Abstract
In order to quantify in vivo the mRNAs of cytokines which play important roles in leptospirosis, we have developed quantitative real-time PCR assays for interleukin-2 (IL-2), IL-4, IL-10, IL-12p40, tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), transforming growth factor β, and two housekeeping genes (encoding β-actin and hypoxanthine phosphoribosyltransferase). We used a lethal hamster model reflecting severe leptospirosis in humans. The LightCycler system was used to quantify the gene expression levels with the SYBR green I detection format using external standard curves for each target. We compared the expression levels of cytokine mRNA in the peripheral blood mononuclear cells of both control (uninfected) hamsters and Leptospira interrogans-inoculated hamsters from 1 to 24 h and then 1 to 4 days postinfection. In this kinetic study, there was pronounced expression of Th1 cytokine mRNA (TNF-α, IFN-γ, and IL-12), with transcripts being detected as early as 1 h postinfection. Expression of anti-inflammatory cytokines, such as IL-4 and IL-10, was prominent in delayed samples from 1 to 4 days postinfection in response to infection with Leptospira interrogans. Our data are the first to establish that pathogenic leptospires can stimulate in vivo the production of type 1 cytokines involved in cellular immunity by using this informative animal model. Measuring and assessing cytokine profiles may provide a useful method for accurate study of the mechanisms of anti-Leptospira immunity, indications of prognosis factors, and prospective evaluation of leptospirosis vaccine efficacy in humans.
Leptospirosis is a widespread zoonosis caused by members of the genus Leptospira, with the incidence of the disease being higher in tropical climates (10). Human leptospirosis is highly variable in its clinical expression. The course of the disease ranges from mild flu-like symptoms to rapidly fatal forms, clinically resembling sepsis with multiple organ failure (mainly kidney and liver) and shock-like syndrome.
Despite socioeconomic consequences, such as significant production losses in cattle and greater risk of infection with increasing fatality rates in humans, there is a lack of knowledge regarding specific immune response leading to protection against Leptospira interrogans infection and immune mechanisms elicited after immunization with vaccine candidates. Establishing the correlation between a Th1 and Th2 profile for dissecting the mechanisms of anti-Leptospira immunity is of major importance in obtaining a successful vaccine against leptospirosis as well as in understanding the pathogenesis of natural or induced infection (20). Regarding innate immune response, interesting results were obtained by Werts et al. (32) when they demonstrated that activation of macrophages by leptospiral lipopolysaccharide occurred through CD14 and the Toll-like receptor 2. Considering induced immune response, despite a serovar specificity, protective immunity to leptospirosis is not exclusively humoral (10). This paradigm was reexamined by Naiman et al. (21, 22), Baldwin et al. (2), and Brown et al. (3). Indeed, by evaluating the cellular immune response induced by a protective monovalent serovar Hardjo vaccine in cattle, a potent Th1-type immune response and the involvement of γδ T cells were clearly in evidence. At the same time, there was also evidence that antilipopolysaccharide antibodies were not the only mechanism playing a role in naturally acquired protective immunity (29).
The mechanism by which leptospires activate the immune system has been pointed out in several studies, mainly in vitro, highlighting the importance of cytokines (17, 33). Indeed, the role of cell-mediated immunity in host defense to Leptospira remains poorly understood in both animal and human diseases. Briefly, after skin or mucosal penetration and a bacteremic stage, virulent leptospires reach and colonize the target tissues of the host organism (29, 30). Primarily, vasculitis is the characteristic lesion found in leptospirosis, leading to major cellular damage (10). Fluid and cell leakage occur in the presence of few leptospires, suggesting the involvement of factors from either the spirochete or the host. As an example, limited studies have reported a significant increase of tumor necrosis factor alpha (TNF-α) in human patients with leptospirosis (9) and levels of expression in plasma were associated with severity of disease and mortality (28). However, these results represent a global response of the host. The natural history of the disease is often unknown, and producing cells were not characterized (type and kinetics of production), nor were molecular events (cascade of activation) investigated. Concerning the pathogen, the glycolipoprotein is a toxic Leptospira interrogans cellular component able to induce cellular activation in vitro, as assessed by cytokine secretion and cell surface antigen expression (7). Preliminary results were obtained in 1980 by Cinco et al. (5) with the in vitro release of TNF-α from human monocytes by Leptospira peptidoglycans. However, no link with virulence could be established (11). In a later study, Klimpel et al. (12) showed that Leptospira induced the in vitro production of Th1 cytokines by peripheral blood mononuclear cells (PBMCs) and cell proliferation in both αβ T cells and γδ T cells. These γδ T cells recognized Leptospira without antigen-processing or antigen-presenting cells. Knowledge of molecular mechanisms implied in the in vivo response to leptospires would shed new light on the delineation between severe and mild forms of the disease.
The rapid and efficient production of cytokines occurs by the accurate control of their gene transcription and RNA stability. A current method to analyze cytokine production is to quantify their corresponding mRNA by real-time (RT)-PCR. Cytokine mRNA quantification is widely used to investigate cytokine profiles, particularly in small samples where transcripts such as cytokine mRNAs are lowly expressed (14, 16, 26, 27). This technique allows the quantification of a larger pattern of cytokines than quantification at the protein level, which is limited to a smaller number of cytokines (15) and does not fully reflect the expression profile.
After experimental infection of hamsters with a virulent strain of Leptospira, the purpose of this study was to perform the accurate quantification of peripheral blood cytokine mRNA levels, based on the combination of tubes containing an mRNA stabilizer for blood collection and the real-time PCR methodology on the LightCycler. In this application, we have developed sensitive and reproducible SYBR green I RT-PCR protocols which allow measurement of various cytokine mRNAs in hamsters (interleukin-2 [IL-2], IL-4, IL-10, IL-12p40, TNF-α, gamma interferon [IFN-γ], and transforming growth factor β [TGF-β]). We used both absolute quantification, to determine a numerical value for the target concentrations, and relative quantification, to describe the change in expression of the target genes in relation to two housekeeping (reference) genes (encoding β-actin and hypoxanthine phosphoribosyltransferase [HPRT]). This approach should provide greater insight into the immunological processes underlying protection after immunization with leptospirosis vaccine candidates.
(This work was presented in part at the 2005 Roche LightCycler User Group meeting, Taupo, New Zealand.)
MATERIALS AND METHODS
Leptospira strain and cultivation.
The virulent L. interrogans serovar Icterohaemorrhagiae strain Verdun was obtained from the Reference Collection of the Institut Pasteur in Paris, France. Leptospires were grown in liquid Ellinghausen McCullough Johnson and Harris (EMJH) medium at 30°C under aerobic conditions (8) to a density of about 108 bacteria per ml and counted in a Petroff-Hausser counting chamber. Virulence was maintained by deep-freeze preservation and iterative lethal passages in Syrian golden hamsters (Mesocricetus auratus). Briefly, specific pathogen-free animals were purchased from Charles River Laboratories (Charles River Wiga GmbH, Sulzfeld, Germany) and bred at the Institut Pasteur of New Caledonia. Two animals weighing 50 to 60 g were lethally infected intraperitoneally with 108 leptospires of a virulent culture in EMJH medium. Another group of two animals was inoculated by the same route with sterile EMJH medium and used as a control. When prostration and anorexia appeared between 4 and 6 days after inoculation, blood from infected hamsters was collected by cardiac puncture and cultured in EMJH medium. After 7 to 10 days of culture (at least 108 leptospires per ml), this highly virulent strain was used for experimental infections.
Experimental infections.
All in vivo studies were carried out using 6- to 8-week-old outbred golden hamsters individually identified by subcutaneous-implanted transponders (Virbac, France) and handled according to French national regulations. Each experimental set was composed of a group comprising three inoculated animals and one noninfected hamster per checking point. Hamsters were lethally infected with 108 leptospires of a virulent culture in EMJH medium by subcutaneous injection to reproduce as much as possible by the conventional way of infection (29), whereas noninfected hamsters received by the same route sterile EMJH medium. Each set of experiments, including control animals, was conducted in duplicate. Blood samples were collected by cardiac puncture under sublethal anesthesia via Vacutainer collection tubes (15% K3E; Becton Dickinson, France) or PAXgene blood RNA tubes (PreAnalytiX; QIAGEN, Australia).
In vitro cell stimulation.
External DNA standards were generated after in vitro stimulation in order to obtain marked concentrations of all target genes. A maximum amount of blood (2 to 3 ml) was collected from healthy hamsters on Vacutainer collection tubes as described above. PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation (Histopaque; Sigma-Aldrich, St Louis, MO). Then, PBMCs were plated in a 24-well plate in duplicate cultures at 106 cells/ml in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated calf serum and 1% penicillin-streptomycin, according to the following stimulation schedule: (i) medium only, (ii) 10 ng/ml of phorbol myristate acetate plus 500 ng/ml inomycin, and (iii) 1 μg/ml Escherichia coli lipopolysaccharide (all from Sigma). Overnight cultures were performed at 37°C in a 5% CO2 atmosphere prior to harvesting the cells and carrying out RNA isolation.
Total RNA isolation and cDNA synthesis.
Total RNA extracted from in vitro-stimulated cells was obtained using the High Pure RNA isolation kit (Roche Applied Science, Auckland, New Zealand), whereas total RNA from in vivo experiments, with larger amount of cells, was isolated using the PAXgene blood RNA system (PreAnalytiX), both according to the manufacturers' instructions. cDNA was synthesized using the Transcriptor first strand cDNA synthesis kit (Roche Applied Science) following the manufacturer's protocol, with the supplied random hexamers as the priming strategy.
Oligonucleotides.
Sequences of all primers used in this study are listed in Table 1. They were designed according to Melby et al. (18) with the LightCycler Primer Probe Design Software 2.0 (Roche Applied Science) and synthesized by Proligo Singapore Pte Ltd. (Biopolis, Singapore). The following criteria were applied during the design: product size from 200 to 350 bp (357 bp for β-actin), primer size from 17 to 30 bp, and a mean melting temperature of 60°C. Proposed oligonucleotides were selected according to intron spanning to prevent amplification of genomic DNA and percentage of G+C.
TABLE 1.
Detailed primers and conditions used for real-time PCR assays
| Gene product | Primer name | Primer sequence | Annealing temp (°C) | Tm (°C)a | Amplicon length (bp) |
|---|---|---|---|---|---|
| TNF-α | TNFα-F | AACGGCATGTCTCTCAA | 60 | 89.6 | 278 |
| TNFα-R | AGTCGGTCACCTTTCT | ||||
| IFN-γ | IFNγ-F | GACAACCAGGCCATCC | 60 | 84.6 | 226 |
| IFNγ-R | CAAAACAGCACCGACT | ||||
| IL-2 | IL-2-F | CACCCACTTCAAGCTCT | 61 | 85.2 | 349 |
| IL-2-R | TCCACCACAGTTACTGTC | ||||
| IL-4 | IL-4-F | CTCCTATCACTGACGGT | 61 | 87.8 | 342 |
| IL-4-R | ATTCACATTGCAGCTCT | ||||
| IL-10 | IL-10-F | TGGACAACATACTACTCACTG | 61 | 87.8 | 308 |
| IL-10-R | GATGTCAAATTCATTCATGGC | ||||
| IL-12p40 | IL12p40-F | AGATCCTAAAAATAAGACCTT | 61 | 89.9 | 308 |
| IL12p40-R | AGTTCTCGTATTTATACTTGT | ||||
| TGF-β | TGFβ-F | ACGGAGAAGAACTGCT | 60 | 90.0 | 245 |
| TGFβ-R | ACGTAGTACACGATGGG | ||||
| β-Actin | βactin-F | TCTACAACGAGCTGCG | 60/61 | 90.0/90.4 | 357 |
| βactin-R | CAATTTCCCTCTCGGC | ||||
| HPRT | HPRT-F | TGCGATGTCATGGTAGAG | 60/61 | 82.4/82.9 | 242 |
| HPRT-R | ATCAAGACATTCTTTCCAGT |
Melting temperature.
Standard curve construction.
For all cytokine and reference genes, a standard curve from serial dilutions of a known concentration of purified DNA was achieved (24). This quantified DNA consisted of the target PCR product prepared by conventional PCR from cDNA positive for the corresponding target mRNA. The copy number of each standard was calculated by standard methods using the Avogadro constant as described by Overbergh et al. (23). The log ranges of the different standard curves were from 107 copies to 1 copy. Threefold measurements for each standard dilution point over the whole standard curve range were produced to generate a reliable standard curve.
Real-time PCR and amplification protocol.
PCR amplification and analysis were achieved using a LightCycler 2.0 instrument (Roche Applied Science) with software version 4.0. All reactions were performed with the LightCycler FastStart DNA master SYBR green I (Roche Applied Science) by using a 20-μl volume in each reaction capillary. For quantification of the cytokines, 2 μl DNA standard dilution or 2 μl cDNA was added before capillaries were capped, centrifuged, and placed in the LightCycler sample carousel. Amplification conditions consisted of an initial preincubation at 95°C for 10 min (FastStart Taq DNA polymerase activation), followed by amplification of the target DNA for 45 cycles (95°C for 8 s, 60°C or 61°C for 5 s, and a variable extension time at 72°C). Melting curve analysis was performed immediately after amplification at a linear temperature transition rate of 0.1°C/s from 65 to 95°C with continuous fluorescence acquisition.
Result expression.
In our protocol, results were systematically normalized to expression levels of two different reference genes in order to correct variations in nucleic acid quality and quantity (26). The concentration of an unknown sample was calculated by comparing its crossing point (Cp) with the corresponding standard curve. The cycle number at the Cp (y axis) was plotted versus the log of the initial template amount (x axis) of the standards. After amplification, three analyses were sequentially performed with the data: (i) absolute quantification resulted in an absolute value (e.g., copies/μl), (ii) relative quantification was expressed as a ratio between a cytokine gene and a reference gene, and (iii) results were normalized to a calibrator sample. Finally, a calibrator-normalized ratio was obtained, providing indirect information on cytokine mRNA increases and decreases and taking into account corrections for experimental variations in different samples (23).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the Syrian golden hamster cytokines and housekeeping gene cDNAs are as follows: IFN-γ, AF034482; IL-2, AF046212; IL-4, AF046213; IL-10, AF046210; IL-12p40, AF046211; TNF-α, AF046215; TGF-β, AF046214; HPRT gene, AF047041; and β-actin gene, AJ312092.
RESULTS AND DISCUSSION
We compared the expression levels of cytokine mRNAs in the PBMCs of both control (uninfected) hamsters and Leptospira-inoculated hamsters from 1 to 24 h and then 1 to 4 days postinfection (p.i.). In this study, administration of 108 leptospires by subcutaneous inoculation resulted in a bacteremia averaging 104 leptospires/ml during the first 24 h postinfection, before reaching a peak of more than 106 leptospires/ml at 4 days p.i (29). We hypothesized that the immune mechanisms related to changes over time in the density of virulent leptospires would be demonstrative during the first 24 h and then following days of infection.
Real-time PCRs were developed for accurate cytokine mRNA quantification, first from cultured cells (i.e., PBMCs and hepatocytes) and second from whole blood (i.e., blood samples from experimentally infected laboratory animals). Whole blood presents a key advantage compared to real-time PCR performed from in vitro-purified cultured cells: it is more representative of the in vivo immune response of the host. One of the difficulties encountered using whole blood is that the cell lysis generates large amounts of proteins. All methods that isolate RNA from whole blood involve purification steps before RNA extraction. This time-consuming protocol can be avoided using commercially available reagents, such as PAXgene tubes. This method was successfully applied by Stordeur et al. (26, 27). This prompted us to combine the use of PAXgene and real-time PCR with the LightCycler to monitor systemic inflammatory responses during leptospirosis.
First, the linearity of cDNA synthesis was verified. When small RNA amounts were used, absorbance values at 260 nm were not reliable enough to allow accurate amounts of total RNA to be added to the cDNA synthesis reaction (14). Therefore, for the cDNA synthesis, a constant volume (2 μl) of total RNA was used. Isolated RNA was serially diluted and used for cDNA synthesis. Then, all cDNAs were amplified in real-time PCR for all tested cytokines and housekeeping genes. A linear relationship of the cDNA synthesis as a function of various RNA contents was observed, ensuring that various quantities of RNA did not affect the reverse transcription (data not shown).
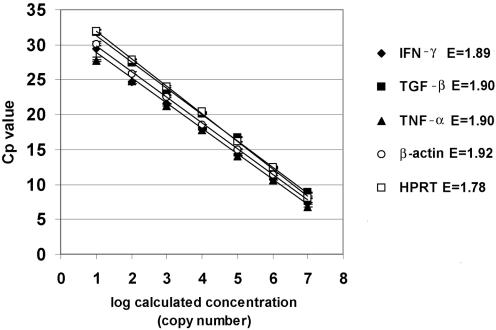
The appropriate amplicons corresponding to each cytokine and reference gene were purified, quantified, and serially diluted, as described in Materials and Methods. Figure 1 shows the standard curves for TNF-α, TGF-β, IFN-γ, HPRT, and β-actin. The curves were obtained by plotting the mean values of log-calculated concentration (copy number) versus the Cp. The amplification efficiencies varied between 1.77 and 1.94 for the different amplicons. As expected, the standard deviation increased with decreasing copy number. The regression coefficient was −1.00 in all cases.
FIG. 1.
Standard curves for TNF-α, TGF-β, and IFN-γ as well as the two housekeeping genes HPRT and β-actin. Curves were constructed using 10-fold dilutions ranging from 107 copies to 1 copy/μl. The curves were obtained by plotting the mean values of log-calculated concentration (copy number) versus the Cp. The error bars show the standard deviations (n = 3). The error (E) values of the standard curves are the mean squared errors calculated by the LightCycler software. The regression coefficient was −1.00 in all cases.
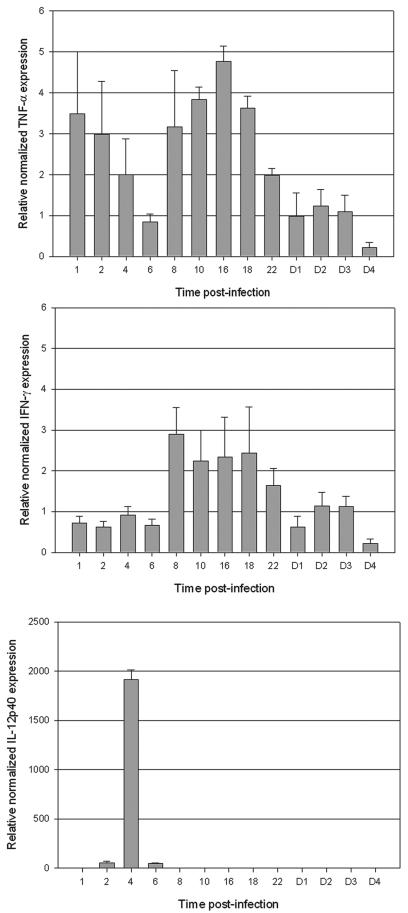
After relative quantification and normalization, we found a significant increase (as soon as 1 h p.i.) in the expression of TNF-α, a proinflammatory cytokine that can have both protective and damaging consequences for the host. Basically, two peak levels were observed during the first 24 h p.i. (Fig. 2). The maximum value, with a 4.8-fold increase compared to the baseline expression, was observed at 16 h p.i. The expression of TNF-α then remained at the baseline level from 24 h p.i. At the same time, we detected a significant increase in the mRNA for IFN-γ. Indeed, a 2.9-fold increase was found as early as 8 h p.i., followed by decreasing levels of expression over the next 16 h (Fig. 2). Interestingly, the first peak of TNF-α was detected just before the increase in the mRNA for IFN-γ, thus indicating that TNF-α seems to act synergistically with IFN-γ. Whether this early expression of TNF-α could stimulate the production of IFN-γ in the course of infection cannot be determined until reagents are available for neutralization of the first cytokine. Our findings are consistent with existing clinical data. Indeed, elevated plasma concentrations of TNF-α have been observed in patients affected by leptospirosis, and correlations were established between high levels of TNF-α and the worsening of symptoms (9, 25, 28).
FIG. 2.
Normalized relative quantification of mRNA expression by real-time PCR for TNF-α, IFN-γ, and IL-12. Shown are the means ± standard errors of the mean of the ratio of the cytokine to HPRT. The same ratios were obtained with β-actin. Hours (from 1 to 22) and days (from D1 to D4) p.i. are indicated on the x axis.
There was a considerable increase in the IL-12 mRNA in infected hamsters, with a reproducible 1,900-fold amplification at 4 h p.i. A very limited production of transcripts was visible 2 h before and after the main peak. However, this 1,900-fold induction is not likely to be accurate as little or no expression of IL-12 transcripts corresponding to the background was detected in uninfected hamsters. Compared to that for TNF-α and IFN-γ, the profile of production was consistently different. Where a progressive expression of TNF-α and IFN-γ was observed over a 6- to 16-h period, production of IL-12p40 was sudden, rapid, and strong. IL-12 is released from antigen-presenting cells and the subunit p35 is ubiquitously produced, whereas the subunit p40 is inducible and detected only in cells producing biologically active IL-12 (13). Considering leptospirosis, interestingly, de Fost et al. (6) evidenced the production of IFN-γ, IL-12p40, and TNF-α in human whole blood after incubation with heat-killed pathogenic Leptospira interrogans. Experiments were performed in vitro, and cytokines were measured by specific enzyme-linked immunosorbent assays (ELISAs). Their data established that the production of IFN-γ was largely dependent on IL-12p40. In our in vivo kinetic study using live pathogenic leptospires, a similar observation was made, confirming these previous in vitro results. Indeed, IL-12p40 was found to be released 4 h before the secretion of IFN-γ. Unfortunately, at present, no reagents for the neutralization of IL-12p40 in hamsters are readily available. Our data establish that pathogenic leptospires can stimulate in vivo, in this informative animal model reflecting severe leptospirosis in humans, the production of type 1 cytokines involved in cellular immunity. Immunological mechanisms have to be further dissected to provide comprehensive interactions between (i) TNF-α and IL-12p40 and (ii) IFN-γ and its potent inducers, TNF-α and IL-12p40, in our model. Data from Chierakul et al. (4) evidenced high plasma concentrations of IL-12p40 and TNF-α in adult Thai patients with leptospirosis. However, depending on the pathogen investigated, the few studies that suggest a direct relationship between these two cytokines are conflicting (13). Therefore, the relationship between TNF-α and IL-12 remains complex, as it strongly relies on the differentiation stage of producing cells.
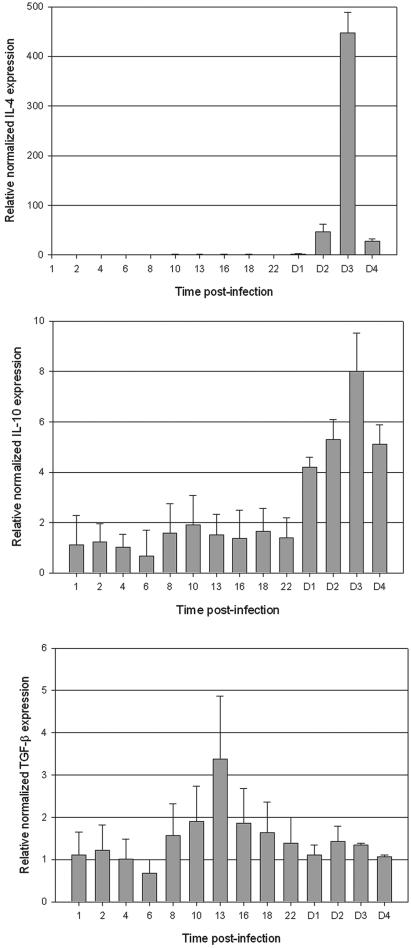
Prominent expression of IL-4 mRNA (Fig. 3 was observed for PBMC samples obtained at 3 days p.i. (450-fold increase). Interestingly, this increase in IL-4 transcripts was observed very late during the course of infection. Similar results were obtained with IL-10. A significant expression of IL-10 mRNA was observed only in delayed specimens from 2 days p.i., with a maximum at day 3 p.i. (eightfold increase). In a previous study (7), it was demonstrated that glycolipoprotein of Leptospira interrogans could induce in vitro and ex vivo lymphocyte and monocyte activation, leading to TNF-α and IL-10 secretion. IL-10 production showed typically delayed kinetics compared to TNF-α production. In our present experiments, we observed in vivo the same sequence of events, showing a cytokine pattern with a delayed induction of anti-inflammatory cytokines, such as IL-10. This cytokine is essential in the control of a sepsis-like infection and regeneration of tissue lesion (1). Further experiments should be performed to show the correlation between IL-10 and TNF-α, as a high IL-10/TNF-α ratio seems to be a good prognosis in human leptospirosis (28). In our hamster model, the average ratio is 1.7 (8.0/4.8) and was not consistent with recovery and healing because all infected hamsters died from 6 to 8 days p.i (30). However, this animal model cannot be fully compared to the natural course of leptospirosis in humans. Indeed, a high number of virulent leptospires is needed to develop acute or carrier leptospirosis, and the model description applies to young animals. In a previous study (29), we evidenced a critical threshold (density of 104 leptospires/ml of blood) for the vital prognosis by using a quantitative PCR assay during the course of human leptospirosis. According to our results here, the suitability of this quantitative method should also include a specific pro- and anti-inflammatory cytokine ratio.
FIG. 3.
Normalized relative quantification of mRNA expression by real-time PCR for IL-4, IL-10, and TGF-β. Shown are the means ± standard errors of the mean of the ratio of the cytokine to HPRT. The same ratios were obtained with β-actin. Hours (from 1 to 22) and days (from D1 to D4) p.i. are indicated on the x axis.
A low-level basal expression of IL-2 was detected in uninfected hamsters but did not increase in response to infection with L. interrogans. A prominent baseline TGF-β expression was also observed in uninfected animals. As reported by Melby et al. (19), the constitutive expression level of this cytokine is not surprising as TGF-β is strictly transcriptionally regulated. However, a single peak was reached at 13 h p.i., followed by a gradual progressive decrease in Leptospira-infected hamsters. Remembering that TGF-β is a potent inhibitor of macrophage function, the role of this cytokine in this leptospire-hamster model needs to be confirmed with analysis of protein expression.
Real-time PCR is advantageous for cytokine transcript quantification, because it can be used to accurately monitor the simultaneous expression of an array of cytokines from a single sample and requires only small quantities of template. Up to now, studies investigating cytokine expression in leptospirosis were performed by ELISA. However, such studies at the protein level are not always possible due to the lack of sensitivity of ELISA kits. Processing of hamster samples is also impaired by the lack of commercialized ELISA kits. Indeed, high-sensitivity ELISA kits are available for rats and mice, but one should be extremely cautious when analyzing results as such animal models are not relevant for studying experimental leptospirosis. We improved our real-time PCR assay to enable large increases (n-fold) (IL-12p40 and IL-4) as well as slight increases (TNF-α, IFN-γ, TGF-β, and IL-10) in cytokines to be accurately measured.
Little is known about the cellular immune response to pathogenic leptospires, either in vitro or in vivo, by PBMCs. The experiments presented here provide a constructive basis for the proinflammatory and anti-inflammatory cytokine responses in in vivo disease outcome. This informative hamster model, which does not need immunosuppressive treatment such as cyclophosphamide (10), mimics severe human leptospirosis with jaundice, hemorrhages, hepatitis, and nephritis. The innovative findings from this study indicate that in vivo interactions of hamster PBMCs with a virulent strain of Leptospira interrogans result in the production of cytokines associated with a Th1-type immune response. Measurement and assessment of cytokine profiles may provide a useful method for accurate study of the mechanisms of anti-Leptospira immunity, indications of prognosis factors, and prospective evaluation of leptospirosis vaccine efficacy. Signaling pathways involved in the production of cytokines should be studied. Therefore, genetic background of the host needs also to be investigated as innate immunity is essential in early infection by pathogenic leptospires. Indeed, the recent observations of Viriyakosol et al. (31) demonstrated that intact Toll-like receptor 4 signaling contributes to the control of the tissue burden of Leptospira in nonlethal infection by using a mouse model. According to the diversity of symptoms in humans, a study of cytokine gene polymorphisms in the host would provide key findings for understanding human predisposition to leptospiral infection.
Acknowledgments
We thank Françoise Charavay for expert technical assistance in virulence procedures and infection of animals. We are indebted to John F. Mackay for critical reading of the manuscript and editorial contribution.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Adib-Conquy, M., P. Moine, K. Asehnoune, A. Edouard, T. Espevik, K. Miyake, C. Werts, and J. M. Cavaillon. 2003. Toll-like receptor-mediated tumor necrosis factor and interleukin-10 production differ during systemic inflammation. Am. J. Respir. Crit. Care Med. 168:158-164. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, C. L., T. Sathiyaseelan, B. Naiman, A. M. White, R. Brown, S. Blumerman, A. Rogers, and S. J. Black. 2002. Activation of bovine peripheral blood gammadelta T cells for cell division and IFN-gamma production. Vet. Immunol. Immunopathol. 87:251-259. [DOI] [PubMed] [Google Scholar]
- 3.Brown, R. A., S. Blumerman, C. Gay, C. Bolin, R. Duby, and C. L. Baldwin. 2003. Comparison of three different leptospiral vaccines for induction of a type 1 immune response to Leptospira borgpetersenii serovar Hardjo. Vaccine 21:4448-4458. [DOI] [PubMed] [Google Scholar]
- 4.Chierakul, W., M. de Fost, Y. Suputtamongkol, R. Limpaiboon, A. Dondorp, N. J. White, and T. van der Poll. 2004. Differential expression of interferon-gamma and interferon-gamma-inducing cytokines in Thai patients with scrub typhus or leptospirosis. Clin. Immunol. 113:140-144. [DOI] [PubMed] [Google Scholar]
- 5.Cinco, M., E. Vecile, R. Murgia, P. Dobrina, and A. Dobrina. 1996. Leptospira interrogans and Leptospira peptidoglycans induce the release of tumor necrosis factor alpha from human monocytes. FEMS Microbiol. Lett. 138:211-214. [DOI] [PubMed] [Google Scholar]
- 6.de Fost, M., R. A. Hartskeerl, M. R. Groenendijk, and T. van der Poll. 2003. Interleukin 12 in part regulates gamma interferon release in human whole blood stimulated with Leptospira interrogans. Clin. Diagn. Lab. Immunol. 10:332-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diament, D., M. K. Brunialti, E. C. Romero, E. G. Kallas, and R. Salomao. 2002. Peripheral blood mononuclear cell activation induced by Leptospira interrogans glycolipoprotein. Infect. Immun. 70:1677-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellinghausen, H. C., and W. G. McCullough. 1965. Nutrition of Leptospira pomona and 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45-51. [PubMed] [Google Scholar]
- 9.Estavoyer, J. M., E. Racadot, G. Couetdic, J. Leroy, and L. Grosperrin. 1991. Tumor necrosis factor in patients with leptospirosis. Rev. Infect. Dis. 13:1245-1246. [DOI] [PubMed] [Google Scholar]
- 10.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis. Medisci, Melbourne, Australia.
- 11.Isogai, E., H. Isogai, N. Fujii, and K. Oguma. 1990. Macrophage activation by leptospiral lipopolysaccharide. Zentbl. Bakteriol. 273:200-208. [DOI] [PubMed] [Google Scholar]
- 12.Klimpel, G. R., M. A. Matthias, and J. M. Vinetz. 2003. Leptospira interrogans activation of human peripheral blood mononuclear cells: preferential expansion of TCR gamma delta+ T cells vs TCR alpha beta+ T cells. J. Immunol. 171:1447-1455. [DOI] [PubMed] [Google Scholar]
- 13.Koth, M., and T. Calandra. 2003. Cytokine and chemokines, p. 151-161. In M. Koth and T. Calandra (ed.), Infectious diseases handbook. Academic Press, New York, N.Y.
- 14.Kühne, B. S., and P. Oschmann. 2002. Quantitative real-time RT-PCR using hybridization probes and imported standard curves for cytokine gene expression analysis. BioTechniques 33:1078-1089. [DOI] [PubMed] [Google Scholar]
- 15.Listvanova, S., S. Temmerman, P. Stordeur, V. Verscheure, S. Place, L. Zhou, C. Locht, and F. Mascart. 2003. Optimal kinetics for quantification of antigen-induced cytokines in human peripheral blood mononuclear cells by real-time PCR and by ELISA. J. Immunol. Methods 281:27-35. [DOI] [PubMed] [Google Scholar]
- 16.Loeffler, J., P. Swatoch, D. Akhawi-Araghi, H. Hebart, and H. Einsele. 2003. Automated RNA extraction by MagNA Pure followed by rapid quantification of cytokine and chemokine gene expression with use of fluorescence resonance energy transfer. Clin. Chem. 49:955-958. [DOI] [PubMed] [Google Scholar]
- 17.Marangoni, A., R. Aldini, V. Sambri, L. Giacani, K. Di Leo, and R. Cevenini. 2004. Production of tumor necrosis factor alpha by Treponema pallidum, Borrelia burgdorferi s.l., and Leptospira interrogans in isolated rat Kupffer cells. FEMS Immunol. Med. Microbiol. 40:187-191. [DOI] [PubMed] [Google Scholar]
- 18.Melby, P. C., V. V. Tryon, B. Chandrasekar, and G. L. Freeman. 1998. Cloning of Syrian hamster (Mesocricetus auratus) cytokine cDNAs and analysis of cytokine mRNA expression in experimental visceral leishmaniasis. Infect. Immun. 66:2135-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melby, P. C., B. Chandrasekar, W. Zhao, and J. E. Coe. 2001. The hamster as a model of human visceral leishmaniasis: progressive disease and impaired generation of nitric oxide in the face of a prominent Th1-like cytokine response. J. Immunol. 166:1912-1920. [DOI] [PubMed] [Google Scholar]
- 20.Merien, F., G. Baranton, and P. Perolat. 1997. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect. Immun. 65:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naiman, B. M., D. Alt, C. A. Bolin, R. Zuerner, and C. L. Baldwin. 2001. Protective killed Leptospira borgpetersenii vaccine induces potent Th1 immunity comprising responses by CD4 and γδ T lymphocytes. Infect. Immun. 69:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naiman, B. M., S. Blumerman, D. Alt, C. A. Bolin, R. Brown, R. Zuerner, and C. L. Baldwin. 2002. Evaluation of type 1 immune response in naïve and vaccinated animals following challenge with Leptospira borgpetersenii serovar Hardjo: involvement of WC1+ γδ and CD4 T cells. Infect. Immun. 70:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbergh, L., A. Giulietti, D. Valckx, B. Decallonne, R. Bouillon, and C. Mathieu. 2003. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J. Biomol. Tech. 14:33-43. [PMC free article] [PubMed] [Google Scholar]
- 24.Peinnequin, A., C. Mouret, O. Birot, A. Alonso, J. Mathieu, D. Clarençon, D. Agay, Y. Chancerelle, and E. Multon. 2004. Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green. BMC Immunol. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petros, S., U. Leonhardt, and L. Engelmann. 2000. Serum procalcitonin and proinflammatory cytokines in a patient with acute severe leptospirosis. Scand. J. Infect. Dis. 32:104-105. [DOI] [PubMed] [Google Scholar]
- 26.Stordeur, P., L. F. Poulin, L. Craciun, L. Zhou, L. Schandene, A. de Lavareille, S. Goriely, and M. Goldman. 2002. Cytokine mRNA quantification by real-time PCR. J. Immunol. Methods 259:55-64. [DOI] [PubMed] [Google Scholar]
- 27.Stordeur, P., L. Zhou, B. Byl, F. Brohet, W. Burny, D. de Groote, T. Van der Poll, and M. Goldman. 2003. Immune monitoring in whole blood using real-time PCR. J. Immunol. Methods 276:69-77. [DOI] [PubMed] [Google Scholar]
- 28.Tajiki, H., and R. Salomao. 1996. Association of plasma levels of tumor necrosis factor alpha with severity of disease and mortality among patients with leptospirosis. Clin. Infect. Dis. 23:1177-1178. [DOI] [PubMed] [Google Scholar]
- 29.Truccolo, J., O. Serais, F. Merien, and P. Perolat. 2001. Following the course of human leptospirosis: evidence of a critical threshold for the vital prognosis using a quantitative PCR assay. FEMS Microbiol. Lett. 204:317-321. [DOI] [PubMed] [Google Scholar]
- 30.Truccolo, J., F. Charavay, F. Merien, and P. Perolat. 2002. Quantitative PCR assay to evaluate ampicillin, ofloxacin, and doxycycline for treatment of experimental leptospirosis. Antimicrob. Agents Chemother. 46:848-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viriyakosol, S., M. A. Matthias, M. A. Swancutt, T. N. Kirkland, and J. M. Vinetz. 2006. Toll-like receptor 4 protects against lethal Leptospira interrogans serovar Icterohaemorrhagiae infection and contributes to in vivo control of leptospiral burden. Infect. Immun. 74:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werts, C., R. I. Tapping, J. C. Mathison, T. H. Chuang, V. Kravchenko, I. Saint Girons, D. A. Haake, P. J. Godowski, F. Hayashi, A. Ozinsky, D. M. Underhill, C. J. Kirschning, H. Wagner, A. Aderem, P. S. Tobias, and R. J. Ulevitch. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2:346-452. [DOI] [PubMed] [Google Scholar]
- 33.Yang, C. W., M. S. Wu, M. J. Pan, J. J. Hong, C. C. Yu, A. Vandewalle, and C. C. Huang. 2000. Leptospira outer membrane protein activates NF-kappaB and downstream genes expressed in medullary thick ascending limb cells. J. Am. Soc. Nephrol. 11:2017-2026. [DOI] [PubMed] [Google Scholar]