Abstract
Autoinducer 2 (AI-2) produced by the oral pathogen Actinobacillus actinomycetemcomitans influences growth of the organism under iron limitation and regulates the expression of iron uptake genes. However, the cellular components that mediate the response of A. actinomycetemcomitans to AI-2 have not been fully characterized. Analysis of the complete genome sequence of A. actinomycetemcomitans (www.oralgen.lanl.gov) indicated that the RbsB protein was related to LuxP, the AI-2 receptor of Vibrio harveyi. To determine if RbsB interacts with AI-2, the bioluminescence of the reporter strain V. harveyi BB170 (sensor 1−, sensor 2+) was determined after stimulation with partially purified AI-2 from A. actinomycetemcomitans or conditioned medium from V. harveyi cultures in the presence and absence of purified six-His-tagged RbsB. RbsB efficiently inhibited V. harveyi bioluminescence induced by both A. actinomycetemcomitans AI-2 and V. harveyi AI-2 in a dose-dependent manner, suggesting that RbsB competes with LuxP for AI-2. Fifty percent inhibition occurred with approximately 0.3 nM RbsB for A. actinomycetemcomitans AI-2 and 15 nM RbsB for V. harveyi AI-2. RbsB-mediated inhibition of V. harveyi bioluminescence was reversed by the addition of 50 mM ribose, suggesting that A. actinomycetemcomitans AI-2 and ribose bind at the same site of RbsB. The RbsB/AI-2 complex was thermostable since A. actinomycetemcomitans AI-2 could not be recovered by heating. This was not due to heat inactivation of A. actinomycetemcomitans AI-2 since signal activity was unaffected by heating in the absence of RbsB. Furthermore, an isogenic A. actinomycetemcomitans mutant that was unable to express rbsB was deficient in depleting A. actinomycetemcomitans AI-2 from solution relative to the wild-type organism. Inactivation of rbsB also influenced the ability of the organism to grow under iron-limiting conditions. The mutant strain attained a cell density of approximately 30% that of the wild-type organism under iron limitation. In addition, real-time PCR showed that the expression of afuABC, encoding a major ferric ion transporter, was reduced by approximately eightfold in the rbsB mutant. This phenotype was similar to that of a LuxS-deficient mutant of A. actinomycetemcomitans that is unable to produce AI-2. Together, our results suggest that RbsB may play a role in the response of A. actinomycetemcomitans to AI-2.
Quorum sensing is a mechanism that allows a bacterial population to monitor its cell density through the action of soluble signal molecules called autoinducers, which may be acylated homoserine lactones (15, 24), peptides (8), quinolones (25), or furan derivatives (40). The concentration of autoinducer increases as the microbial population grows and eventually reaches a critical threshold that triggers changes in gene expression. It is thought that this process may allow organisms to synchronize the expression of genes that may be required for community survival (2, 13, 38).
Autoinducer 2 (AI-2) is a furan-like quorum-sensing signal that was initially identified in Vibrio harveyi (3) and is produced by the luxS gene (28). The enzyme encoded by luxS cleaves S-ribosylhomocysteine to produce homocysteine and 4,5-dihydroxy-2,3-pentanedione (39), which in turn may undergo further rearrangement to produce AI-2. At least two structural forms of AI-2 have currently been identified: Salmonella enterica serovar Typhimurium produces 2R,4S-2,3,3,4-methyltetrahydroxytetrahydrofuran (R-THMF), whereas V. harveyi produces the borate diester form of 2S,4S,-2,3,3,4-methyltetrahydroxytetrahydrofuran (S-THMF) (6, 22). In addition, Sperandio et al. suggest that LuxS may be required to produce another signal in Escherichia coli, designated AI-3 (32). However, the structure of AI-3 and its similarity to the known AI-2 isoforms are not currently known. AI-2 regulates the expression of a variety of genes in Vibrio species, including the lux operon of V. harveyi. Detection of AI-2 by V. harveyi is mediated by LuxP, a periplasmic AI-2 receptor (6) which activates the LuxQ sensor kinase/phosphatase and initiates a phosphotransfer cascade involving LuxU (12) and the response regulator LuxO (11). LuxO in turn influences the expression of small RNAs that are bound by Hfq, an RNA chaperone. This complex destabilizes the luxR transcript encoding the activator of the lux operon (17).
Interestingly, luxS is highly conserved in a wide range of gram-positive and gram-negative bacteria, and many, if not all, of these organisms produce an AI-2-like signal that is capable of inducing the expression of the lux operon of V. harveyi. As a result, AI-2 has been suggested to represent a universal signal that is recognized by many organisms (40). AI-2 has been reported to influence a wide variety of cellular processes, including type III secretion (31), cell motility (18, 30), the development of biofilms (4, 7, 14, 20, 21, 41), the expression of virulence factors (10, 16, 19), and iron uptake (9). However, in organisms outside of the genus Vibrio, the mechanism that governs the response to AI-2 is not well understood since some of the downstream proteins that are involved in AI-2-dependent signal transduction in Vibrio spp. are absent or not well conserved in these organisms.
Our studies have focused on the oral pathogen Actinobacillus actinomycetemcomitans, a gram-negative organism that is associated with aggressive forms of early-onset periodontitis and other systemic infections (5, 23, 29, 42). We previously showed that A. actinomycetemcomitans produces an AI-2-like signal that induces Vibrio harveyi bioluminescence (10) and regulates growth of the organism under iron limitation and expression of a variety of iron storage and uptake genes (9). However, A. actinomycetemcomitans appears to lack direct homologs of the AI-2 sensor kinase/phosphatase (LuxQ) and phosphotransfer protein (LuxU) that are dedicated to the Vibrio harveyi response to AI-2. However, this organism does possess two genes that encode polypeptides (RbsB and LsrB) related to LuxP, the receptor for AI-2 in V. harveyi. The A. actinomycetemcomitans RbsB protein is similar to the E. coli K-12 and S. enterica serovar Typhimurium LT2 periplasmic ribose binding proteins (76% sequence identity to each) encoded by the rbs operon (rbsDABCK), which functions to transport ribose into the cell. LsrB is homologous (80% sequence identity) to the LsrB polypeptide of S. enterica serovar Typhimurium (35), which is part of an ABC transporter involved in the uptake of AI-2. Presumably, LsrB functions in a similar manner in A. actinomycetemcomitans since the lsr operon is highly conserved with the S. enterica serovar Typhimurium gene cluster. Since AI-2 is derived from the ribose moiety of S-ribosylhomocysteine, we hypothesized that the rbsDABCK operon may also function in the transport of AI-2 in A. actinomycetemcomitans. The focus of this study was to determine if the periplasmic ribose binding protein RbsB interacts with AI-2 and represents a potential receptor for AI-2 in A. actinomycetemcomitans. We show that purified RbsB protein inhibits AI-2-mediated induction of V. harveyi bioluminescence. An A. actinomycetemcomitans mutant that does not express RbsB was deficient in depleting AI-2 from solution relative to the parent strain. The mutant also failed to attain high cell density under iron-limiting growth conditions and exhibited reduced expression of a ferric ABC transport system. Together, these results suggest that RbsB may play a role in the response of A. actinomycetemcomitans to AI-2.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A. actinomycetemcomitans strains were cultured on brain heart infusion medium (Difco, Detroit, MI) containing 0.5% yeast extract and supplemented with 40 mg of NaHCO3 per liter. Cultures were maintained at 37°C in an atmosphere of 5% CO2. Growth of the rbsB mutant strain of A. actinomycetemcomitans was carried out in the medium described above, which was further supplemented with 25 μg per ml kanamycin. E. coli strains were grown in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) with aeration at 37°C. E. coli strains containing plasmids pQE60 and pRE4 were cultured as described above in LB supplemented with 100 μg per ml ampicillin and 25 μg per ml kanamycin. V. harveyi BB170 (sensor 1−, sensor 2+) was a gift from B. Bassler (Princeton University) and was grown overnight with aeration in AB medium (34) at 30°C. AB medium consists of 10 mM potassium phosphate, pH 7.0, 0.3 M NaCl, 0.05 M MgSO4, 0.2% vitamin-free Casamino Acids (Difco), 2% glycerol, 1 mM l-arginine, 1 μg per ml thiamine, and 0.01 μg per ml riboflavin.
Expression and purification of A. actinomycetemcomitans RbsB.
A. actinomycetemcomitans rbsB was identified from the complete genome sequence of A. actinomycetemcomitans HK1651 (26; www.oralgen.lanl.gov) and amplified from A. actinomycetemcomitans genomic DNA using primers rbsNS5 (5′-GGAGATATACCATGGAAAAACTAACCACATTAGCC-3′) and rbs3B (5′-CCGGATCCTTCGCTGATGACTTT-3′). The underlined sequences in the primers represent NcoI and BamHI restriction sites used to facilitate cloning into the expression plasmid pQE60. Amplification was carried out using the following profile: 95°C for 10 min followed by 30 cycles of 95°C for 1 min, 60°C for 2 min, and 72°C for 3 min. For some reactions, an overnight soak at 4°C followed the profile described above. The resulting PCR product was ligated with pGEM-T Easy, and the ligation product was transformed into E. coli DH5α to produce strain pGEMrbsB. Recombinant clones were confirmed by restriction digestion with EcoRI to release the rbsB insert. Plasmid pGEMrbsB was subsequently digested with NcoI and BamHI (specified by the primers listed above), and the resulting fragment was isolated from a 1% agarose gel and ligated with pQE60 digested with NcoI and BamHI. After transformation into E. coli DH5α (which contains a mutation in luxS and does not produce AI-2), recombinant clones were confirmed by restriction analysis. The resulting E. coli strain, QE60rbsB, was used for expression of the RbsB polypeptide.
E. coli QE60rbsB was grown to early exponential phase in 100 ml LB supplemented with ampicillin (100 μg per ml) and kanamycin (25 μg/ml), and expression of rbsB was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Growth was continued with aeration for an additional 3 to 4 h, after which cells were harvested, suspended in 4 ml of loading buffer (20 mM sodium phosphate, pH 7.5, 0.5 M NaCl, 10 mM imidazole), lysed, and centrifuged at 10,000 × g to remove cellular debris. RbsB was purified from the resulting supernatant by affinity chromatography on nickel resin (HisTrap resin; Amersham Pharmacia Biotech.) The column was first washed with distilled H2O (10 ml), followed by equilibration with an equal volume of loading buffer. The protein extract was loaded by gravity, and the column was subsequently washed with 20 ml of loading buffer. Bound RbsB was eluted with 10 ml of elution buffer (20 mM sodium phosphate, pH 7.5, 0.5 M NaCl, 0.5 M imidazole). The eluted protein was dialyzed overnight against 10 mM sodium phosphate, pH 7.5, and lyophilized. Prior to use, RbsB was suspended in distilled H2O and protein concentration was determined using a Micro bicinchoninic acid assay kit from Pierce according to the manufacturer's instructions. Purity was assessed by visualizing the polypeptide by Coomassie blue staining after polyacrylamide gel electrophoresis (PAGE) on 4 to 15% gradient gels and by Western blotting using polyclonal anti-six-His antibodies.
Determination of V. harveyi bioluminescence.
Induction of V. harveyi bioluminescence was carried out essentially as described by Surette and Bassler (34). Preparations of V. harveyi AI-2 were obtained from conditioned medium of overnight V. harveyi cultures grown in AI broth (34). After cells were harvested, the medium was filtered through a 0.22-μm-pore-size filter and used immediately or stored at −70°C. AI-2 from A. actinomycetemcomitans was partially purified by chromatography of conditioned medium from an exponential-phase culture on a Sep-Pak C18 column (Waters) as described by Sperandio et al. (32). Briefly, cells were harvested from 4 ml of culture and the supernatant was filtered through a 0.22-μm-pore-size filter and subsequently through a Centricon YM-3 1-kDa exclusion filter (Millipore Co.). The filtered solution was lyophilized and concentrated by resuspension in 200 μl of cold 5 mM sodium phosphate, pH 6.2. Chromatography on a C18 Sep-Pak column was carried out according to the instructions of the manufacturer. AI-2 activity was present in the column flowthrough fraction.
For the determination of V. harveyi bioluminescence, an aliquot of an overnight V. harveyi culture was diluted 1:5,000 into fresh sterile AB medium and 80 μl of cells was added to wells on a 96-well microtiter dish. Positive-control wells received cell-free conditioned medium from V. harveyi or partially purified A. actinomycetemcomitans AI-2 (20 μl). Negative-control wells received 20 μl sterile AB medium. Experimental wells received 20 μl of V. harveyi or A. actinomycetemcomitans AI-2 that was preincubated for 30 min at 30°C with various concentrations of purified RbsB protein. Samples were then added to wells containing the diluted V. harveyi cells to give a final concentration of 0.1 to 5,000 ng per ml RbsB. The microtiter plate was shaken on a rotary shaker at 500 rpm at 30°C, and bioluminescence was measured at half-hour or hourly intervals by use of a Wallac Victor3 1420 MultiLabel counter (PerkinElmer). Bioluminescence of each well was initially calculated as induction (n-fold) of light relative to that for the negative-control reaction (V. harveyi BB170 cells incubated in sterile AI medium). Bioluminescence of the positive-control reaction was subsequently normalized to a value of 1.0, and experimental data were calculated as percentages of the positive-control value.
Ribose-dependent reversal of RbsB inhibition of V. harveyi bioluminescence was determined as follows. V. harveyi or A. actinomycetemcomitans AI-2 was added to diluted V. harveyi BB170 cells (in the absence of RbsB), and the reaction mixtures were incubated as described above until the onset of the induction of bioluminescence in the positive control (3-h time point). At that time point, all reaction mixtures were removed from the shaker and experimental wells received purified RbsB protein (to a final concentration of 2.5 to 10 μg per ml) or RbsB protein (2.5 μg per ml final concentration) in the presence of ribose (0 to 500 mM final reaction concentration). After the addition of RbsB or RbsB-ribose, plates were incubated with shaking for an additional 1 to 2 h to allow the induction of V. harveyi bioluminescence to continue, and then light production was determined as described above. Data were analyzed as already described.
The heat stability of RbsB, AI-2, and the RbsB/AI-2 complex was determined by heating the reactants at the desired temperature (37°C to 100°C) for 15 min prior to addition to diluted V. harveyi BB170 cells. Light production by the reporter strain was monitored as described above.
Inactivation of A. actinomycetemcomitans rbsB.
Inactivation of the rbsB gene was carried out with suicide plasmid pGEMrbsKan, which does not replicate in A. actinomycetemcomitans. A portion of the A. actinomycetemcomitans rbsB operon spanning the rbsC-rbsB genes was amplified from A. actinomycetemcomitans genomic DNA by using oligonucleotides R5 (5′-ATTCTCAACATTTTGCGCC-3′) and R3 (5′-CGGCAACGTTGTCGGATGC-3′). R5 anneals in the rbsC gene upstream from the rbsB coding region in the rbs operon (rbsDACBK). The reverse primer R3 anneals within rbsB. The 1,250-bp fragment was ligated into pGEM-T and transformed into E. coli DH5α. The resulting plasmid was isolated, digested with PstI, and subsequently modified by inserting the kanamycin resistance marker obtained by PstI digestion of pUC4K (Promega). Kanr Ampr clones were selected, and pGEMrbsKan was confirmed by restriction digestion.
pGEMrbsKan (2 μg) was introduced into A. actinomycetemcomitans by electroporation essentially as described by Sreenivasan et al. (33), and Kanr Ampr clones were selected. To confirm Campbell-type integration of the plasmid into the A. actinomycetemcomitans genome, genomic DNA was isolated and used as a template for PCR amplification with primers rbsC5 (5′-TGCTTATTGGCGTCGTGTCGG-3′) and kan3 (5′-GTGCAATGTAACATCAGAG-3′). Primer rbsC5 anneals in rbsC immediately upstream from the annealing site of primer R5 and therefore does not anneal to the suicide plasmid. Primer kan3 anneals within the kanamycin resistance gene of pUC4K. Together, these primers will generate a 1.4-kbp product from A. actinomycetemcomitans genomic DNA only if pGEMrbsKan is integrated in the rbs operon. Clones exhibiting the desired 1.4-kbp product were chosen for further study. The resulting mutant contains intact rbsDAC genes but is unable to express rbsB and the downstream rbsK gene.
To complement the rbsB mutation, a plasmid-borne copy of rbsB under the control of the A. actinomycetemcomitans leukotoxin promoter was transformed by electroporation into the mutant strain. The rbsB gene was amplified from the A. actinomycetemcomitans genome by use of primers RC5 (5′-CCGGATCCAAAAACTAACCACATTAGCC) and RC3 (5′-CCTCTAGATTATTCGCTGATGACTTTGAG-3′), which possess restriction sites for BamHI and XbaI, respectively. The resulting fragment contains the entire rbsB open reading frame except for the start codon and also contains the translational stop codon. After digestion with BamHI and XbaI, the fragment was ligated in frame into pBluescript-ltx, which contains a fragment encompassing the A. actinomycetemcomitans leukotoxin promoter, its ribosome binding site, and the start codon of the leukotoxin operon. The desired plasmid, designated pBSrbsB, was identified by restriction analysis after transformation of E. coli DH5α. Plasmid pBSrbsB was subsequently cleaved with KpnI and XbaI, and the ltx-rbsB fragment was subsequently ligated into the A. actinomycetemcomitans shuttle vector pYGS (9). After transformation by electroporation into the rbsB knockout strain, clones exhibiting resistance to both kanamycin and streptomycin were selected. The presence of pYGSrbsB in these clones was confirmed by restriction analysis.
Depletion of AI-2 by wild-type and RbsB-deficient strains of A. actinomycetemcomitans.
Overnight cultures of A. actinomycetemcomitans strains were harvested by centrifugation, washed twice with sterile AB medium, and suspended in 0.5 ml of partially purified A. actinomycetemcomitans AI-2 at a cell density of approximately 4 × 108 cells per ml. Cell suspensions were incubated at various times up to 30 min at 37°C. The bacteria were then removed by centrifugation, and the supernatant was filtered through a 0.22-μm-pore-size filter. Filtered supernatants were subsequently analyzed in triplicate for AI-2 activity by using the V. harveyi BB170 reporter strain. Bioluminescence was measured and analyzed as described above.
Growth of A. actinomycetemcomitans strains under iron limitation.
The growth of A. actinomycetemcomitans strains under iron-limiting conditions was determined by use of brain heart infusion medium supplemented with 100 μM ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) essentially as previously described (9). EDDHA was incubated overnight with medium prior to inoculation with A. actinomycetemcomitans. All cultures were inoculated at an identical cell density (2 × 108 CFU/ml) and were incubated at 37°C for 6 h. The optical density at 600 nm of each culture was determined. Cell density, expressed in CFU per milliliter, was calculated as previously described (9) by using standard curves of the optical density at 600 nm versus CFU (per ml) for each strain.
RNA isolation and real-time PCR.
A. actinomycetemcomitans total RNA was isolated from cells by using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. The RNA preparations were subsequently digested with RQ RNase-free DNase I (Promega) to remove contaminating genomic DNA. Prior to conducting real-time reactions, samples were assayed for the presence of genomic DNA by standard PCR using primers afuA5 (5′-GGAAGCCTTTACTGCTGTGCG-3′) and afuA3 (5′-TCCTGCCTGTTCAATCAATT-3′) and A. actinomycetemcomitans genomic DNA as a template. Samples that produced the 150-bp product were discarded or treated again with DNase until a negative result was obtained by normal PCR. For real-time reverse transcription-PCR, first-strand cDNA was prepared by using Ready-To-Go You-Prime first-strand beads (Amersham Pharmacia) as described by the manufacturer, with primer afuA3 and 2 μg total RNA. The resulting cDNA was amplified using a SmartCycler system (Cepheid) with a final reaction volume of 25 μl that contained 8 μl of first-strand cDNA mix, primer mix (0.3 μM each primer), 0.5× SYBR green dye (Roche Applied Science), and 2.5 U of Taq polymerase (Roche Applied Science). Amplification conditions for real-time PCR were as follows: denaturation at 95°C for 30 s, annealing at 62°C for 30 s, and elongation at 72°C for 30 s for 40 cycles. The cycle threshold was determined from a second derivative plot of total fluorescence as a function of cycle number by using the analysis software supplied with the SmartCycler system. Real-time PCRs were carried out at least twice using independently isolated RNA samples with consistent results. Control reactions were carried out as described above but using the primers rpoA5 (5′-TGAAGTAGAGATTGATGG-3′) and rpoA3 (5′-GACAGATTACATGCTCCGG-3′).
RESULTS
A. actinomycetemcomitans RbsB inhibits AI-2-dependent induction of V. harveyi bioluminescence.
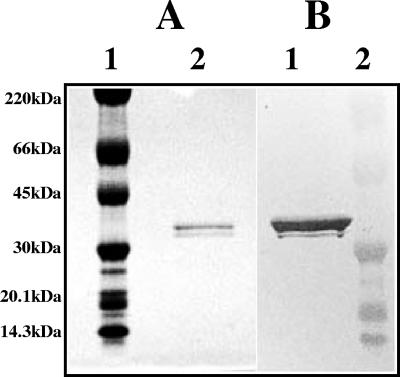
To determine if RbsB interacts with A. actinomycetemcomitans AI-2, purified protein was incubated with partially purified A. actinomycetemcomitans AI-2 for 30 min at 30°C and the mixture was subsequently added to the AI-2 indicator strain V. harveyi BB170. The RbsB polypeptide was expressed with a C-terminal six-His tag in E. coli DH5α, which cannot produce AI-2 (34), and was purified by affinity chromatography on nickel resin (HisTrap resin). The purified protein migrated as a doublet with a molecular mass of approximately 34 kDa (consistent with the value of 32 kDa predicted from the deduced amino acid sequence of rbsB) (Fig. 1A) and reacted with polyclonal anti-six-His antibodies (Fig. 1B). The doublet likely arises from incomplete processing of the RbsB signal sequence in the E. coli host. Incubation of purified RbsB with V. harveyi BB170 and A. actinomycetemcomitans AI-2 potently inhibited the induction of V. harveyi bioluminescence in a dose-dependent manner (Fig. 2A). The level of V. harveyi bioluminescence that was induced in the positive-control reaction was approximately 1,300-fold greater than light production by V. harveyi BB170 incubated with sterile AB medium. The concentration of RbsB required to inhibit A. actinomycetemcomitans AI-2-mediated induction of V. harveyi bioluminescence by 50% was approximately 1 ng per ml (0.35 nM). Interestingly, RbsB was also an effective inhibitor of bioluminescence induced by AI-2 from V. harveyi (Fig. 2B), with 50% inhibition of light induction occurring in the presence of approximately 40 ng per ml of RbsB protein. Light induction by the positive control in these reactions was approximately 1,100-fold greater than that achieved by the negative control. Addition of bovine serum albumin instead of RbsB to V. harveyi BB170 cells stimulated with A. actinomycetemcomitans AI-2 or V. harveyi AI-2 had no effect on the production of light (data not shown). These results suggest that exogenously added A. actinomycetemcomitans RbsB effectively competes with V. harveyi LuxP for AI-2 and that RbsB is capable of interacting not only with its cognate signal but also with a heterologous signal produced by an unrelated organism.
FIG. 1.
Expression of A. actinomycetemcomitans RbsB polypeptide. The rbsB gene was amplified from A. actinomycetemcomitans genomic DNA by use of primers derived from the complete genome sequence of A. actinomycetemcomitans HK1651 (26). Cloning of the amplified product into the vector pQE60, expression, and affinity purification of RbsB protein were done as described in Materials and Methods. (A) Coomassie-stained gel subjected to PAGE. Lane 1, size markers; lane 2, affinity-purified RbsB. (B) Western blot of gel subjected to PAGE (from panel A). RbsB was visualized using anti-six-His monoclonal antibody. The doublet likely arises from incomplete processing of the RbsB signal sequence in the E. coli host. Molecular size markers are labeled on the left.
FIG. 2.
RbsB inhibits V. harveyi BB170 bioluminescence induced by A. actinomycetemcomitans AI-2 (A) and V. harveyi AI-2 (B) in a dose-dependent manner. Purified RbsB protein (0.3 to 5,000 ng per ml) was incubated with partially purified AI-2 from A. actinomycetemcomitans or with conditioned AI medium from an overnight V. harveyi culture for 30 min at 30°C. After subsequent addition of V. harveyi BB170 cells, bioluminescence was measured at hourly intervals by using a Wallac Victor3 microtiter plate reader. The data shown represent the 6-h time point after the addition of AI-2, at which time the positive control (V. harveyi BB170 cells exposed to conditioned AI medium from an overnight V. harveyi culture) exhibited approximately 1,100- to 1,300-fold induction of bioluminescence over that of the negative-control reaction mixture containing sterile AI medium. Reactions were performed in triplicate.
RbsB-mediated inhibition of V. harveyi bioluminescence is reversed by ribose.
Since RbsB is a periplasmic ribose binding protein and is a component of an ABC-type transporter of ribose, we next determined if the addition of ribose could reverse RbsB-mediated inhibition of V. harveyi bioluminescence by competing with A. actinomycetemcomitans AI-2 or V. harveyi AI-2 for the RbsB binding site. These reactions were carried out as direct competitions, in which RbsB (or RbsB mixed with ribose) was added to V. harveyi BB170 cells that were already stimulated with AI-2 rather than preincubated with signal prior to addition to the reporter cells. As shown in Fig. 3A, the addition of RbsB at the onset of induction of V. harveyi AI-2-mediated bioluminescence (e.g., the 3-h time point in the experiment shown in Fig. 3A) resulted in a significant inhibition of bioluminescence of cells at later time points. The addition of 2.5 μg per ml RbsB (0.08 μM) inhibited light production by 87%, and complete inhibition of bioluminescence occurred using 10 μg per ml (0.3 μM) RbsB. Consistent with the results presented in Fig. 2, RbsB was a more potent inhibitor of V. harveyi bioluminescence induced by A. actinomycetemcomitans AI-2 than of that induced by V. harveyi AI-2. Addition of 2.5 μg per ml RbsB inhibited light production induced by A. actinomycetemcomitans AI-2 by approximately 97% (Fig. 3B). However, inclusion of 50 mM ribose with RbsB (2.5 μg per ml) dramatically reversed RbsB-mediated inhibition and increased light production by A. actinomycetemcomitans AI-2-stimulated cells to 47% of the level for the positive control (Fig. 3B). This suggests that ribose competes with A. actinomycetemcomitans AI-2 for RbsB and that both AI-2 and ribose bind to the same site on the RbsB polypeptide. As shown in Fig. 3C, ribose itself had no effect on A. actinomycetemcomitans AI-2-mediated induction of V. harveyi bioluminescence at concentrations up to 50 mM.
FIG. 3.
(A) Direct competition of RbsB with V. harveyi BB170 cells induced with V. harveyi AI-2. RbsB protein (2.5 μg per ml, •; 5 μg per ml, ▾; or 10 μg per ml, ▪) was added to V. harveyi BB170 cells at the onset of induction of AI-2-mediated bioluminescence (e.g., the 3-h time point in the experiment shown), and light production was monitored at various time points after the addition of RbsB. The positive-control reaction mixture (⧫) did not receive RbsB and exhibited ∼1,200-fold induction of bioluminescence at the 5.5-h time point. (B) RbsB-mediated inhibition of V. harveyi BB170 bioluminescence induced by A. actinomycetemcomitans AI-2 is reversed by the addition of ribose. V. harveyi BB170 cells were mixed with partially purified AI-2 from A. actinomycetemcomitans and incubated at 30°C with shaking. At the onset of the induction of bioluminescence (3 h after the addition of A. actinomycetemcomitans AI-2), RbsB protein alone (2.5 μg per ml, left bar), RbsB (2.5 μg per ml) in the presence of 10 mM ribose (middle bar), or 50 mM ribose (right bar) was added. Light production was subsequently determined at the 5.5-h time point. Positive-control reaction mixtures did not receive RbsB and exhibited approximately 1,000-fold induction of light over levels for the negative control (V. harveyi BB170 induced with sterile AI broth). (C) Influence of ribose on induction of V. harveyi BB170 bioluminescence in the absence of RbsB. V. harveyi BB170 cells were induced with partially purified A. actinomycetemcomitans AI-2 or with A. actinomycetemcomitans AI-2 in the presence of ribose at a final concentration of 0 to 1,000 mM. The induction of bioluminescence was determined for each sample as described in Materials and Methods. All reactions were performed in triplicate.
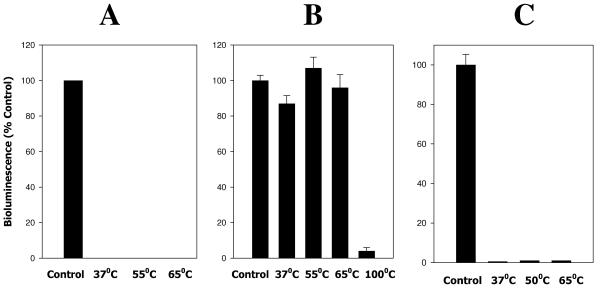
A. actinomycetemcomitans AI-2 and the RbsB/A. actinomycetemcomitans AI-2 complex are heat stable.
Chen et al. (6) previously reported that thermal denaturation of the V. harveyi LuxP/AI-2 complex released active AI-2. To determine if bound AI-2 could be recovered from the RbsB/AI-2 complex, RbsB protein was preincubated with A. actinomycetemcomitans AI-2 for 30 min at 30°C and the complex was subsequently heated at 37°C, 55°C, and 65°C prior to addition to V. harveyi BB170 cells. Samples were then assayed for AI-2 activity by monitoring induction of V. harveyi BB170 bioluminescence. As shown in Fig. 4A, no A. actinomycetemcomitans AI-2 activity was recovered from these samples, suggesting that the interaction of A. actinomycetemcomitans AI-2 with RbsB may differ from the previously characterized interaction of V. harveyi AI-2 with LuxP. To exclude the possibility that A. actinomycetemcomitans AI-2 was heat labile, samples of A. actinomycetemcomitans AI-2 were pretreated at various temperatures prior to addition to BB170 cells. As shown in Fig. 4B, A. actinomycetemcomitans AI-2-mediated induction of V. harveyi bioluminescence was not affected by exposing the signal to temperatures up to 65°C, but A. actinomycetemcomitans AI-2 lost activity when heated at 100°C. Thus, our inability to recover A. actinomycetemcomitans AI-2 from the RbsB complex did not arise from heat inactivation of A. actinomycetemcomitans AI-2. We next examined the thermostability of the RbsB polypeptide. Samples of RbsB protein were subjected to increasing temperatures (37°C to 65°C) prior to incubation with A. actinomycetemcomitans AI-2 for 30 min at 30°C. The RbsB/A. actinomycetemcomitans AI-2 mixtures were then tested for induction of V. harveyi BB170 bioluminescence. As shown in Fig. 4C, exposure of RbsB to temperatures up to 65°C did not significantly alter its ability to compete with V. harveyi LuxP for A. actinomycetemcomitans AI-2. Together, these results suggest that the RbsB/A. actinomycetemcomitans AI-2 complex is thermostable or, alternatively, that RbsB may be capable of refolding to an active conformation upon cooling to physiologic temperatures after thermal denaturation.
FIG. 4.
RbsB, AI-2, and the RbsB/AI-2 complex are heat stable. (A) A. actinomycetemcomitans AI-2 was incubated for 30 min at 30°C with purified RbsB (2.5 μg) and then heated for 10 min at 37°C, 55°C, or 65°C. Samples were subsequently tested for induction of V. harveyi BB170 bioluminescence. (B) To assess the heat stability of AI-2, partially purified A. actinomycetemcomitans AI-2 was heated for 10 min at 37°C, 55°C, 65°C, or 100°C and then added to V. harveyi BB170 cells. Bioluminescence was determined as described above. (C) For the RbsB protein, purified RbsB (2.5 μg) was heated for 10 min at 37°C, 50°C, or 65°C and then incubated with partially purified A. actinomycetemcomitans AI-2 for 30 min at 30°C. Induction of V. harveyi BB170 bioluminescence was determined as described in Materials and Methods.
Inactivation of rbsB influences AI-2 transport by A. actinomycetemcomitans.
Taga et al. (36) showed that the S. enterica serovar Typhimurium lsr operon encoded a transporter that functioned to internalize AI-2. To determine if the A. actinomycetemcomitans rbs operon may function as a transporter of AI-2, we constructed an isogenic mutant deficient in RbsB and compared its ability to deplete signal from solutions of partially purified A. actinomycetemcomitans AI-2 with that of the wild-type strain. As shown in Fig. 5, wild-type A. actinomycetemcomitans depleted AI-2 activity by approximately 85% after incubation of cells with A. actinomycetemcomitans AI-2 for 10 min, and almost-complete depletion of AI-2 (>95%) was observed after 15 min. In contrast, incubation of the RbsB-deficient mutant with A. actinomycetemcomitans AI-2 for 10 min resulted in only a 10% loss of AI-2 activity, and approximately 50% of the AI-2 activity still remained after 15 min. However, both the wild-type and mutant strains completely depleted AI-2 activity after longer incubation with A. actinomycetemcomitans AI-2 (e.g., 30 min). Thus, inactivation of rbsB influences the rate of depletion of AI-2 but does not completely inhibit the ability of the mutant strain to deplete AI-2 from solution. These results suggest that the rbs operon may function to internalize AI-2 but also suggest that other redundant mechanisms may exist in A. actinomycetemcomitans to transport AI-2. This is consistent with the findings of Taga et al. (36), who reported that inactivation of the S. enterica serovar Typhimurium Lsr transporter did not completely inhibit AI-2 transport.
FIG. 5.
Inactivation of the A. actinomycetemcomitans rbs operon influences the kinetics of depletion of AI-2 from solution by bacterial cells. Wild-type (•—•) and Rbs-deficient (○—○) strains of A. actinomycetemcomitans were incubated at 37°C with a solution of partially purified A. actinomycetemcomitans AI-2. At various times up to 30 min, bacterial cells were removed by centrifugation and the supernatant was analyzed in triplicate for the induction of V. harveyi bioluminescence as described in Materials and Methods.
Inactivation of rbsB influences aerobic growth of A. actinomycetemcomitans under iron limitation.
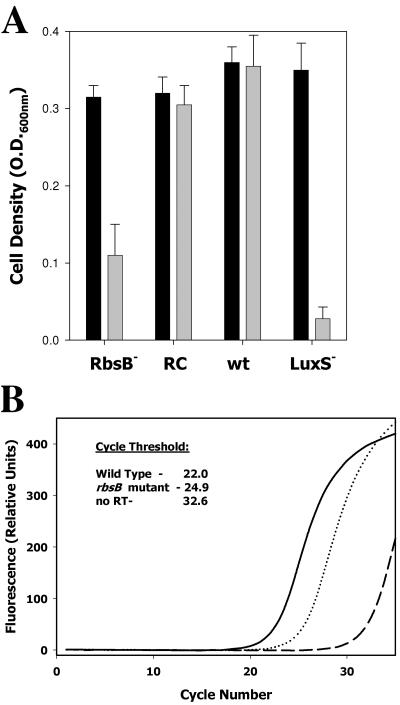
The interaction of RbsB with A. actinomycetemcomitans AI-2 suggests that the RbsB protein may play a role in the response of A. actinomycetemcomitans to AI-2. If so, the loss of RbsB function would be predicted to generate a phenotype similar to that of a LuxS-deficient A. actinomycetemcomitans strain that cannot produce AI-2. Fong et al. (9, 10) previously showed that inactivation of luxS affected aerobic growth of A. actinomycetemcomitans under conditions of iron limitation and influenced several genes involved in iron uptake, transport, and storage. To determine the potential role of RbsB in the response of A. actinomycetemcomitans to AI-2, we examined the growth of the RbsB-deficient organism under iron limitation. As shown in Fig. 6A, the levels of growth of wild-type and RbsB-deficient A. actinomycetemcomitans strains were similar under iron-replete conditions. As expected, iron limitation did not affect the growth of the wild-type strain, but growth of the RbsB mutant was only approximately 30% that of the wild-type strain under iron-limiting conditions. Complementation of the rbsB mutation with a plasmid-borne copy of rbsB restored growth to near-wild-type levels under iron limitation. The LuxS-deficient A. actinomycetemcomitans mutant exhibited a cell density of only 8% that of the control.
FIG. 6.
Inactivation of A. actinomycetemcomitans rbsB influences aerobic growth under iron limitation and expression of afuA, encoding a ferric iron transporter. (A) Levels of growth of wild-type A. actinomycetemcomitans (wt), isogenic rbsB and luxS mutants (RbsB− and LuxS−, respectively), and the rbsB mutant complemented with a plasmid-borne copy of rbsB (RC) were determined under iron-replete conditions (black bars) and in the presence of 100 μM EDDHA (gray bars). Cultures were analyzed at mid-exponential to late exponential phases of growth by determining optical density at 600 nm (O.D.600nm). The results presented are averages from three independent experiments. (B) Expression of afuA was monitored by real-time PCR using gene-specific primers and the fluorophore SYBR green. Fluorescence as a function of cycle number was plotted for reactions using RNA from wild-type A. actinomycetemcomitans (solid line) and an isogenic rbsB mutant (dotted line). Results from a negative-control reaction using RNA from the mutant strain but without reverse transcriptase (RT) are shown by the dashed line. The cycle threshold was calculated from the raw fluorescence data by using the onboard analysis software supplied with the SmartCycler system.
To determine if inactivation of rbsB influenced the expression of iron uptake genes, real-time PCR was used to follow the expression of afuA, which encodes a major ferric iron binding protein of A. actinomycetemcomitans that was previously shown to be regulated by AI-2 (9). As shown in Fig. 6B, expression of afuA is reduced by approximately eightfold in the rbsB mutant relative to expression in the wild-type strain. This is consistent with the eightfold reduction in afuA expression that was previously observed for the LuxS-deficient strain of A. actinomycetemcomitans (9). Together, these results suggest that RbsB may play a role in the response of A. actinomycetemcomitans to AI-2 and may function as a receptor for AI-2 in A. actinomycetemcomitans.
DISCUSSION
Many gram-positive and gram-negative bacterial species express LuxS and secrete and respond in various ways to a signal that is related to autoinducer 2 of V. harveyi (4, 7, 9, 10, 14, 16, 18, 19, 20, 21, 30, 31, 41). However, few of these organisms possess the dedicated two-component circuit that mediates the cell density-dependent response of V. harveyi to AI-2. Our previous studies showed that AI-2 secreted by the oral pathogen A. actinomycetemcomitans was capable of inducing V. harveyi bioluminescence (10) and also influenced aerobic growth of A. actinomycetemcomitans under iron-limiting conditions (9) These studies also suggested that AI-2-mediated regulation of iron uptake and storage genes contributed to optimal growth of A. actinomycetemcomitans under iron limitation. However, the mechanism by which A. actinomycetemcomitans responds to AI-2 is not fully understood. Analysis of the complete genome sequence of A. actinomycetemcomitans HK1651 (26; www.oralgen.lanl.gov) indicates that this organism lacks direct homologs of the dedicated sensor kinase/phosphatase (LuxQ) and the phosphotransfer polypeptide (LuxU) that exist in Vibrio spp. However, A. actinomycetemcomitans does possess genes that exhibit significant similarity to the V. harveyi RNA chaperone Hfq and the receptor for AI-2, LuxP. Indeed, the A. actinomycetemcomitans genome contains two genes that are related to LuxP, one encoding a protein similar to the periplasmic ribose binding protein RbsB of E. coli and the second encoding a polypeptide that is homologous to the periplasmic LsrB protein of S. enterica serovar Typhimurium. The LsrB polypeptide of S. enterica serovar Typhimurium has recently been shown to bind to AI-2 and to function as part of a transport system encoded by the lsrACDBFGE operon, which internalizes AI-2 (35, 36). However, inactivation of the Lsr transporter did not completely inhibit the internalization of AI-2 by S. enterica serovar Typhimurium (36), suggesting that other AI-2 transport mechanisms may exist. Our current studies show that A. actinomycetemcomitans RbsB interacts with AI-2 and plays a role in the cellular response to AI-2, suggesting that RbsB may also function to transport AI-2.
RbsB protein that was purified from an AI-2-deficient background inhibited V. harveyi bioluminescence induced by both A. actinomycetemcomitans AI-2 and V. harveyi AI-2 in a dose-dependent manner, suggesting that RbsB competes with LuxP (the periplasmic receptor for AI-2 in V. harveyi) for AI-2. Furthermore, A. actinomycetemcomitans AI-2 appeared to interact with the ribose binding fold of the RbsB protein, since RbsB-mediated inhibition of V. harveyi bioluminescence was reversed in the presence of ribose. This observation is consistent with the structural similarity that exists between ribose and the different known isoforms of AI-2. Furthermore, the precursor of AI-2 (4,5-dihydroxy-2,3-pentanedione) is generated by LuxS from the ribose moiety of S-ribosylhomocysteine (15). Interestingly, RbsB was a more effective competitive inhibitor of V. harveyi bioluminescence that was induced by A. actinomycetemcomitans AI-2 than of that induced by V. harveyi AI-2. Fifty percent inhibition of light production induced by A. actinomycetemcomitans AI-2 occurred with the addition of 1 ng per ml (0.03 nM) of RbsB, whereas approximately 40 ng per ml (1.3 nM) RbsB was required for 50% inhibition of bioluminescence stimulated by V. harveyi AI-2. This may reflect a difference in binding affinity for A. actinomycetemcomitans AI-2 and V. harveyi AI-2 by RbsB, possibly arising from structural variation of the A. actinomycetemcomitans and V. harveyi AI-2 signals. Indeed, multiple forms of AI-2 are known to exist. For example, the structure of the LsrB/AI-2 complex reported by Miller et al. (22) showed that S. enterica serovar Typhimurium produces a form of AI-2 that differs in stereochemistry from and lacks the borate diester that is present in AI-2 produced by V. harveyi. Since boron is present at much lower levels in terrestrial environments than in seawater, RbsB and LsrB may have evolved to interact at higher affinity with the AI-2 isoform that lacks boron, whereas marine organisms expressing LuxP bind to the borate diester form of AI-2 at higher affinity.
An alternative explanation for the different levels of RbsB-mediated inhibition observed is that the concentration of A. actinomycetemcomitans AI-2 in conditioned medium from A. actinomycetemcomitans is significantly lower than the level of AI-2 that is present in conditioned medium from V. harveyi. We feel that this explanation is unlikely since the A. actinomycetemcomitans AI-2 that was used in these experiments was partially purified and concentrated 20-fold from A. actinomycetemcomitans conditioned medium. However, a difference in AI-2 levels cannot be excluded because a direct biochemical assay to quantify the levels of AI-2 in conditioned medium is not currently available. Nonetheless, our results clearly show that A. actinomycetemcomitans RbsB interacts with both its cognate signal and AI-2 produced by V. harveyi. Thus, the activity of RbsB is consistent with the hypothesis that AI-2 functions as a universal signal that is recognized by a broad and diverse group of bacteria (1, 27). Furthermore, if structural variation of AI-2 extends beyond the two known isoforms (S. enterica serovar Typhimurium and V. harveyi), it is possible that the affinity of the receptor interaction with AI-2 may allow an organism to distinguish and respond to AI-2 signals produced by different organisms.
Our previous results showed that inactivation of luxS affected aerobic growth of A. actinomycetemcomitans under iron limitation and influenced the expression of iron uptake/transport genes, e.g., afuA (9). Interestingly, inactivation of rbsB resulted in a similar phenotype. Growth of the RbsB-deficient strain was inhibited under iron limitation, attaining only approximately 30% of the cell density exhibited by wild-type A. actinomycetemcomitans during late exponential growth. In addition, real-time PCR indicated that expression of afuA was reduced by eightfold in the mutant. Together, these results suggest that RbsB contributes to the cellular response of A. actinomycetemcomitans to AI-2. However, the molecular mechanism whereby RbsB contributes to the response to AI-2 remains to be determined. In E. coli, RbsB is the periplasmic binding component of an ABC-type ribose transport complex that functions to internalize ribose and subsequently phosphorylate the internalized sugar via the kinase RbsK (ribokinase). Our findings that RbsB competes with LuxP (Fig. 2 and 3) and that inactivation of rbsB inhibits the ability of the mutant strain to deplete AI-2 from solution (Fig. 5) suggest that the RbsB protein may bind to and deliver AI-2 to the membrane permease (RbsC), facilitating internalization of AI-2 in A. actinomycetemcomitans. Thus, our hypothesis is that the RbsB protein represents a periplasmic receptor for AI-2 and that the rbs operon of A. actinomycetemcomitans may function to internalize AI-2, similarly to the activity of the lsr operon which internalizes AI-2 in S. enterica serovar Typhimurium (35).
Inactivation of rbsB did not completely inhibit the ability of the mutant to deplete AI-2 from solution (Fig. 5). This result is of interest since Taga et al. (36) showed that some internalization of AI-2 by S. enterica serovar Typhimurium still occurred in a strain that was unable to express a functional Lsr transporter. However, Taga et al. (36) also noted that the S. enterica serovar Typhimurium genome contains the rbsDACBKR operon, and our analysis of the A. actinomycetemcomitans genome indicates the presence of an operon that encodes a potential Lsr-like transporter (open reading frames AA02215 to AA02226 in the A. actinomycetemcomitans HK1651 genome sequence). Indeed, the lsr operons of A. actinomycetemcomitans and S. enterica serovar Typhimurium are highly conserved, with the exception of lsrE, which does not appear to function in AI-2 transport in S. enterica serovar Typhimurium (36). This raises the possibility that multiple transport mechanisms exist to internalize AI-2. Our results suggest that the residual transport activity observed by Taga et al. (36) in the Lsr-deficient strain of S. enterica serovar Typhimurium may arise from internalization of AI-2 by the rbsDACBKR operon. Furthermore, the ability of the A. actinomycetemcomitans rbsB knockout strain to deplete AI-2 during longer incubations could occur via the Lsr transporter, which presumably is functional in the rbsB mutant. It is also interesting to note that the growth of the RbsB mutant under iron limitation was not stunted to the extent of the LuxS mutant of A. actinomycetemcomitans that we previously analyzed (9). This phenotype might arise if the putative A. actinomycetemcomitans Lsr transporter partially complements the loss of Rbs-mediated AI-2 transport. Studies to inactivate the lsr operon of A. actinomycetemcomitans, both alone and in combination with the rbsB mutation, and to compare the kinetics of the interaction of the LsrB and RbsB proteins with AI-2 are underway.
The physiologic role of AI-2 remains controversial since it has been shown to function as a quorum-sensing signal in Vibrio spp. (1, 2, 37) and yet is suggested to be a metabolic by-product that is possibly consumed by many other organisms (37). A. actinomycetemcomitans does not possess the two-component phosphorelay signal transduction pathway that is coupled to LuxP (or related proteins) of Vibrio spp., suggesting that it does not perform a quorum-sensing role at least in the context of the Vibrio paradigm. Yet, in the presence of a rich nutrient source, A. actinomycetemcomitans requires AI-2 for optimal growth under iron limitation and for the development of biofilms (D. R. Demuth, unpublished). This suggests that AI-2 does not function solely as a consumable metabolite. To understand the physiologic role of AI-2, it will be necessary to determine the structure of the A. actinomycetemcomitans AI-2 and to clarify the molecular pathway that governs the response of the organism to AI-2. Our studies provide some of the first evidence that the rbs operon may be involved in the recognition of AI-2 by A. actinomycetemcomitans. However, additional studies are required to determine if redundant transporters of AI-2 exist in A. actinomycetemcomitans and to determine the downstream events that occur within the bacterium after internalization of signal.
Acknowledgments
This work was supported by Public Health Service Grant DE14605 from the National Institute of Dental and Craniofacial Research.
Editor: V. J. DiRita
REFERENCES
- 1.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 4.Blehert, D. S., R. J. Palmer, J. B. Xavier, J. S. Ameida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block, P. J., A. C. Fox, C. Yoran, and A. J. Kaltman. 1973. Actinobacillus actinomycetemcomitans endocarditis: report of a case and review of the literature. Am. J. Med. Sci. 276:387-392. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X., S. Schauder, N. Potier, A. Van Dorsselear, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in Gram positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 9.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 14.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 15.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S. J., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 17.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 18.Loh, J. T., M. H. Forsyth, and T. L. Cover. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 20.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, S. T., K. B. Xavier, S. R. Campaign, M. E. Tag, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 23.Page, M. I., and E. O. King. 1966. Infection due to Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. N. Engl. J. Med. 275:181-188. [DOI] [PubMed] [Google Scholar]
- 24.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roe, B. A., F. Z. Najar, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer. Actinobacillus genome sequencing project. University of Oklahoma, Norman, Okla. [Online.] http://www.oralgen.lanl.gov.
- 27.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 28.Schauder, S., K. Showkat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 29.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion on enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreenivasan, P. K., D. J. LeBlanc, L. N. Lee, and P. Fives-Taylor. 1991. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect. Immun. 59:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 36.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 37.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. M. Hardie. 2005. Making “sense” of metabolism: auto-inducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 38.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. G. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 40.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambon, J. J., J. Slots, and R. J. Genco. 1983. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect. Immun. 41:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]