Abstract
Cells of an attenuated live vaccine strain (LVS) of F. tularensis grown under iron-restricted conditions were found to contain increased quantities of several proteins relative to cells of this same strain grown under iron-replete conditions. Mass spectrometric analysis identified two of these proteins as IglC and PdpB, both of which are encoded by genes located in a previously identified pathogenicity island in F. tularensis LVS. Regions with homology to the consensus Fur box sequence were located immediately in front of the iglC and pdpB open reading frames (ORFs), and in silico analysis of the F. tularensis Schu4 genome detected a number of predicted 5′ untranslated regions that contained putative Fur boxes. The putative Fur box preceding Francisella iron-regulated gene A (figA) had the highest degree of identity with the consensus Fur box sequence. DNA microarray analysis showed that nearly 80 of the genes in the F. tularensis LVS genome were up- or down-regulated at least twofold under iron-restricted growth conditions. When tested for possible siderophore production by means of the Chrome Azurol S assay, a wild-type F. novicida strain produced a large reaction zone whereas its figA mutant produced very little reactivity in this assay. In addition, a cross-feeding experiment demonstrated that this siderophore-like activity produced by the wild-type F. novicida strain could enhance the ability of the F. novicida figA mutant to grow under iron-restricted conditions. This study provides the first identification of iron-regulated genes in F. tularensis LVS and evidence for the production of a siderophore-like molecule by F. novicida.
Francisella tularensis is a gram-negative coccobacillus that causes tularemia in both humans and animals (28, 33, 40). F. tularensis subspecies tularensis (type A) and F. tularensis subspecies holarctica (type B) are highly infectious for humans. Inoculation or inhalation of as few as 10 CFU of F. tularensis type A (i.e., F. tularensis Schu4) may cause disease in humans (37, 38). Because of its extremely high infectivity, this bacterium is listed as a class A biological warfare agent (7, 28). Type B strains of F. tularensis are less virulent than type A strains for higher mammals and typically cause nonfatal disease in humans. An attenuated live vaccine strain (LVS) of F. tularensis which was derived from a virulent type B strain by passage in vitro (37, 38) has been widely used in studies of F. tularensis. Francisella novicida, which is closely related to F. tularensis (12, 17), is highly virulent in mice but has relatively low virulence for humans (28). F. novicida is more easily manipulated genetically and has less fastidious growth requirements than F. tularensis strains (13). Recently, F. novicida was used to identify and characterize a pathogenicity island involved in intracellular growth (27). In addition, the MglA protein was shown to be involved in the regulation of bacterial gene products necessary for intracellular growth of this pathogen (21, 36). With these few exceptions, however, little is known about the F. tularensis gene products involved in virulence.
For several decades, investigators have been aware of the importance of iron in microbial physiology (4). In this context, the human body provides an environment of extreme iron limitation from the standpoint of the bacterium because free iron is not normally present in the bloodstream or tissues (42). As a consequence, successful commensal organisms and pathogens express high-affinity iron uptake systems by which they can obtain iron in vivo. Bacterial genes that encode factors involved in the iron acquisition process in vivo are considered virulence factors and have been subjected to intensive study. However, compared with the well-documented iron acquisition systems of other facultative intracellular pathogens, including Legionella pneumophila (8), Yersinia pestis (32), and Mycobacterium species (34), virtually nothing is known about how F. tularensis acquires iron from its mammalian hosts.
In a study of the effect of stress conditions on the virulence of F. tularensis LVS, it was reported that a few cell envelope proteins appeared to be up-regulated when iron was limiting (2). However, these proteins were not identified and there have been no subsequent reports on this subject to date. In the present study, we used two different methods to identify F. tularensis LVS genes whose expression was up-regulated during growth under iron-restricted conditions. Francisella iron-regulated gene A (figA) was among those most highly expressed under these conditions and was shown, by mutant analysis and cross-feeding experiments, to encode a protein necessary for the synthesis and/or secretion of a siderophore-like activity by F. novicida U112.
MATERIALS AND METHODS
Bacterial strains and culture media.
F. tularensis LVS no. 11 (11) and F. novicida U112 (27) have been described previously. These strains and the others used in this study are listed in Table 1. The basal medium for these studies was Mueller-Hinton agar (Difco, Detroit, MI) supplemented with 2% (vol/vol) IsoVitaleX (Becton-Dickinson, Sparks, MD), 0.1% (wt/vol) glucose, and 1% (vol/vol) fetal bovine serum (FBS); this medium was designated MH−. For routine cultivation of Francisella strains, ferric pyrophosphate (Fe-PPi) was added to MH− to a final concentration of 250 μg/ml (330 μM); the resultant medium was designated MH+. For iron-restricted growth, deferoxamine mesylate (DF; Desferal; Novartis, East Hanover, NJ) was added to MH− to a final concentration of 100 μM; this medium was designated MH−/DF. Broth-based media of all three types lacked FBS. All strains were routinely grown at 37°C in an atmosphere of 95% air-5% CO2 or in broth at 37°C with aeration.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Host strain for cloning experiments | 34 |
| HB101 | Host strain essential for propagating plasmids carrying mutated Francisella genes used for electroporation of Francisella | 34 |
| Francisella tularensis strains | ||
| LVS no. 11 | F. tularensis LVS | 10 |
| LVS figA::np-kan | Mutant of LVS with promoterless kan cartridge inserted in figA gene | This study |
| LVS figA::np-kan(pKD107) | LVS figA::np-kan containing pKD107 with F. novicida U112 figA gene; expresses FigA protein | This study |
| Francisella novicida strains | ||
| U112 | Wild-type strain of F. novicida | 26 |
| U112 figA::np-kan | Mutant of U112 with promoterless kan cartridge inserted in figA gene | This study |
| U112 figA::np-kan(pKD107) | U112 figA::np-kan containing pKD107 with F. novicida U112 figA gene; expresses FigA protein | This study |
| U112 figA::np-kan(pKD108) | U112 figA::np-kan containing pKD108 with F. tularensis LVS figA gene; expresses FigA protein | This study |
| U112 figA::np-kan(pFNLTP-CAT) | U112 figA::np-kan containing vector pFNLTP-CAT | This study |
| Plasmids | ||
| pACYC184 | Broad-host-range cloning vector; Chlorr Tetr | New England Biolabs |
| pKD101 | pACYC184 with 2.2-kb fragment containing figA flanking regions | This study |
| pKD101-np-kan | pKD101 with promoterless kan cartridge inserted into KpnI site | This study |
| pFNLTP6 | Shuttle vector for Francisella; Kanr Ampr | 24 |
| pKD105 | pFNLTP6 with F. tularensis LVS figA gene inserted into BamHI site | This study |
| pKD108 | pKD105 with ΔEcat cartridge inserted into XhoI site; this vector backbone containing the ΔEcat cartridge is the same as that of pFNLTP-CAT | This study |
| pFNLTP-CAT | pFNLTP6 with ΔEcat cartridge inserted in XhoI site; Kanr Ampr Chlorr | This study |
| pKD107 | pFNLTP-CAT carrying F. novicida U112 figA gene | This study |
Growth of F. tularensis LVS under iron-replete and iron-restricted conditions.
F. tularensis LVS was streaked from a frozen stock onto MH+ agar and grown at 37°C for 24 h. For iron-replete growth, bacteria from this agar plate were inoculated into 1 ml of MH+ broth and grown at 37°C overnight. This overnight broth culture was then used to inoculate 10 ml of MH+ broth to an optical density at 600 nm (OD600) of 0.05. For iron-restricted growth, the overnight culture was obtained by inoculating the bacteria from the aforementioned agar plate into 1 ml of MH− broth. This overnight broth culture was used to inoculate 10 ml of MH−/DF broth to an OD600 of 0.05. For iron add-back experiments, an overnight culture of the F. tularensis LVS in MH− medium was used to inoculate 20 ml of MH−/DF broth to an OD600 of 0.05. When the culture reached an OD600 of 0.4, the culture was divided into two cultures of 10 ml each. Fe-PPi was added to one culture, whereas an equivalent volume of sterile H2O was added to the other culture. Both cultures were then grown as described above.
Preparation of whole-cell lysates.
F. tularensis LVS or F. novicida U112 cells grown on MH+ plates were inoculated into 5 ml of MH+ or MH−/DF medium for iron-replete or iron-restricted growth, respectively. After overnight incubation, the cells were harvested by centrifugation at 5,800 × g for 10 min at 4°C. The cell pellet was resuspended in 3 ml of phosphate-buffered saline (PBS)/g (wet weight) of cells. A portion of 3×-concentrated sodium dodecyl sulfate (SDS) sample buffer (30) equivalent to half the volume of these suspended cells was then added, and the samples were heated at 100°C for 5 min. Western blot analysis was performed as previously described (10), with polyclonal antisera to various F. tularensis antigens; these antisera were diluted 1:3,000.
Preparation of cell envelopes of F. tularensis LVS.
Cultures (100 ml) of the F. tularensis LVS were grown into stationary phase under iron-replete or iron-restricted conditions. After harvesting of the cells by centrifugation, the cells were washed twice in phosphate-buffered saline (PBS) and disrupted by sonication. Unbroken cells and cell debris were removed by centrifugation at 10,000 × g for 20 min at 4°C. The supernatant fluid was then subjected to centrifugation at 150,000 × g for 1 h at 4°C. The resultant cell envelope pellet was washed twice with PBS and resuspended in 0.5 ml PBS. The protein concentration of the cell envelope preparation was determined by the Bradford method with the Protein Assay reagent (Bio-Rad, Hercules, CA), and approximately 20 μg of protein was loaded in each well for SDS-polyacrylamide gel electrophoresis (PAGE).
Protein identification.
Proteins in cell envelope preparations were resolved by SDS-PAGE and stained with Coomassie blue. Selected protein bands were excised and identified by nano-high-performance liquid chromatography-tandem mass spectrometry performed by the Protein Chemistry Technology Center at the University of Texas Southwestern Medical Center.
Generation of polyclonal mouse antibodies against F. tularensis proteins.
The predicted amino acid sequences of the F. tularensis Schu4 FigA and PdpB proteins were derived from the genome of this organism (19). A 21-amino-acid (aa) sequence (KDNNPQINHDDWQQFEYELDN) from FigA and a 24-aa sequence (KLQHFYGEITKLNKQNNTNDQNDK) from PdpB were synthesized with a cysteine residue added to the N terminus of each peptide. These peptides were covalently coupled to Imject maleimide-activated mariculture keyhole limpet hemocyanin (KLH; Pierce, Rockford, IL) and used to immunize mice.
Nucleotide sequence analysis.
PCR was used to amplify a 7-kb fragment from F. novicida that contained the fur gene and the figABCD gene cluster. Nucleotide sequence analysis of this DNA fragment showed that these five open reading frames (ORFs) from F. novicida (GenBank accession number DQ497185) had 97 to 100% identity with the same ORFs from both F. tularensis LVS and F. tularensis Schu4. The nucleotide sequences of these ORFs from the latter two strains are available from GenBank.
RNA isolation from F. tularensis LVS.
F. tularensis LVS cells were grown under both iron-replete and iron-restricted conditions into the early logarithmic phase (OD600, ∼0.3). The cultures were harvested by centrifugation at 6,000 × g for 15 min at 4°C. Total RNA was isolated with the RNeasy Midi kit (QIAGEN, Valencia, CA) by following the manufacturer's protocol. During RNA extraction, treatment with the RNase-free DNase set (QIAGEN) was used to remove any DNA contamination. RNA concentration was determined by spectrophotometry (OD260), and RNA integrity was verified by agarose gel electrophoresis.
DNA microarray development.
Preliminary annotation of the incomplete F. tularensis Schu4 genome was accomplished with a combination of automated ORF prediction and homology searching. The collection of 48 contigs in the August 2002 release of the F. tularensis Schu4 genome was obtained from http://artedi.ebc.uu.se/Projects/Francisella/. The Glimmer 2.02 software was obtained from The Institute for Genomic Research and was used to predict most of the ORFs in the incomplete genome (9). Because Glimmer requires a complete, intact genome, the 48 contigs from the incomplete genome were concatenated into one contiguous sequence. The sequence 5′-TTAACTAACTAG-3′, which contains stop codons in all six reading frames, was inserted between contigs to prevent Glimmer from erroneously predicting false ORFs that spanned contig junctions. Glimmer was then used to predict putative ORFs (encoding proteins containing at least 50 aa) in this contiguous sequence. 2,051 putative ORFs were predicted and were translated with the standard genetic code. Seventy-mer oligonucleotides designed to anneal specifically to a region within the 5′ 70% of each of these 2,051 annotated ORFs were synthesized by QIAGEN and spotted in triplicate on Corning UltraGapII slides by Microarrays, Inc. (Nashville, TN). Subsequent to the preparation of these microarray slides, the definitive annotation of the F. tularensis Schu4 genome was published (19) and this more recent annotation was used in this study to identify relevant ORFs.
DNA microarray analysis.
Genome-directed primers (3) were designed to anneal within the 3′ 30% of each ORF. These 47 genome-directed primers were synthesized by QIAGEN and used to prime the mRNA in total RNA preparations for cDNA synthesis. Equal quantities of total RNA from cells grown under iron-replete and iron-restricted conditions were used to generate cDNA with the CyScribe Post-Labeling kit (Amersham Biosciences, Piscataway, NJ). After reverse transcription, residual RNA was removed by alkaline treatment, followed by neutralization. The cDNAs were purified with a QIAquick gel extraction kit (QIAGEN). For each DNA microarray slide, one sample was labeled with Cy3-dCTP while another sample was labeled with Cy5-dCTP. Dye swap experiments were performed for each pair of samples to compensate for the different labeling efficiencies of Cy3 and Cy5. The labeling mixtures were cleaned by using YM-30 Microcon centrifugal filter devices (Millipore, Bedford, MA). Equal amounts of labeled cDNA from cells grown under both conditions were used to hybridize microarray slides in the hybridization buffer provided with the CyScribe Post-Labeling kit. Hybridization was carried out at 50°C for 16 h in the dark. After hybridization, the slides were washed in saline-sodium phosphate-EDTA buffer and scanned with GenePix scanner 4100A and GenePix Pro 5.0 software (Axon Instruments Inc., Union City, CA). Data from three independent RNA preparations hybridized on a total of seven slides were normalized and then analyzed with Acuity 4.0 software (Axon). Genes that were differentially regulated more than twofold were considered to be differentially transcribed. Selected results from these analyses are contained in Table 2.
TABLE 2.
F. tularensis LVS genes whose expression was maximally altered by growth under iron-restricted conditionsa
| F. tularensis Schu4 ORF no. | Gene identity or function | Avg fold differenceb |
|---|---|---|
| Genes up-regulated under iron-restricted growth conditions | ||
| 29c | Hypothetical protein (FigA) | 19.70 |
| 27c | Diaminopimelate decarboxylase (FigC) | 15.13 |
| 26c | Hypothetical protein (FigD) | 14.78 |
| 28c | Hypothetical protein (FigB) | 9.78 |
| 1565c | Glycosyl hydrolase, family 3, pseudogene | 4.08 |
| 1702 | Conserved hypothetical protein | 3.87 |
| 1701 | Hypothetical protein | 3.62 |
| 1707 | Conserved hypothetical protein | 3.41 |
| 1542c | Outer membrane protein | 3.29 |
| 1714c | Intracellular growth locus, subunit A (IglA) | 3.26 |
| 383 | Hypothetical protein | 3.22 |
| 989 | Hypothetical protein | 3.19 |
| 1709 | Conserved hypothetical protein | 3.09 |
| 980 | Aminotransferase, class II | 3.08 |
| 1711c | Intracellular growth locus, subunit D (IglD) | 3.06 |
| 1703 | Conserved hypothetical protein | 3.01 |
| 1700 | Conserved hypothetical protein (PdpB) | 2.99 |
| 1706 | Conserved hypothetical protein | 2.99 |
| 1717 | Major facilitator superfamily transport protein | 2.98 |
| 1712c | Intracellular growth locus, subunit C (IglC) | 2.96 |
| Genes down-regulated under iron-restricted growth conditions | ||
| 1214c | Haloacid dehalogenase-like hydrolase family protein | 0.32 |
| 881c | Amino acid permease | 0.39 |
| 265 | ABC transporter, membrane protein | 0.42 |
| 1269c | Chaperone protein, DnaK (heat shock protein family 70) | 0.43 |
| 1696 | Chaperone protein, GroEL | 0.43 |
| 1624c | Hypothetical protein | 0.44 |
| 1695 | Chaperone protein GroES | 0.44 |
| 938 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | 0.46 |
| 331 | 30S ribosomal protein S3 | 0.47 |
| 172 | Hypothetical membrane protein, fragment | 0.48 |
| 327 | 50S ribosomal protein L23 | 0.49 |
| 413c | 1,4-α-Glucan branching enzyme | 0.49 |
| 328 | 50S ribosomal protein L2 | 0.49 |
| 140 | 50S ribosomal protein L11 | 0.50 |
| 326 | 50S ribosomal protein L4 | 0.50 |
| 1048c | Hypothetical protein | 0.51 |
| 266 | ABC transporter, ATP-binding protein | 0.51 |
| 551 | Conserved hypothetical protein, pseudogene | 0.52 |
| 999c | ZIP metal transporter family protein, pseudogene | 0.52 |
| 339 | 30S ribosomal protein S8 | 0.52 |
For detailed DNA microarray data, see Table S1 in the supplemental material.
The average fold difference indicates the gene expression level under iron-restricted conditions relative to that obtained under iron-replete conditions.
Quantitative real-time reverse transcription-PCR (qRT-PCR).
Twenty-one genes were selected for analysis of their transcription levels by qRT-PCR. For each of the genes tested, primers (Table 3) were designed with Primer Express software (Applied Biosystems, Foster City, CA). Total RNA was isolated from F. tularensis LVS cells grown under iron-replete and iron-restricted conditions as described above. To remove possible DNA contamination, an additional step of DNase treatment of the RNA samples was performed with the MessageClean kit (GeneHunter, Nashville, TN). Equal quantities of RNA from cells grown under both conditions were used in qRT-PCRs with SYBR Green PCR Master Mixture (Applied Biosystems) and MultiScribe reverse transcriptase (Applied Biosystems). Each reaction was carried out with a 25-μl volume containing 50% (vol/vol) SYBR master mix, the two oligonucleotide primers at a final concentration of 50 nM each, and 10 ng of RNA. PCR amplification was accomplished with a model 7500 real-time PCR system (Applied Biosystems). The relative levels of gene expression were calculated with 7500 System SDS software (Applied Biosystems) by the ΔΔCt method. The F. tularensis LVS fopA gene was chosen as an internal control because this gene apparently did not alter its level of expression between iron-replete and iron-restricted growth conditions as measured by DNA microarray and Western blot analyses (data not shown). Both no-reverse transcriptase reaction mixtures and no-template reaction mixtures were always included as negative controls.
TABLE 3.
qRT-PCR analysis of gene expressiona
| Schu4 ORF no. | Putative function | Fold differenceb
|
qRT-PCR primer pair (5′-3′)d | |
|---|---|---|---|---|
| Microarray | qRT-PCR | |||
| 26c | Hypothetical protein (FigD) | 14.8 | 13.4 | F: AAGCCTAATGGTAGCTGGTGAATC |
| R: TTTTGGTGAGACTCCGTAAGTTTTT | ||||
| 29c | Hypothetical protein (FigA) | 19.7 | 24.7 | F: GGCTACTAAAGACAAAACACTAATGCC |
| R: CAGACTCAGGCGATACTGTTCTTG | ||||
| 27c | Diaminopimelate decarboxylase (FigC) | 15.1 | 13.0 | F: GGTTTAGCAGGAATAAGCCCTACTC |
| R: TTGTCGCGGCATATAGTTCTAAATC | ||||
| 28c | Hypothetical protein (FigB) | 9.9 | 19.1 | F: GAGTTTTTTAGCTCAGACCCGATAATC |
| R: ATCGGCGCTGAAAATAGCA | ||||
| 651c | Proton-dependent oligopeptide family protein | 2.4 | 0.6 | F: CTCGCAAAAAAGACATTAGTATAAAATTAAAT |
| R: AAGTTGCAAATTGTCCCCAAA | ||||
| 1712c | Intracellular growth locus, subunit c (IglC) | 3.0 | 2.6 | F: AAAAAGGAGAATGATTATGAGTGAGATG |
| R: TGCAGTAGGATCAGTTCTCACATG | ||||
| 403 | Peptide deformylase | 2.8 | 3.2 | F: CAGCTATAATATTTCAGCATGAATTTAATCA |
| R: AGCAAATTTAGCTTGTAGTTCTTCGTT | ||||
| 1699 | Conserved hypothetical protein (PdpA) | 2.6 | 2.6 | F: TGAGTTAATTTCAAACTCTGCCATATC |
| R: GTTTGGGTATATGCCATTTCACAG | ||||
| 1780 | Putative transposase | 1.5 | 1.1 | F: AAGAACCGGAGATTATAGTTCAAAGC |
| R: AAATACTGTTCAATCAATGTTTTATCGG | ||||
| 651c | Proton-dependent oligopeptide family protein | 1.3 | 0.7 | F: CGTTGATAGTAAAACTATTGGTTTTGCA |
| R: GGAAGAGCAACCACTTGATTTAATAGA | ||||
| 664c | Histidine decarboxylase | 1.0 | 1.1 | F: CATTTGATGGTGCTTTTCTACCG |
| R: ATATTCCTGAAGGCATCGGATTAC | ||||
| 583 | Outer membrane-associated protein | 0.9 | 1.0 | F: CAAATCTAGCAGGTCAAGCAACAG |
| R: AACACTTGCTTGAACATTTCTAGATAGTTC | ||||
| 700 | Conserved hypothetical protein | 0.8 | 0.6 | F: AAAATGGTGTTCGCCTTTTGG |
| R: CCTTCGACATCATCTTTGTAGTCAAC | ||||
| 445 | ABC transporter, ATP-binding, pseudogene | 0.9 | 1.2 | F: TTAGAAGAGTTTGTTGGAGGATATGATG |
| R: ATTTTTCGACAACTCACTGTTATTTTTAGT | ||||
| 881c | Amino acid permease | 0.4 | 0.6 | F: CCAAGGTTCAGAAATTATCGGACTA |
| R: GTGCTTAGCGATTCTTGGCAT | ||||
| 50 | Translation initiation factor IF2 | 0.6 | 0.8 | F: TTGGCGCGTATTCTGTTAAGAC |
| R: GCTCGCATTGAAGTAAAAGCCT | ||||
| 140 | 50S ribosomal protein L11 | 0.5 | 0.8 | F: ACGCCACCTGCTTCTTACTTAATT |
| R: CGCACGAGTTATAGTTCCAACAA | ||||
| 350 | DNA-directed RNA polymerase, alpha subunit | 0.7 | 0.7 | F: TTGACAAACCAAAGAGCATTTAGC |
| R: TCTCAAGCTCGGTTGGTATCG | ||||
| 1695 | Chaperone protein, GroES | 0.4 | 0.5 | F: GCTCAAGAGAAACCTAGCCAAGG |
| R: ACATCCATAGGTAGCGTAGTGCC | ||||
| 326 | 50S ribosomal protein L4 | 0.5 | 0.7 | F: AGTTTCTGGTGGCGGTGC |
| R: GGTGAACGGATAGTACCCGCT | ||||
| 1696 | Chaperone protein, GroEL | 0.4 | 0.7 | F: TTCAAAGACAGCGGATGTTGC |
| R: GCGACAGCTTTTAGACCCTCTG | ||||
qRT-PCR for each gene was performed with two individually prepared RNA samples, each in triplicate.
The fold difference indicates the gene expression level under iron-restricted conditions relative to that obtained under iron-replete conditions.
ORF 651 in the F. tularensis Schu4 genome was found to be fragmented into two separate ORFs in the F. tularensis LVS genome by the deletion of a single nucleotide.
F, forward; R, reverse.
RT-PCR.
RT-PCR was performed with the SuperScript III kit (Invitrogen, Carlsbad, CA). Total RNA was isolated from F. tularensis LVS cells grown in iron-replete medium as described above. RNA (1 μg) was incubated with SuperScript III reverse transcriptase and appropriate oligonucleotide primers. PCR amplification with Taq DNA polymerase (New England Biolabs, Beverly, MA) was carried out at temperatures of 52°C for annealing and 72°C for extension.
Construction of figA mutants of F. tularensis LVS and F. novicida U112.
All of the oligonucleotide primers used in this study were derived from the F. tularensis Schu4 genome (19). An ∼1.0-kb fragment corresponding to the 5′ upstream region of the F. tularensis LVS figA gene was PCR amplified from chromosomal DNA with Pfu DNA polymerase (Stratagene, La Jolla, CA), primer 1 (5′-GCGGATCCGCGCCAACAATCACTGATAAAAC-3′), and primer 2 (5′-GGTACCGCATGCATGCATTTTAAAATCCCTACATGATAATG-3′). Another ∼1.2-kb fragment corresponding to the 3′ downstream region of the F. tularensis LVS figA gene was amplified with primer 3 (5′-ATGCATGCATGCGGTACCGTAAAAGGTGAAATACACCAAAG-3′) and primer 4 (5′-GCGGATCCCAAAGGTAACTCCAAGTATG-3′). Primers 2 and 3 shared an 18-nucleotide (nt) complementary sequence (in bold) at their 5′ ends, and a KpnI site (bold and underlined) was contained in these sequences. One-microliter portions of the two PCR amplicons were then mixed and used as the template in a bridging PCR (i.e., PCR sewing) (24, 29) with primers 1 and 4. This second PCR generated an ∼2.2-kb product corresponding to the 5′ and 3′ flanking regions of the figA ORF with a KpnI site in the middle of this amplicon and BamHI sites (underlined in primers 1 and 4) at both ends. This fragment was digested with BamHI and ligated into BamHI-cut pACYC184 (New England Biolabs) to obtain pKD101.
A promoterless 850-bp kanamycin resistance cartridge (np-kan) (26) was amplified with Pfu DNA polymerase together with primer 5 (5′-ATAGGTACCGGGTGACTAACTA GGAGG-3′) and primer 6 (5′-ATAGGTACCGGGTCGCATTATTCCCTCCA-3′) (KpnI sites underlined). The np-kan amplicon was digested with KpnI and ligated with KpnI-digested pKD101 to obtain pKD101-np-kan. This plasmid was used to transform Escherichia coli HB101 (35); plasmid purified from this recombinant strain by means of a QIAGEN Midi-prep kit was used to electroporate F. tularensis LVS. For electroporation, cell paste was scraped from two confluent MH+ agar plates and washed three times in 50 ml of 0.5 M sucrose by centrifugation at 5,800 × g for 10 min at 10°C. The washed cells were resuspended in 250 μl of 0.5 M sucrose and divided into individual 80-μl portions. A 7-μg portion of pKD101-np-kan (in a 10-μl volume) was added to an 80-μl portion of these cells, and the mixture was incubated at room temperature for 10 min. The mixture was then transferred into an electroporation cuvette (1-mm electrode gap) and electroporated at 2.0 kV, 10 μF, and 600 Ω. A 200-μl portion of MH+ medium was added to the cuvette, and the entire content of the cuvette was spread onto a chocolate II agar (CA II) plate (Becton Dickinson), which was incubated at 37°C in 95% air-5% CO2 for 6 h. Next, growth on this CA II plate was scraped into 500 μl MH− medium and the surface of the CA II plate was washed once with another 500-μl portion of this medium. The liquid was pooled and spread onto two MH− plates containing kanamycin (15 μg/ml) and incubated as described above for 36 to 48 h. Kanamycin-resistant colonies were patched onto MH−/DF agar, and figA mutants were identified by colony blot analysis with mouse antiserum to FigA. For construction of an F. novicida U112 figA mutant, the method used was the same as that described above, with two exceptions; the electroporated cells were incubated on the CA II agar plate for only 3 h, and the concentration of kanamycin for selection on MH− agar plates was increased to 30 μg/ml.
Colony blot analysis.
F. tularensis LVS or F. novicida U112 colonies grown on MH− plates were patched onto MH−/DF plates. After incubation for 24 h at 37°C, the patches were blotted with a sterile nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The membrane was then placed for 5 min on Whatman filter paper that had been saturated with 20% (wt/vol) SDS, transferred to another filter paper saturated with denaturing solution (0.5 M NaOH, 1.5 M NaCl), incubated for 5 min, and finally placed on a filter paper saturated with neutralizing solution (1.3 M NaCl, 0.3 M Tris-HCl, pH 8.0) for 5 min. After drying at room temperature for 30 min, the membrane was gently agitated in PBS with 0.05% (vol/vol) Tween 20 (PBST) for 5 min with one change of PBST buffer. Acetic acid (0.1 N) was used to wash the membrane twice to remove residual agar and cell debris. Residual acid was removed by washing with PBST twice, and the membrane was blocked with 3% (wt/vol) skim milk in PBST for 1 h. Mouse antiserum to FigA was used as the primary antibody, and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G was used as the secondary antibody. Final color development was accomplished by using the 3,3′,5,5′-tetramethylbenzidine liquid substrate system (Sigma) according to the manufacturer's protocol.
Complementation analysis.
The primers used to PCR amplify a 2.9-kb fragment of F. tularensis LVS chromosomal DNA containing the figA ORF and some flanking DNA were primer 7 (5′-CGGGATCCTTAATCAGTTTGAGTCAGCAGG-3′) and primer 8 (5′-CGGGATCCTAACTAAGTAATGTGGCTAAAACACC-3′). Both primers contained a BamHI site (underlined) at their 5′ ends, and the PCR amplicon was ligated into the BamHI site in the vector pFNLTP6 (25) to obtain pKD105. A chloramphenicol resistance cassette (ΔEcat) (15) was then ligated into the XhoI site of this plasmid to obtain pKD108. Digestion of this plasmid with BamHI yielded three fragments; these were the vector backbone, the cat cartridge, and the cloned F. tularensis LVS DNA insert. The gel-purified cat cartridge and vector backbone were religated to yield pFNLTP-CAT. The primers used to PCR amplify a 3.2-kb fragment containing the F. novicida U112 figA ORF and some flanking DNA were primer 9 (5′-CAGCTAGCTGTTGAGATACTTAATTTATTTG-3′) and primer 10 (5′-CTGCTAGCCAAAGGACCTATAGCGATAC-3′). Both primers contained an NheI site (underlined) at the 5′ end, and the PCR amplicon was cloned into this site in pFNLTP-CAT to construct pKD107. Both pKD107 and pKD108 were purified from E. coli DH5α and used to electroporate F. novicida U112 as described above. The desired transformants were selected with kanamycin (30 μg/ml) on MH+ plates. Plasmids pKD107 and pKD108 purified from F. novicida U112 were then used to electroporate the F. novicida U112 figA::np-kan mutant. Plasmid pKD107 purified from E. coli DH5α was used to electroporate the F. tularensis LVS figA::np-kan mutant. Transformants were selected with chloramphenicol (10 μg/ml) on MH+ plates.
Chrome Azurol S (CAS) assay.
CAS/CDM− plates were prepared by incorporating CAS/Fe(III)/hexadecyltrimethylammonium (HDTMA) into Chamberlain's chemically defined medium (6) lacking FeSO4 (CDM−). Briefly, 250 ml of sterile CAS-HDTMA solution was prepared as described by Payne (31). Four grams of the amino acid mixture (6) was mixed with 20 mg of spermine phosphate and dissolved in 150 ml of H2O. This solution was extracted with 150 ml of 3% (wt/vol) hydroquinoline in chloroform to remove contaminating iron. Traces of residual hydroquinoline were removed by another extraction with an equal volume of chloroform. Thiamine-HCl, l-calcium pantothenate, glucose, MgSO4, KH2PO4, K2HPO4, and NaCl were added to this deferrated amino acid solution to the final concentrations described (6). The pH of this CDM− solution was adjusted to 6.2 to 6.4 before the volume was brought to 250 ml with H2O. The CDM− solution was then filter sterilized and warmed to 55°C. A 250-ml portion of 2% (wt/vol) agar (in water) was sterilized and cooled to 55°C. This agar solution, the CDM− solution, and 50 ml of CAS-HDTMA were mixed gently and dispensed into petri dishes. F. tularensis LVS and F. novicida U112 strains were inoculated onto the CAS-CDM− plates and incubated as described above for either 2 to 5 days (for F. tularensis LVS) or overnight (for F. novicida U112). Siderophore (enterobactin)-producing E. coli strain W3110N and its enterobactin-deficient entF mutant were used as positive and negative controls, respectively, in the CAS assay.
Cross-feeding experiment.
F. novicida U112 and its figA mutant U112 figA::np-kan were streaked in parallel in various combinations onto both a CAS/CDM− plate and a CDM+ plate which was supplemented with FeSO4 (2 μg/ml) (6). These plates were incubated at 37°C for 36 h.
Intracellular growth assay.
Wild-type and mutant bacterial strains were used to infect mouse macrophage cell line J774A.1 (American Type Culture Collection, Manassas, VA) at a multiplicity of infection of ∼10. Briefly, approximately 1 × 105 J774A.1 cells were seeded into the wells of a 12-well tissue culture plate containing Dulbecco's modified Eagle's medium (Cellgro-Mediatech, Inc., Herndon, VA) supplemented with 4 mM GlutaMax (Invitrogen), 10% (vol/vol) heat-inactivated FBS (HyClone, Logan, UT), and 1 mM sodium pyruvate 24 h before infection. Bacteria grown in MH− broth to an OD600 of 0.3 were washed with this growth medium three times and added to these monolayers (which were ∼75% confluent) in triplicate. After 1 h incubation at 37°C in an atmosphere of 95% air-5% CO2, the monolayers were washed three times with tissue culture medium, gentamicin was added to a final concentration of 100 μg/ml, and the plate was incubated for 30 min to kill extracellular bacteria. After gentamicin treatment, the monolayers were washed and the macrophages were incubated for 2 to 48 h. At various time points, the tissue culture medium was removed, the monolayers were washed once with tissue culture medium, and then 0.5% (wt/vol) octyl-β-d-glucopyranoside in PBS was added to lyse the macrophages. These lysates were plated onto MH+ agar to determine the number of bacteria recovered from the macrophages. The F. novicida U112 iglC mutant KKF24 (ΔiglC::ermC), which is deficient in the ability to grow intracellularly (20), was used as a negative control in these assays.
Accession number for DNA microarray data.
The raw data from the DNA microarray experiments described here (Table 2) are located at the National Center for Biotechnology Information's GEO database under series accession number GSE3622.
RESULTS
Growth of F. tularensis LVS under iron-replete and iron-restricted conditions.
To study iron-related gene regulation in F. tularensis LVS, it was necessary to determine growth conditions under which iron was limiting. The iron chelator DF was used to bind free ferric ions in the growth medium. In initial experiments, it was observed that F. tularensis LVS cells grown on MH+, when washed and inoculated into MH−/DF, continued to grow normally (data not shown). However, if these cells were first grown overnight in MH− and then subcultured into MH−/DF, both the rate and extent of growth were severely limited (Fig. 1). Confirmation that this growth restriction was iron specific was obtained by adding iron to the MH−/DF medium and showing that both the rate and extent of growth were similar to those obtained in MH+ (Fig. 1). This latter experiment was necessary because it has been reported that DF can bind other metal ions in addition to iron (5).
FIG. 1.
Growth of F. tularensis LVS under iron-replete and iron-restricted conditions. F. tularensis LVS was grown overnight on an MH+ agar plate. For iron-replete growth (filled circles), bacteria from the plate were inoculated into 1 ml of MH+ broth and grown at 37°C overnight. This overnight culture was then used to inoculate 10 ml MH+ broth to an OD600 of 0.05. For iron-restricted growth, the overnight culture was obtained by inoculating the bacteria from the plate into 1 ml MH− broth. This overnight culture was used to inoculate 20 ml of MH−/DF broth. When the culture reached an OD600 of 0.4, the 20-ml MH−/DF culture was divided into two cultures of 10 ml each. Fe-PPi was added to a final concentration of 330 μM in one culture (filled triangles), whereas an equivalent volume of sterile H2O was added to the other culture (open triangles).
Cell envelope protein profiles of F. tularensis LVS grown under iron-replete and iron-restricted conditions.
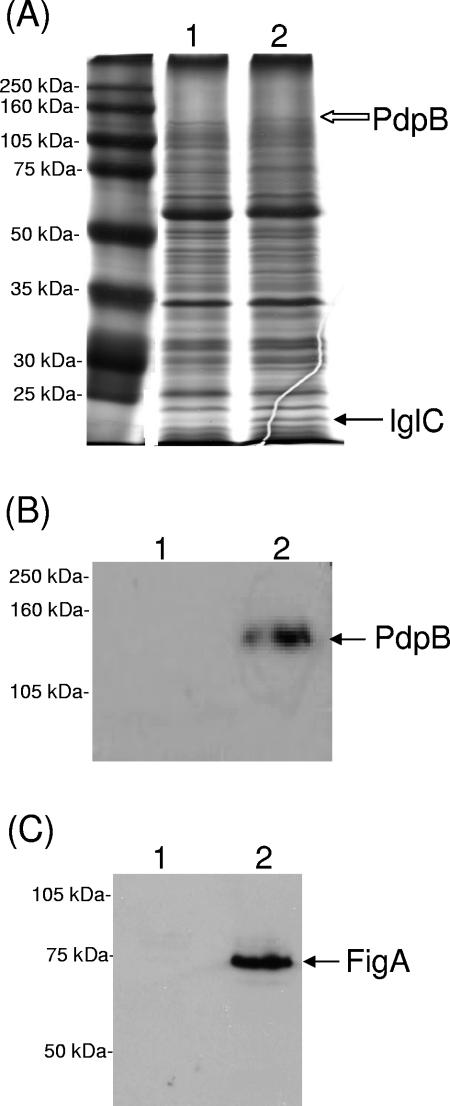
To identify proteins whose expression was increased under iron-restricted conditions, SDS-PAGE was used to resolve proteins in cell envelopes prepared from bacteria grown under both iron-replete (Fig. 2A, lane 1) and iron-restricted (Fig. 2A, lane 2) conditions. Two proteins, with apparent sizes of approximately 23 kDa and 140 kDa, were more abundant in envelopes from cells grown under iron-restricted conditions (Fig. 2A, lane 2, black arrow and white arrow, respectively). Mass spectrometric analysis identified the 23-kDa protein as IglC and the 140-kDa protein as PdpB. Both the iglC and pdpB genes are located in the previously identified pathogenicity island of F. tularensis (27). Mouse antiserum raised against a PdpB peptide-KLH conjugate was used in a Western blot analysis to probe these same sets of cell envelopes, and it was found that these PdpB-reactive antibodies bound a 140-kDa protein in the iron-restricted sample (Fig. 2B, lane 2) and were unreactive with the iron-replete sample (Fig. 2B, lane 1).
FIG. 2.
Identification of F. tularensis LVS proteins with increased expression under iron-restricted conditions. F. tularensis LVS cells were grown to stationary phase under both iron-replete (lane 1) and iron-restricted (lane 2) conditions. Cell envelopes and whole-cell lysates were prepared as described in Materials and Methods. (A) Proteins in cell envelopes were resolved by SDS-PAGE and stained with Coomassie blue. Molecular mass standards are present in the far left lane. The black arrow indicates the IglC protein, and the white arrow indicates the PdpB protein. Proteins in cell envelopes (B) or in whole-cell lysates (C) were resolved by SDS-PAGE, electrophoretically transferred to nitrocellulose membranes, and probed in a Western blot analysis with polyclonal mouse antiserum to PdpB (B) or FigA (C).
Identification of putative Fur boxes in F. tularensis.
Transcription of bacterial genes involved in iron acquisition is usually controlled by the ferric uptake regulator (Fur) protein, which binds ferrous ion and then itself binds to a DNA regulatory sequence (i.e., the Fur box), causing repression of transcription. The typical Fur box is located immediately upstream from the iron-regulated ORF, and a 19-nt consensus Fur box has been defined (Fig. 3) (22). When the iglC and pdpB genes of F. tularensis Schu4 (19) were examined, sequences with similarity to the consensus Fur box were found (11/19 matches for iglC and 12/19 matches for pdpB) (Fig. 3). In addition, when the F. tularensis Schu4 genome was searched for consensus Fur box sequences, a number of ORFs were identified that had putative 5′ untranslated regions with possible Fur boxes (data not shown). The gene whose putative Fur box had the highest level of identity (17/19 matches) with this consensus sequence was figA. Interestingly, this same region contained a second putative Fur box with a slightly lower degree of identity with the consensus sequence (Fig. 3). The figA gene is located 253 nt downstream from the fur gene in the F. tularensis Schu4 genome (Fig. 4A) and encodes a predicted 74-kDa protein with 36% identity to the predicted FrgA protein (accession number NP_819459.1) from Coxiella burnetii and 34% identity to the LbtA (1) and FrgA (16) proteins from L. pneumophila. Expression of both of these Legionella proteins has been shown to be regulated by iron (1), and an lbtA mutant is deficient in expression of the Legionella siderophore legiobactin (1). Regulation of expression of the F. tularensis LVS FigA protein by iron availability was confirmed by the finding that mouse antibodies raised against a FigA peptide-KLH conjugate bound a 74-kDa protein in a whole-cell lysate of iron-restricted cells (Fig. 2C, lane 2) but did not react with an equivalent whole-cell lysate of iron-replete cells (Fig. 2C, lane 1).
FIG. 3.
Putative Fur boxes associated with the F. tularensis Schu4 pdpB, iglC, and figA ORFs. The ClustalW alignment was accomplished with the MacVector 6.5.3 software. The consensus Fur box is outlined. Identical bases in each pair are underlined and bold; nonidentity is indicated by italics.
FIG. 4.
F. tularensis Schu4 figA and adjacent ORFs. (A) Schematic drawing. (B) RT-PCR analysis of possible transcriptional linkage between figA and adjacent ORFs. RT-PCR was carried out as described in Materials and Methods, with oligonucleotide primer pairs spanning intergenic regions 1 (5′-GTGCTGAAATGATTGACTATAGTCTC-3′ and 5′-CTTGTCATTGTTTATATTGTAGAAGAATA-3′), 2 (5′-AGTGCGATAAAGATGACATGC-3′ and 5′-TAGCTGCTATGATTATAAGC-3′), 3 (5′-TATCTATTTGACCTTAAATC-3′ and 5′-TGAATAATTGTTTCAATTTG-3′), and 4 (5′-TCAATACCAAGTTCTCAGAG-3′ and 5′-ACCTAAAAACCATGTTAAAAG-3′), respectively. Lanes 1, 4, 7, and 10, PCR products derived from F. tularensis LVS chromosomal DNA. The sizes of these PCR products are 547 bp (lane 1), 135 bp (lane 4), 120 bp (lane 7), and 121 bp (lane 10). Lanes 2, 5, 8, and 11, RT-PCR negative controls lacking reverse transcriptase. Lanes 3, 6, 9, and 12, products obtained when these same primers were used in an RT-PCR with F. tularensis LVS total RNA. Size markers are present on the left side of panel B.
DNA microarray analysis of F. tularensis LVS gene expression in response to iron availability.
Having both established conditions for iron-restricted growth and identified several genes which appear to be iron regulated, DNA microarray analysis was used to provide a more comprehensive picture of the effect of iron limitation on gene expression by F. tularensis LVS. These DNA microarrays contained probes derived from the nucleotide sequence of the F. tularensis Schu4 genome (19). Approximately 80 of the F. tularensis LVS genes were found to be either up- or down-regulated at least twofold by iron limitation (see Table S1 in the supplemental material). Forty of the genes most affected by iron limitation, either positively or negatively, are listed in Table 2.
Among these genes, figA was found to be up-regulated nearly 20-fold by iron limitation. In the F. tularensis Schu4 genome, figA is located just downstream from the predicted fur gene and 5′ to a set of three consecutive ORFs (figB, figC, and figD; Fig. 4A) whose expression was also up-regulated as determined by DNA microarray analysis (Table 2). The predicted protein encoded by figB in the Schu4 genome is listed as a hypothetical protein and is 31% identical to a multidrug resistance efflux pump from L. pneumophila (accession number YP_095354.1). The predicted protein encoded by the Schu4 figC gene is described as a diaminopimelate decarboxylase and has 31% identity with a diaminopimelate decarboxylase from Magnetospirillum magnetotacticum (accession number ZP_00054090.1). The protein encoded by the Schu4 figD ORF is annotated as a hypothetical protein and is 38% identical to a drug resistance transporter from L. pneumophila (accession number YP_095353.1). RT-PCR was used to determine possible transcriptional linkage of these five ORFs. RNA was isolated from the F. tularensis LVS grown in MH+ medium and reverse transcribed with four pairs of oligonucleotide primers designed to span the four intergenic regions (Fig. 4A). Positive RT-PCRs were observed for all four sets of primers (Fig. 4B), indicating that these five ORFs are likely transcribed together to yield a polycistronic mRNA in F. tularensis LVS.
Validation of DNA microarray results.
The qRT-PCR method was used to validate the DNA microarray data in Table 2. Transcript levels were determined for 21 genes, including 9 whose expression, as measured by DNA microarray analysis, appeared to be up-regulated, 8 with apparent down-regulation, and 4 whose expression appeared relatively unaffected by growth under iron-limiting conditions. For most of the genes tested, the values obtained by qRT-PCR (Table 3) correlated well (R2 = 0.94) with those obtained from DNA microarray analysis (Table 2).
Generation of figA mutants of the F. tularensis LVS and F. novicida U112.
Expression of the figA gene was shown to be substantially increased under iron-restricted growth conditions by both DNA microarray analysis and qRT-PCR. To determine the relevance of the FigA protein to iron acquisition, figA deletion mutants were constructed in both F. tularensis LVS and F. novicida U112. PCR was used to amplify the F. tularensis LVS chromosomal DNA region immediately upstream from the figA gene and, separately, a small portion of the 3′ end of the figA gene and some flanking DNA (Fig. 5A). A promoterless kan cartridge (26) was ligated between these two fragments to create pKD101-np-kan as described in Materials and Methods. This plasmid was used to electroporate both F. tularensis LVS and F. novicida U112, and kanamycin-resistant transformants were tested by colony blot analysis with the mouse antiserum to FigA. Chromosomal DNA was purified from putative figA mutants and tested in PCR with oligonucleotide primers 5′-GCAAAGAGTCCCAGCAACAATAAG-3′ and 5′-GAATAATAGTGTTTAGAGATTGAG-3′ (which bind outside of the chromosomal DNA insert in pKD101) (Fig. 5B). A second PCR was performed with primers for the kan cartridge to confirm its presence in these two mutants, designated F. tularensis LVS figA::np-kan and F. novicida U112 figA::np-kan (Fig. 5C). Finally, Western blot analysis was used to confirm that these two mutants did not express FigA (Fig. 5D).
FIG. 5.
Construction and characterization of figA mutants. (A) Schematic of PCRs used to amplify and sew flanking regions of the F. tularensis LVS figA gene. The promoterless kan cartridge was ligated into this cloned PCR product at the KpnI site. (B) PCR products obtained from the chromosomal DNAs of wild-type and figA mutant strains with oligonucleotide primers (described in Results) binding 1.5 kb from the 5′ and 3′ ends of the figA gene. Lane 1, wild-type F. tularensis LVS; lane 2, F. tularensis LVS figA::np-kan; lane 3, wild-type F. novicida U112; lane 4, F. novicida U112 figA::np-kan. The positions of 4.0-kb and 5.0-kb size markers are indicated on the left. (C) PCR products obtained from chromosomal DNAs of wild-type and mutant strains with primers binding within the kan cartridge; lanes are the same as in panel B. The position of an 850-bp size marker is shown on the left. (D) Western blot analysis of whole-cell lysates of these wild-type and mutant strains grown in MH−/DF broth. Lanes 1 to 4 contain whole-cell lysates of the same strains as in panel B; lane 5 contains a whole-cell lysate of F. novicida U112 figA::np-kan (pKD107). The primary antibody was mouse FigA antiserum. (E) Western blot analysis of these same lysates with rat polyclonal F. tularensis LVS FopA antiserum (i.e., a loading control).
Assessment of potential siderophore production by Francisella species.
The predicted FigA proteins of F. tularensis strains Schu4 and LVS have 34% identity with the L. pneumophila LbtA protein, which has been shown to be necessary for siderophore production by this pathogen (1). Although siderophore production has not been reported to date for F. tularensis, it was appropriate to test whether these figA mutants differed from their parent strains in the production of possible siderophore activity. To accomplish this, the CAS assay was used with CDM− agar plates. The CAS assay is a universal method to detect siderophores independent of their structure (39).
The CAS/CDM− agar plates were tested first with a well-studied E. coli strain and its mutant deficient in enterobactin synthesis. E. coli W3110N produces the siderophore enterobactin, which results in removal of Fe3+ from the CAS/Fe(III)/HDTMA complex and release of the free (orange) dye. Orange halos were present around each colony of E. coli W3110N grown on CAS/CDM− agar (data not shown). In contrast, the entF mutant of W3110N did not show any siderophore activity (data not shown). This CAS/CDM− medium was shown to be iron limiting for growth of F. tularensis LVS (data not shown). When wild-type F. tularensis LVS was grown on this medium, a modest orange zone appeared around the bacterial growth after 2 to 5 days (Fig. 6A, sector 1). The F. tularensis LVS figA::np-kan mutant formed an orange zone that appeared to be only slightly smaller (Fig. 6A, sector 2). In contrast, a much larger orange zone formed around F. novicida U112 (Fig. 6B, sector 1) after 24 h of growth and its figA mutant produced little or no orange zone (Fig. 6B, sector 2).
FIG. 6.
CAS assay. Wild-type F. tularensis LVS and F. novicida U112 and mutant and complemented mutant strains of both were grown on CAS/CDM− agar plates. (A) Wild-type F. tularensis LVS (sector 1) and F. tularensis LVS figA::np-kan (sector 2) grown for 5 days. (B) Wild-type F. novicida U112 (sector 1), F. novicida U112 figA::np-kan (sector 2), F. novicida U112 figA::np-kan(pKD107) (sector 3), F. novicida U112 figA::np-kan(pFNLTP-CAT) (sector 4), and F. novicida U112 figA::np-kan(pKD108) (sector 5) grown for 24 h.
Complementation of the F. novicida U112 figA mutant.
The significant difference in CAS test results obtained with the wild-type F. novicida U112 strain and its figA mutant facilitated subsequent complementation analysis. This was necessary to eliminate the possibility that an unlinked secondary mutation was responsible for the phenotype of the F. novicida figA mutant in the CAS assay. To accomplish this, the wild-type F. novicida figA gene was cloned into the pFNLTP-CAT vector to construct pKD107 in E. coli DH5α as described in Materials and Methods. Several attempts involving electroporation of E. coli-derived pKD107 into F. novicida U112 figA::np-kan, followed by chloramphenicol selection, were unsuccessful, so this E. coli-derived plasmid was used to electroporate the F. novicida U112 wild-type strain, followed by selection with kanamycin for plasmid-containing transformants. (Kanamycin cannot be used to select this plasmid in this kanamycin-resistant mutant.) When pKD107 propagated in F. novicida U112 was used to electroporate the F. novicida U112 figA::np-kan mutant, chloramphenicol-resistant transformants containing this plasmid were obtained. This same mutant was transformed with the vector pFNLTP-CAT (propagated first in F. novicida U112). When introduced into F. novicida U112 figA::np-kan, the wild-type F. novicida figA gene in trans restored both expression of the FigA protein (Fig. 5D) and production of a large orange zone in the CAS assay (Fig. 6B, sector 3). Similarly, the presence of the wild-type F. tularensis LVS figA gene (in pKD108) in this F. novicida figA mutant also resulted in the production of a readily detectable orange zone in the CAS assay (Fig. 6B, sector 5). The presence of only the pFNLTP-CAT vector in this same mutant did not affect the phenotype of this strain in the CAS assay (Fig. 6B, sector 4).
Effect of the figA mutation on extracellular growth.
To address whether inactivation of the figA gene in F. tularensis LVS and F. novicida U112 had any effect on extracellular growth, the figA mutants were compared with their respective parent strains for the ability to grow in broth medium under iron-replete and iron-restricted conditions. Chelation of free iron in MH−/DF medium was shown to inhibit growth of the F. novicida U112 wild-type strain (data not shown). However, unlike F. tularensis LVS, which had to be prestarved for iron to obtain iron-restricted growth, F. novicida U112 was more sensitive to iron limitation and did not require prestarvation in MH− medium prior to inoculation into MH−/DF. As shown in Fig. 7A and B, respectively, the F. tularensis LVS and F. novicida U112 wild-type strains and their respective figA mutants grew at similar rates and to the same extent in iron-replete medium. Similarly, both wild-type strains and their figA mutants exhibited the same degree of growth inhibition in MH−/DF medium.
FIG. 7.
Extracellular and intracellular growth of wild-type and mutant strains of F. tularensis LVS and F. novicida U112. For extracellular growth (A and B), bacteria were grown in both iron-replete (filled symbols) and iron-restricted (open symbols) media. The wild-type F. tularensis LVS strain (panel A, circles), its figA mutant (A, triangles), the wild-type F. novicida strain (B, circles), and its figA mutant (B, triangles) were grown as described in Materials and Methods. To assess intracellular growth (C and D), these same strains were grown in the macrophage cell line J774A.1 as described in Materials and Methods. (C) Wild-type F. tularensis LVS (filled columns) and its figA mutant (open columns). (D) Wild-type F. novicida (filled columns) and its figA mutant (open columns). These data are the means from two independent experiments. In panel D, the difference between the F. novicida wild-type and mutant strains was significant only at 24 h postinfection (asterisk; P = 0.0096, Student's t test).
When the F. novicida wild-type strain and its figA mutant, which exhibited the greatest difference in the CAS assay (Fig. 6), were inoculated separately onto CAS/CDM− agar, only the wild-type strain exhibited abundant growth (data not shown). To determine whether the siderophore-like activity secreted by the F. novicida wild-type strain could enhance the ability of its figA mutant to grow under these iron-restricted conditions, a cross-feeding experiment was performed. When F. novicida U112 figA::np-kan (Fig. 8A, streaks 3 and 4) was streaked in parallel on a CAS/CDM− plate, this mutant grew very poorly compared to this same mutant (Fig. 8B, streaks 3 and 4) streaked in parallel on CDM+ medium. In contrast, when this figA mutant (Fig. 8A, streak 2) was streaked alongside the F. novicida wild-type strain (Fig. 8A, streak 1), this mutant apparently grew as well as the wild-type strain. These results indicated that the siderophore-like activity evidenced by F. novicida U112 in the CAS assay was able to enhance the growth of its figA mutant under iron-restricted conditions.
FIG. 8.
Cross-feeding of the F. novicida figA mutant by the F. novicida U112 wild-type (w.t.) strain. The F. novicida U112 wild-type strain (streak 1) and its figA mutant (streaks 2, 3, and 4) were inoculated onto both CAS/CDM− (A) and CDM+ (B) agar plates and grown for 36 h.
Effect of the figA mutation on intracellular growth.
The intracellular growth abilities of these wild-type and mutant strains were compared in mouse macrophage cell line J774A.1. Over the 48-h experimental period postinfection, there was no significant difference between wild-type F. tularensis LVS and its figA mutant (Fig. 7C). In contrast, the F. novicida U112 figA mutant appeared to grow less well in these macrophages although the difference was statistically significant only at the 24-h time point (Fig. 7D).
DISCUSSION
In marked contrast to the abundance of information about iron acquisition by both enteric and nonenteric gram-negative pathogens, the data available concerning iron uptake by F. tularensis are rudimentary at best. Moreover, much of the published data on this subject date back more than 4 decades. Studies by early workers indicated that in vitro growth of small inocula of the virulent SCHU-S4 strain of F. tularensis was stimulated by ferric chloride, ferric sulfate (41), and two ferric hydroxamates, ferrichrome and ferrioxamine B (14). In addition, F. tularensis has been shown to produce a growth-stimulating compound whose function could be at least partially replaced by iron salts or siderophores (14). This latter finding raised the possibility that F. tularensis could produce some type of siderophore. Other facultative intracellular pathogens, including Y. pestis (32) and Mycobacterium species (18), have been shown to produce siderophores. Moreover, L. pneumophila, long thought to be unable to produce a siderophore, was recently shown to release an iron chelator into culture supernatant fluid under certain growth conditions (23) and gene products involved in the expression of this siderophore have been identified (1).
In the present study, a combination of genomic screening, DNA microarray technology, and mutant analysis was used to identify F. tularensis genes encoding products whose expression was affected by iron limitation. Prior to this study, there was a single published report in which a rifampin-resistant mutant of the F. tularensis LVS was grown under conditions of limited iron availability (2). These authors subjected this organism to a number of stresses, including increased temperature, low pH, and iron limitation, independently. SDS-PAGE analysis of cell envelopes obtained from these bacteria grown under iron-replete and iron-restricted conditions showed apparent increases in the expression of a few proteins, but these proteins were not identified (2).
To obtain conditions under which iron was the limiting factor for growth, F. tularensis LVS had to be starved for iron by growth in MH− broth overnight prior to inoculation into MH−/DF medium. Interestingly, prestarvation of F. novicida was not necessary to achieve iron-limited growth, a result which suggests that these two Francisella species may have different iron storage capabilities. Under iron-limiting conditions, a few F. tularensis LVS proteins detectable in cell envelope preparations were apparently up-regulated and at least two of these (IglC and PdpB) were identified by mass spectrometry. Examination of the nucleotide sequence of the iglC and pdpB genes in the F. tularensis Schu4 genome (19) revealed the presence of putative Fur boxes immediately upstream of both of these ORFs (Fig. 3). Both iglC and pdpB are located in possible operons (iglABCD and pdpAB, respectively) in the Schu4 genome, and examination of the nucleotide sequences immediately upstream from both iglA and pdpA also showed the presence of putative Fur boxes (data not shown). When the consensus Fur box sequence was used to search the entire Schu4 genome, the putative Fur box that had the highest degree of identity (17/19) with the consensus sequence was located in front of the figA gene and overlapped with a second predicted Fur box. In L. pneumophila, the iron-regulated gene lbtA, which encodes a protein that may be functionally similar to FigA, also has two overlapping Fur boxes (1).
Subsequent DNA microarray analysis confirmed that expression of the figA gene, as well as that of about 80 other F. tularensis LVS ORFs, was up- or down-regulated at least twofold by growth under iron-restricted conditions (Table S1 in the supplemental material). These iron-regulated ORFs included both the iglC and pdpB genes previously shown by protein analysis to be up-regulated under these growth conditions (Fig. 2). Genes that were repressed under iron starvation were mainly those involved in protein synthesis (Table 2). These DNA microarray data were verified for several ORFs by the use of qRT-PCR (Table 3). The genes located in the recently identified F. tularensis pathogenicity island involved in intracellular growth (27), including pdpA, pdpB, pdpC, iglA, iglB, iglC, and iglD, were all up-regulated at least twofold under iron-restricted conditions (Table S1 in the supplemental material). It is possible that, under this iron-restricted growth condition in vitro, the bacteria may sense an environment similar to that encountered within macrophages. The relatively low concentrations of iron encountered in the host cell may function as a signal to the bacterium to increase the production of virulence or survival factors that would be beneficial in this location. It must be noted that MglA has been shown to be a transcriptional activator for these genes (21), and additional studies are necessary to determine the relative importance of iron limitation in affecting expression of these genes.
Among the genes that were up-regulated by iron restriction, the tightly linked figABCD genes had the greatest increases in expression as assessed by both DNA microarray and qRT-PCR analyses. The figA gene was found to be located ∼250 nt downstream from the predicted fur gene in the Schu4 genome. The figABCD ORFs, together with the predicted fur gene, likely constitute an operon on the basis of RT-PCR analysis (Fig. 4). Interestingly, expression of the fur gene was apparently not affected substantially by iron limitation (Table S1 in the supplemental material). Exactly how fur and these other four genes are differentially regulated by iron restriction remains to be determined.
F. tularensis LVS and F. novicida U112 figA mutants were constructed with a kanamycin resistance cartridge (26) designed to prevent or minimize polar effects on downstream ORFs. Under iron-restricted conditions in broth, these two figA mutants grew at a rate and to a final density that were similar, if not identical, to those obtained with the respective wild-type parent strains (Fig. 7). The intracellular growth ability of the F. tularensis LVS figA mutant was the same as that of its parent wild-type strain, whereas the figA mutant of F. novicida seemed to be slightly deficient in the ability to grow in macrophages (Fig. 7). Further investigation of these mutants did reveal a striking difference between the F. novicida U112 figA mutant and its wild-type parent strain in an assay originally designed to detect siderophore production by enteric bacteria (31). Interestingly, an L. pneumophila mutant unable to express the LbtA protein, which resembles FigA, was also deficient in siderophore production as assessed in a CAS-based assay (1). A difference between the F. tularensis LVS wild-type strain and its figA mutant (Fig. 6A) was detected in the CAS assay but was very modest compared to that observed with the F. novicida wild-type-mutant pair. This latter pair of strains was also used in a cross-feeding experiment (Fig. 8) which demonstrated that the siderophore-like activity released by F. novicida in the CAS assay could enhance the growth of the F. novicida figA mutant under these iron-restricted conditions on CAS/CDM− agar.
The role of the FigA protein in the CAS plate phenotype was confirmed by the use of complementation analysis (Fig. 6B). When the cloned F. novicida figA gene in pKD107 was expressed in the F. tularensis LVS figA mutant, this complementation in trans did not increase the reactivity of this mutant in the CAS assay even though the FigA protein could be detected by Western blot analysis (data not shown). However, when we cloned and expressed the wild-type F. tularensis LVS figA gene in the F. novicida figA mutant, this recombinant did express a readily detectable reaction zone on this same medium (Fig. 6B). Taken together, these results raise the possibility that the very weak reactivity of the wild-type F. tularensis LVS in the CAS assay is the result of a secondary mutation elsewhere in the genome that affects the synthesis or release of the siderophore-like activity and is not caused by a nonfunctional F. tularensis LVS FigA protein. It is also noteworthy that expression of the cloned F. novicida figA gene in F. novicida was subject to regulation by iron availability (data not shown).
This study provides the first identification of iron-regulated genes in F. tularensis LVS and evidence for the production of a siderophore-like molecule by F. novicida U112. Whether this siderophore-like molecule, the FigA protein, or both are necessary for virulence expression by F. tularensis remains to be determined.
Supplementary Material
Acknowledgments
This study was supported by U.S. Public Health Service grant PO1 AI55637.
We thank David Rasko for assistance with bioinformatics, Patrick Conley and Michael Norgard for providing rat antiserum to the FopA protein, Wei Wang for assistance with DNA microarray analysis, Simon Daefler for instruction in the use of the macrophage infection assay, Bruce Green for supplying the ΔEcat cartridge, Shelley Payne for providing E. coli W3110N and its entF mutant, Thomas Zahrt for providing plasmid pFNLTP6, and Karl Klose for providing the F. novicida U112 iglC mutant.
Editor: J. L. Flynn
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatnagar, N. B., K. L. Elkins, and A. H. Fortier. 1995. Heat stress alters the virulence of a rifampin-resistant mutant of Francisella tularensis LVS. Infect. Immun. 63:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blick, R. J., A. T. Revel, and E. J. Hansen. 2003. FindGDPs: identification of primers for labeling microbial transcriptomes for DNA microarray analysis. Bioinformatics 19:1718-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. M., V. Stone, P. Findlay, W. MacNee, and K. Donaldson. 2000. Increased inflammation and intracellular calcium caused by ultrafine carbon black is independent of transition metals or other soluble components. Occup. Environ. Med. 57:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronquist, S. D. 2004. Tularemia: the disease and the weapon. Dermatol. Clin. 22:313-320. [DOI] [PubMed] [Google Scholar]
- 8.Crosa, J. H., A. R. Mey, and S. M. Payne. 2004. Iron transport in bacteria. ASM Press, Washington, D.C.
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, K., J. L. Latimer, D. A. Lewis, and E. J. Hansen. 2001. Investigation of the interaction among the components of the cytolethal distending toxin of Haemophilus ducreyi. Biochem. Biophys. Res. Commun. 285:609-615. [DOI] [PubMed] [Google Scholar]
- 11.Elkins, K. L., S. C. Cowley, and C. M. Bosio. 2003. Innate and adaptive immune responses to an intracellular bacterium, Francisella tularensis live vaccine strain. Microbes Infect. 5:135-142. [DOI] [PubMed] [Google Scholar]
- 12.Forsman, M., G. Sandstrom, and A. Sjostedt. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38-46. [DOI] [PubMed] [Google Scholar]
- 13.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215:53-56. [DOI] [PubMed] [Google Scholar]
- 14.Halmann, M., and J. Mager. 1967. An endogenously produced substance essential for growth initiation of Pasteurella tularensis. J. Gen. Microbiol. 49:461-468. [Google Scholar]
- 15.Hansen, E. J., J. L. Latimer, S. E. Thomas, M. Helminen, W. L. Albritton, and J. D. Radolf. 1992. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J. Bacteriol. 174:5442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollis, D. G., R. E. Weaver, A. G. Steigerwalt, J. D. Wenger, C. W. Moss, and D. J. Brenner. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, S. J., P. S. Marshall, R. J. Upton, C. Ratledge, and M. Ewing. 1995. Novel extracellular mycobactins, the carboxymycobactins from Mycobacterium avium. Tetrahedron Lett. 36:4129-4132. [Google Scholar]
- 19.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 20.Lauriano, C. M., J. R. Barker, F. E. Nano, B. P. Arulanandam, and K. E. Klose. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195-202. [DOI] [PubMed] [Google Scholar]
- 21.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 101:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavrrar, J. L., and M. A. McIntosh. 2003. Architecture of a fur binding site: a comparative analysis. J. Bacteriol. 185:2194-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liles, M. R., T. A. Scheel, and N. P. Cianciotto. 2000. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 182:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, S., D. S. Thaler, and A. Libchaber. 2002. Signal and noise in bridging PCR. BMC Biotechnol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 70:7511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967-978. [DOI] [PubMed] [Google Scholar]
- 29.Paabo, S., D. M. Irwin, and A. C. Wilson. 1990. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265:4718-4721. [PubMed] [Google Scholar]
- 30.Patrick, C. C., A. Kimura, M. A. Jackson, L. Hermanstorfer, A. Hood, G. H. McCracken, Jr., and E. J. Hansen. 1987. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypable Haemophilus influenzae. Infect. Immun. 55:2902-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 32.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145(Pt. 5):1181-1190. [DOI] [PubMed] [Google Scholar]
- 33.Petersen, J. M., and M. E. Schriefer. 2005. Tularemia: emergence/re-emergence. Vet. Res. 36:455-467. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, G. M., and I. Smith. 2003. Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol. Microbiol. 47:1485-1494. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969-979. [DOI] [PubMed] [Google Scholar]
- 37.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:134-146. [DOI] [PubMed] [Google Scholar]
- 38.Saslaw, S., H. T. Eigelsbach, H. E. Wilson, J. A. Prior, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107:121-133. [DOI] [PubMed] [Google Scholar]
- 39.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 40.Sjostedt, A. 2005. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes. Infect. 8:561-567. [DOI] [PubMed] [Google Scholar]
- 41.Tresselt, H. B., and M. K. Ward. 1964. Blood-free medium for the rapid growth of Pasteurella tularensis. Appl. Microbiol. 12:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg, E. D. 1993. The iron-withholding defense system. ASM News 59:559-562. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.