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 9:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, M. C., E. Martinez-Carretero, E. Carmelo, A. C. Gonzalez, and B. Valladares. 2004. Molecular characterization of the Leishmania braziliensis L6 ribosomal protein. J. Parasitol. 90:908-913. [DOI] [PubMed] [Google Scholar]
- 28.Zijlstra, E. E., Y. Nur, P. Desjeux, E. A. Khalil, A. M. El-Hassan, and J. Groen. 2001. Diagnosing visceral leishmaniasis with the recombinant K39 strip test: experience from the Sudan. Trop. Med. Int. Health 6:108-113. [DOI] [PubMed] [Google Scholar]