Abstract
Candida biofilms formed on indwelling medical devices are increasingly associated with severe infections. In this study, we used proteomics and Western and Northern blotting analyses to demonstrate that alcohol dehydrogenase (ADH) is downregulated in Candida biofilms. Disruption of ADH1 significantly (P = 0.0046) enhanced the ability of Candida albicans to form biofilm. Confocal scanning laser microscopy showed that the adh1 mutant formed thicker biofilm than the parent strain (210 μm and 140 μm, respectively). These observations were extended to an engineered human oral mucosa and an in vivo rat model of catheter-associated biofilm. Inhibition of Candida ADH enzyme using disulfiram and 4-methylpyrazole resulted in thicker biofilm (P < 0.05). Moreover, biofilms formed by the adh1 mutant strain produced significantly smaller amounts of ethanol, but larger amounts of acetaldehyde, than biofilms formed by the parent and revertant strains (P < 0.0001), demonstrating that the effect of Adh1p on biofilm formation is mediated by its enzymatic activity. Furthermore, we found that 10% ethanol significantly inhibited biofilm formation in vitro, with complete inhibition of biofilm formation at ethanol concentrations of ≥20%. Similarly, using a clinically relevant rabbit model of catheter-associated biofilm, we found that ethanol treatment inhibited biofilm formation by C. albicans in vivo (P < 0.05) but not by Staphylococcus spp. (P > 0.05), indicating that ethanol specifically inhibits Candida biofilm formation. Taken together, our studies revealed that Adh1p contributes to the ability of C. albicans to form biofilms in vitro and in vivo and that the protein restricts biofilm formation through an ethanol-dependent mechanism. These results are clinically relevant and may suggest novel antibiofilm treatment strategies.
Indwelling medical devices (IMDs) infected with Candida represent a major problem, since members of this pathogenic genus, particularly Candida albicans, adhere avidly to biomaterials, including catheters, and form biofilms (a community of microorganisms encased in a self-produced extracellular matrix [ECM]), making antifungal therapy alone insufficient for cure (19, 35, 39). C. albicans is the most common fungal pathogen associated with colonization and biofilm formation on the surfaces of IMDs, such as intravenous catheters (35, 38). Candida biofilms are extremely resistant to commonly used antifungals (7, 37), and as stipulated in the Infectious Diseases Society of America guidelines, removal of the catheter is often necessary to effect a cure (53). However, removal of IMDs may not be feasible in many cases, e.g., in patients with coagulopathy or those with limited vascular access (as often occurs among the pediatric population). Moreover, catheter removal is also associated with increased health care expenses, as well as other complications (49). Therefore, it is critical to devise new approaches to manage biofilm-associated infections, a prerequisite for which is obtaining a better understanding of the biology of the Candida biofilm.
An approach that has been used to understand the biology and gain a global insight into the processes occurring in cells living in a biofilm environment is proteomics. Unlike genomic analyses, the proteomic approach provides information on the functional roles of proteins and protein-protein interactions. This state-of-the-art technique has been used to identify specific proteins expressed differentially in biofilms formed by bacteria (2, 51, 52, 60, 66). A recent proteomic analysis by Ojha et al. (50) identified mycolic acid and GroEL1, a heat shock protein (HSP60) chaperone involved in biosynthesis of mycolic acid, as unique components essential for biofilm formation by Mycobacterium smegmatis. Proteomics has been used to study global protein expression and to identify proteins specific to Candida germination and virulence (56) but has not been applied to Candida biofilms.
The objectives of this study were to (i) use a proteomic approach to identify proteins that are differentially expressed in early-phase biofilms; (ii) use gene disruption and inhibition assays to prove that one of the identified proteins contributes to Candida biofilm formation in vitro; (iii) move our studies closer to the clinical setting by showing that a correlation exists between the ability of isogenic C. albicans strains to form biofilm in vitro, in an engineered human oral mucosa (EHOM) model, and in a rat model of catheter-associated Candida biofilm; and (iv) determine the mechanism by which the identified protein modulates C. albicans biofilm formation.
In this study, using a proteomic approach, we demonstrate that alcohol dehydrogenase (Adh1p) expression is reduced in Candida biofilm relative to planktonic cells (grown in suspension). Targeted disruption of ADH1 or inhibition of the enzyme using specific inhibitors resulted in thicker biofilm in vitro than those of the parent and revertant strains. The effect of Adh1p was also tested in an in vivo-like EHOM model. EHOM tissues infected with the adh1 mutant exhibited increased biofilm formation, tissue damage, and penetration of tissue layers compared to tissues infected with the parent and revertant C. albicans strains. To demonstrate the in vivo relevance of the role of Adh1p in biofilm formation, we tested the ability of an adh1 mutant strain to form biofilm in a rat model of intravenous-catheter-associated Candida biofilm. We found that the adh1 mutant formed more biofilm than the isogenic parental strain, demonstrating that deletion of ADH1 results in increased biofilm formation in vivo. Since Adh1p catalyzes the conversion of acetaldehyde to ethanol, we measured the amounts of these two substances in biofilms and showed a significant increase and decrease in their levels, respectively. Furthermore, we used in vitro and in vivo assays to demonstrate that ethanol inhibits biofilm formation and that the biofilm-inhibitory activity of Adh1p is mediated by its enzymatic activity.
This is the first report that identifies a specific regulator of fungal biofilm, demonstrating its in vivo relevance and associated mechanism. Our findings have important implications for biofilm formation and the coexistence of microbes in a mixed-species environment, as well as wide-ranging clinical implications.
MATERIALS AND METHODS
Candida isolates.
The Candida strains used in this study are described in Table 1. Candida cultures were maintained on Sabouraud dextrose agar (yeast extract, peptone, dextrose [1:2:1]) or kept at −80°C for long-term storage.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BWP17 (DAY1) | ura3Δ::λimm434arg4::hisGhis1::hisG | 67 |
| ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| OSA101 (parent) | ura3Δ::λimm434arg4::hisGhis1::hisG adh1::Tn7-UAU1 | This study |
| ura3Δ::λimm434 arg4::hisG his1::hisG adh1::Tn7-URA3 | ||
| OSA108 (revertant) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1-ADH1 adh1::Tn7-UAU1 | This study |
| ura3Δ::λimm434 arg4::hisG his1::hisG adh1::Tn7-URA3 | ||
| OSA114 (adh1 mutant) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 adh1::Tn7-UAU1 | This study |
| ura3Δ::λimm434 arg4::hisG his1::hisG adh1::Tn7-URA3 | ||
| W303 (AMP 1411) | MATaura3-1 leu2-3,112 his3-11 15 trp1-1 ade2-1 can1-100 | 27 |
Animals.
Rats and rabbits were used in this study to compare the abilities of isogenic strains to form biofilm on catheters and to evaluate the ability of ethanol to inhibit Candida biofilm formation, respectively. Specific-pathogen-free female Sprague-Dawley rats weighing 400 g (Harlan Sprague-Dawley, Indianapolis, Ind.) or female New Zealand White rabbits weighing 2.5 to 3.0 kg (Covance Inc., Kalamazoo, Mich.) were used. The animals were housed in an environmentally controlled room with a 12-h light-dark cycle and were maintained on a standard ad libitum rat diet. Animal studies with rats were in accordance with the American Association for Accreditation of Laboratory Care criteria and approved by the Animal Research Committee of the William S. Middleton Memorial Veterans Administration Hospital, Madison, Wisconsin. All studies with rabbits were performed in accordance with the guidelines for animal health and welfare required by the Institutional Animal Care and Use Committee at Case Western Reserve University School of Medicine, Cleveland, Ohio.
Biofilm formation in vitro.
Candida biofilms were formed and quantified as described previously (7). Briefly, C. albicans cells were grown overnight at 37°C in yeast nitrogen base (YNB) medium (Difco Laboratories, Detroit, MI) supplemented with 50 mM glucose. A standardized cell suspension was prepared from this culture by adjusting the cell density to 1 × 107 cells/ml. Silicone elastomer disks (1.5-cm diameter; Cardiovascular Instrument Corp., Wakefield, MA) were placed in 12-well tissue culture plates and incubated in fetal bovine serum for 24 h at 37°C on a rocker. Next, the disks were immersed in 3 ml of the standardized cell suspension (1 × 107 cells/ml) and incubated for 90 min at 37°C. Subsequently, the disks were immersed in YNB medium with 50 mM glucose and incubated for different times at 37°C on a rocker. Fungal biofilms were quantified as described previously (7) by measuring their biomass and metabolic activity using dry-weight determination and tetrazolium dye [2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] (XTT) reduction assays (by measuring absorbance at 492 nm), respectively. Assays were carried out in four replicates on different days.
Protein extraction.
Candida biofilms are characterized by the presence of cell wall glycoprotein-rich ECM (7, 8, 36). Therefore, we hypothesized that proteins associated with the fungal cell wall play a critical role in biofilm development. To test this hypothesis, C. albicans was grown as biofilm to the early phase (6 h) and as planktonic cells using our in vitro model. Total proteins were extracted and fractionated into different cellular fractions using a modification of the method previously described for Candida cells (48). Similar sodium dodecyl sulfate (SDS) extraction-based methods have routinely been used by our group and other investigators to separate the soluble and insoluble components of C. albicans cell wall proteins (17, 48, 57). Briefly, biofilms and planktonic cells were harvested, washed with prechilled phosphate-buffered saline (PBS), and homogenized with glass beads (0.45-mm diameter; Sigma) in buffer A (10 mM sodium acetate, pH 5.0, 1 mM phenylmethylsulfonyl fluoride). The resulting lysate was centrifuged to remove cytoplasmic material, and the cell wall pellet was incubated with buffer B (10 mM Tris-Cl, pH 7.8, 2% SDS, 100 mM EDTA, 40 mM β-mercaptoethanol) for 10 min at 100°C to release SDS-soluble cell wall (CWS) proteins into the mixture. These released CWS proteins were collected, after a brief centrifugation step, in the supernatant and processed for proteomic analyses.
Proteomic analysis.
Proteomic analyses of the CWS samples isolated from C. albicans biofilm or planktonic cells were performed as described previously (23, 43). A brief description is given below.
(i) Cy dye labeling.
The fractionated CWS protein samples were precipitated by using a Clean-up kit (GE Healthcare). Precipitates were solubilized in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 30 mM Tris), and the concentration was measured by using a 2D Quant kit (GE Healthcare). A 250-μg biofilm sample was labeled with 400 pmol of Cy3, and a 250-μg planktonic sample was labeled with 400 pmol of Cy5. Labeling was performed for 30 min on ice in the dark, after which the reactions were quenched with the addition of 1 μl of 10 mM lysine. The quenched 250-μg Cy3- and Cy5-labeled samples for biofilm and planktonic C. albicans were then combined and mixed together, after which an equal volume of 2× rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 4 mg/ml dithiothreitol) supplemented with 2% immobilized pH gradient (IPG) buffer 4-7 (GE Healthcare) was added.
(ii) Two-dimensional gel electrophoresis and imaging.
The pair-labeled samples for biofilms and planktonic C. albicans (450-μl final volume) were passively rehydrated in 24-cm 4-7 IPG strips (GE Healthcare) for 2 h at 0 V and 12 h at 30 V, followed by isoelectric focusing using a IPGphor II (GE Healthcare) for a total of 65.5 kVh (held at 500 V for 1 h, held at 1,000 V for 1 h, and held at 8,000 V for 8 h). The cysteine sulfhydryls were reduced and carbamidomethylated, while the proteins were equilibrated into the second-dimensional loading buffer by incubating the focused strips in equilibration buffer (30% glycerol, 6 M urea, 2% SDS, 100 mM Tris, pH 8.0, trace bromophenol blue) supplemented with 0.5% dithiothreitol for 10 min at room temperature, followed by 4.5% iodoacetamide in fresh equilibration buffer for an additional 10 min of room temperature incubation. IPG strips were then placed on top of 12% homogeneous polyacrylamide gels that were precast with low-fluorescence glass plates using an Ettan-DALT caster (GE Healthcare). The glass plate was presilanized (Bind-silane; GE Healthcare) to affix the polymerized gel to only one of the glass plates, thereby preventing gel swelling/shrinkage between data acquisition and protein excision. The second-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) was then carried out under the following conditions: 40 mA/gel; ∼130 V starting voltage for 14 h. The Cy3 (biofilm) and Cy5 (planktonic) components of each gel were individually imaged using excitation/emission wavelengths of 532/580 nm for Cy3 and 633/670 nm for Cy5 using a Typhoon 9400 (GE Healthcare). After being imaged for Cy dye components, the nonsilanized glass plate was removed, and the gels were fixed in 10% methanol, 7.5% acetic acid overnight and then washed in wash solution (2.94 g of sodium hydrogen carbonate and 31.8 g of sodium carbonate in 750 ml water brought to 1 liter with water) for 30 min and then incubated in Deep Purple solution (GE Healthcare; 1 in 250 dilution) in the dark for 1 h at room temperature. This poststain visualizes ca. 95% of the unlabeled protein and ensures accurate protein excision, as the molecular weights and hydrophobicity of Cy dyes can influence protein migration during SDS-PAGE. The Deep Purple image was acquired on the same imager using 532/560-nm wavelengths, as well as reimaging postexcision to ensure accurate protein excision.
Data analysis.
DeCyder software (GE Healthcare) differential in-gel analysis was used for simultaneous comparison of abundance changes for pairwise comparisons of individual Cy3 biofilm and Cy5 planktonic samples. The entire signal from each Cy dye channel was normalized prior to the codetection of protein spot boundaries and the calculation of the volume ratio for each protein spot pair. The fold change for each protein spot was reported. Proteins of interest were robotically excised in a 96-well plate format using Ettan Spot Picker (GE Healthcare). The gels were dehydrated and digested in gel with trypsin protease, and tryptic peptides were then extracted from the gel and applied to a matrix-assisted laser desorption ionization (MALDI) target. Peptide mass maps were acquired using MALDI-time of flight/time of flight mass spectrometry performed on a Voyager 4700 (Applied Biosystems). The spot intensity was quantified with the spot normalized volume depending on the whole volume of the difference in-gel electrophoresis (DIGE) image after scanning the image using a Typhoon scanner. Dark-blue spots (Cy5 labeled) indicated proteins identified as upregulated in biofilms, while dark-red spots (Cy3 labeled) indicated proteins downregulated in biofilms. The data were analyzed using the Applied Biosystems GPS Explorer software with Mascot analysis, and a combined peptide mass fingerprinting/tandem mass spectrometry search. An increase in the Cy5/Cy3 ratio indicated proteins upregulated in biofilms, while an increase in the Cy3/Cy5 ratio indicated upregulated proteins in planktonic C. albicans.
Western and Northern blotting analyses.
To confirm the identity and levels of Adh1p during biofilm development, we performed Western and Northern blotting analyses as described previously (7, 46). For Western blot analyses, 10 μg protein was separated by SDS-PAGE and detected using an anti-yeast ADH primary antibody (1:10,000; Calbiochem), a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:5,000; Calbiochem), and a chemiluminescence-based detection kit (ECL; Amersham). For Northern analysis, total RNA was extracted from biofilm and planktonic C. albicans, and Northern analysis of the extracted RNA was performed using a PCR-amplified ADH1-specific probe (primers: forward, 5′-GCAAGCTTATTCAGAATTTTCAGAGGTGC-3′, and reverse, 5′-CAACTGGTGTCCAATACGTATCTACTCAAG-3′).
Disruption and reintroduction of C. albicans ADH1 gene.
To unequivocally confirm the contribution of Adh1p to Candida biofilm formation, we constructed an isogenic strain pair that differed only in the ability to produce Adh1p and a revertant strain in which a functional copy of ADH1 was reintroduced.
Gene disruption.
All Candida albicans strains used in this study were derived from strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG) (67) and are listed in Table 1. The His− homozygous insertion mutant strains were constructed by random-transposon mutagenesis with the UAU1 cassette, as described previously (16). C. albicans strains were grown in YPD plus uridine (1% yeast extract, 2% Bacto Peptone, 2% dextrose, and 80 μg uridine ml−1) at 30°C. Following transformations, selection was done on synthetic medium (2% dextrose, 6.7% yeast nitrogen base plus ammonium sulfate and the auxotrophic requirements).
Gene complementation.
The open reading frame (ORF) of the ADH1 (orf19.3997) gene was identified through the Stanford C. albicans genome database (http://www-sequence.stanford.edu/group/candida). PCR was used to produce a fragment of ADH1 from approximately 1,000 bp upstream of the ATG to approximately 500 bp downstream of the stop codon of the ORF. The primer sequences used were as follows: Forward, 5′-GGTCATAGTTTCATTTGCCCCCG-3′; Reverse, 5′-CCACAACAACAAATACTCTTCTACAGGA-3′. This fragment was inserted into the pGEMT-Easy vector (Promega), which contains NotI or AlwNI and NgoMIV sites flanking the insertion. The insert was then released with AlwNI and NgoMIV and integrated into NotI-digested pDDB78, a TRP1 HIS1 vector (65), by in vivo recombination using Saccharomyces cerevisiae W303 (27), a Trp1− strain (AMP 1411). The recombination within the W303 strain generated plasmid pOSA107 containing the complementing ADH1 insert within the pDDB78 backbone, suitable for integration into the HIS1 locus of C. albicans. The revertant strain (OSA108) was constructed by transforming OSA101 (tADH1 7.4 5), the adh1/adh1 homozygous insertion mutant, to histidine prototrophy with the NruI-digested plasmid pOSA107. The unique NruI site in these plasmids lies in HIS1 sequences; NruI digestion thus directs integration to the HIS1 locus. For comparison to the complemented strains, the mutant strains were made His+ by transforming each mutant to histidine prototrophy with NruI-digested plasmid pDDB78. Strain OSA114 (pDDB78 7.4 5) was derived from mutant OSA101 (tADH1 7.4 5). Disruption and reintroduction of the ADH1 gene was confirmed by PCR analyses of the isogenic strains using the primers mentioned above.
Phenotypic characterization of constructed isogenic strains.
The growth and germination abilities of the constructed isogenic strains were determined as described previously (14, 25, 42). Briefly, growth of the strains was followed at 2-h intervals until the stationary phase of growth was attained. For germination assays, cells were grown in YNB medium overnight. These freshly grown cells were adjusted to a standardized suspension (5 × 107 cells in 50 μl PBS) and then incubated with 10% fetal bovine serum for 180 min. After the incubation period, 10-μl samples were withdrawn for each strain, a wet mount from each sample was prepared, and the number of yeasts that had germ tubes at least 1 blastospore diameter in length was determined microscopically. The percentage of germinating cells for each isogenic strain was determined compared to the total number of cells counted.
Confocal scanning laser microscopy (CSLM) of Candida biofilms.
To determine the effect of Adh1p inhibitor or disruption of the ADH1 gene on the morphology and architecture of Candida biofilms, confocal analyses of the biofilms were performed as described previously (7). Briefly, at various time points during biofilm formation, silicone elastomer disks on which biofilms were developing were transferred to a 12-well plate and incubated for 45 min at 37οC in 4 ml PBS containing the fluorescent stains FUN-1 (10 μM) and concanavalin A (ConA)-Alexa Fluor-488 conjugate (ConA, 25 μg/ml). FUN-1 (excitation wavelength, 543 nm; emission, 560-nm long-pass filter) is converted to orange-red cylindrical intravacuolar structures by metabolically active cells, while ConA (excitation wavelength, 488 nm; emission, 505-nm long-pass filter) binds to glucose and mannose residues of cell wall polysaccharides with green fluorescence. After incubation with the dyes, the silicone elastomer disks were placed on a 35-mm glass-bottom petri dish (MatTek Corp., Ashland, MA) and observed with a Zeiss LSM510 confocal scanning laser microscope (Carl Zeiss, Inc.).
EHOM model.
To move our study closer to the clinical setting, we used an EHOM model and tested the abilities of the isogenic strains to form biofilm and damage and invade EHOM tissue layers. The EHOM model was created as described previously (58). Briefly, fibroblasts and epithelial cells were isolated from human oral biopsy specimens and cultured in a manner allowing the formation of a complex three-dimensional spatial cellular organization, found in normal human oral mucosa. Specimens of normal palatal mucosae were obtained from patients who had attended the dental clinic at Laval University (School of Dental Medicine) to undergo gingival grafts. The sample of normal palatal mucosa removed from the donor site usually exceeds by a few millimeters the size needed for the graft. Usually the excess tissue is thrown away with the biomedical waste products. Following informed consent, the excess tissue was collected and used in the study. The epithelium was separated from the lamina propria using thermolysin treatment. Epithelial cells and fibroblasts were isolated, cultured, and then used to design EHOM. The lamina propria was produced by mixing bovine (skin) type I and type III collagens (Sigma, St. Louis, MO) with gingival fibroblasts and then cultured in fetal calf serum-supplemented culture medium for 4 days. The lamina propria was then seeded with gingival epithelial cells to obtain the EHOM. The tissues were grown under submerged conditions until the total surface of the lamina propria was covered with epithelial cells. To produce a stratified epithelium, the EHOM was raised to an air-liquid interface for five more days to allow the organization of the epithelium into its different strata, including the stratum corneum. To test the abilities of the isogenic Candida strains to form biofilm on EHOM tissue, the oral mucosae were infected with 1 × 106 cells/cm2 of (i) wild-type C. albicans, (ii) the adh1 mutant, or (iii) the revertant strain. Tissues treated with no Candida cells served as uninfected controls for 24 h. The tissues were collected, stained with Masson trichrome, observed with an optical microscope, and photographed.
Quantitation of biofilms formed on EHOM tissues.
To quantitatively assess the difference between biofilm formations by the different C. albicans strains, we collected multiple biopsy specimens from each infected EHOM and stained them with hematoxylin and eosin. All slides were assigned a number, and measurements were made independently by two different observers. The blinded observer measured the thickness of C. albicans biofilms at regular intervals using a calibrated image analysis system (Image Pro-Plus software; Media Cybernetics, MD) as previously described by Chakir et al. (6). Ten measurements were performed on the unfolded area of each slide. The thicknesses of biofilms formed on EHOM tissues infected with the adh1 mutant strain were compared with those of biofilms formed by the wild-type and revertant strains. A P value of <0.05 was considered statistically significant.
Rat model of intravenous-catheter-associated Candida biofilm.
The ability of the adh1 mutant to form biofilm in vivo was tested using a rat model of intravenous-catheter-associated Candida biofilm formation (1), which allows rapid screening of Candida mutants for the ability to form biofilm. The catheter diameter was chosen in an attempt to permit blood flow around the extraluminal catheter surface. To mimic material used in patients, polyethylene tubing (inner diameter, 0.76 mm; outer diameter, 1.52 mm) was chosen. Specific-pathogen-free Sprague-Dawley rats weighing 400 g were used. A heparinized (100-U/ml) catheter was surgically inserted into the external jugular vein and advanced to a site above the right atrium (2-cm length). The catheter was secured to the vein, and the proximal end was tunneled subcutaneously to the midscapular space and externalized through the skin. The catheters were placed 24 h prior to infection to allow a conditioning period for deposition of host protein on the catheter surface. Infection was achieved by intralumenal instillation of 500 μl C. albicans cells (106 cells/ml). After a dwelling period of 4 h, the catheter volume was withdrawn and the catheter was flushed with heparinized 0.15 M NaCl.
The catheters were removed from two animals 24 h after C. albicans infection to determine biofilm development on the internal surfaces of the intravascular devices. The distal 2 cm of the catheter was cut from the entire catheter length for biofilm imaging using both confocal and scanning electron microscopy (SEM). The distal 2 cm of catheter was transected perpendicular to the catheter length into 2-mm-long circular segments.
SEM of catheters inserted in rats.
SEM analysis of catheters placed intravenously in rats was performed as described previously by Andes et al. (1). Briefly, catheter segments were washed with 0.1 M phosphate buffer, pH 7.2, and placed in fixative (1% glutaraldehyde and 4% formaldehyde). The samples were then washed with buffer for 5 min, placed in 1% osmium tetroxide for 30 min, and dehydrated in a series of 10-min ethanol washes (30%, 50%, 70%, 85%, 95%, and 100%). Final desiccation was accomplished by critical-point drying (Tousimis, Rockville, Md.). Specimens were mounted on aluminum stubs and sputter coated with gold. Samples were imaged in a scanning electron microscope (JEOL JSM-6100) in the high-vacuum mode at 10 kV. The images were processed for display using Adobe Photoshop 7.0.1.
Effects of ADH inhibitors on Candida biofilm formation in vitro.
To determine whether biochemical inhibition of ADH enzyme affects Candida biofilm formation, we investigated the effect of treating C. albicans with disulfiram, a specific inhibitor of this enzyme (40, 41), or 4-methylpyrazole (21, 63). Biofilms were grown for 48 h in the presence of 200 μM disulfiram or 10 mM 4-methylpyrazole. The metabolic activities and dry biomasses of the biofilms formed were determined by XTT and dry-weight measurements, respectively.
Determination of levels of extracellular ethanol and acetaldehyde in C. albicans biofilms.
For determining extracellular ethanol and acetaldehyde levels, C. albicans biofilms were scraped into 4 ml of fresh PBS. The resulting suspension was centrifuged at 6,000 × g in a Fisher Scientific centrifuge, and the supernatant was retained. The levels of ethanol and acetaldehyde (g/100 ml) present in these supernatants were then determined using commercially available assay kits (Boehringer Mannheim/R-Biopharm) according to the manufacturers' instructions.
Ethanol lock treatment of biofilm formed by C. albicans on catheters placed in rabbits.
We examined the efficacy of ethanol lock therapy against biofilms formed by C. albicans, Staphylococcus epidermidis, and Staphylococcus aureus in a rabbit model of catheter biofilm, as described previously (62). This model allows the administration and withdrawal of nutrients at different time points and is relevant to the clinical setting of biofilm formation. Briefly, silicone catheters were surgically placed in New Zealand White rabbits. Three groups of 10 rabbits each were infected with C. albicans, S. epidermidis, or S. aureus, respectively. After catheter placement, a standard 300-μl inoculum consisting of 1 × 107 CFU of microorganisms and 100 U of heparin (Abbott Laboratories, North Chicago, Ill.) was “locked” in the internal lumen of the catheter and allowed to dwell for 24 h. The inoculum was removed, and the catheter was flushed daily with 300 μl of heparinized saline (100 U) or 50% ethanol. Daily heparin flushes were performed for the first 3 days. Five animals in each group were treated with 300 μl of 50% ethanol locked in the catheter lumen for 30 min/day for 7 days. The remaining 5 animals in each group were treated with daily heparin flushes. At the end of the treatment period, the rabbits were anesthetized, and 5 ml of blood was obtained for blood culture (BD Bactec; Becton Dickinson) through the catheter and via cardiac puncture. The animals were euthanized with an intracardiac injection of pentobarbital. Next, using sterile technique, the catheter was removed and divided into proximal (subcutaneous tunnel) and distal (intravenous) 4-cm segments. Each segment was evaluated by SEM.
Statistical analysis.
All statistical analyses were performed using StatView version 5.0.1 software (SAS Institute Inc., Cary, NC). For paired comparisons, Students t test was used, while analysis of variance was performed for comparisons involving more than two groups. A P value of <0.05 was considered statistically significant.
RESULTS
Proteomic analysis demonstrated that alcohol dehydrogenase is downregulated in Candida biofilm.
Studies performed with whole-cell proteins result in high background and also make it difficult to detect proteins with low abundance (17, 57). To avoid these limitations, and to identify differentially expressed proteins in early-phase Candida biofilms, we performed fractionation of crude lysates from biofilm and planktonically grown cells to obtain proteins enriched in different cellular components (cell wall, membranes, and cytoplasm). Since Candida biofilms are characterized by the presence of polysaccharide-rich ECM, which is commonly associated with fungal cell walls (7, 47), in this study we focused on the cell wall-enriched fraction of C. albicans biofilm proteins. Crude lysates of biofilm and planktonic C. albicans grown to 6 h were separated by differential centrifugation and detergent fractionation, resulting in a CWS fraction. Using two-dimensional gel electrophoresis, we found that 33 spots were downregulated while 38 spots were upregulated in biofilms. Mass spectrometry revealed the identities of 24 proteins, with molecular masses ranging from 19 kDa to 86 kDa and isoelectric points ranging from 4 to 7 (Fig. 1; see Table S1 in the supplemental material).
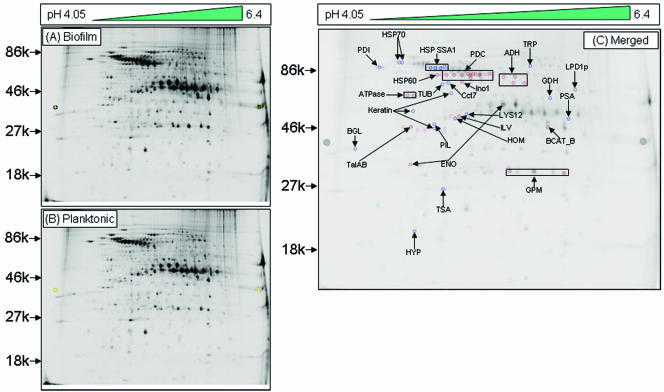
FIG. 1.
Alcohol dehydrogenase is downregulated in cell walls in the early phase of C. albicans biofilms. Two-dimensional DIGE analysis was performed with SDS-soluble cell wall proteins of C. albicans grown as a (A) biofilm (Cy-5 labeled) or (B) planktonic (Cy-3 labeled) form, followed by MALDI-time of flight mass spectrometry analysis. (C) Merged image (Cy5 and Cy3 overlap) showing 24 differentially expressed spots, including Adh1p, in C. albicans biofilms. An increase in the Cy5/Cy3 ratio (dark-blue spots) indicated proteins upregulated in biofilms, while an increase in the Cy3/Cy5 ratio (dark-red spots) indicated upregulated proteins in planktonic C. albicans. Proteins were identified by mass spectrometry analysis of the CWS fraction of C. albicans biofilm. The spot intensity was quantified with the spot normalized volume depending on the whole volume of the DIGE image after scanning the image using a Typhoon scanner. Sequence information from mass spectrometry analysis was matched with the online protein database NCBInr using the MatrixScience Mascot search site (http://www.matrixscience.com). Protein scores of greater than 58 were considered significant (P < 0.05).
The differentially expressed proteins included alcohol dehydrogenase (Adh1p), which was downregulated in biofilms. This protein is known to catalyze the reversible conversion of acetaldehyde to ethanol (4). Our results showed that the level of Adh1p in C. albicans biofilm was ≥1.5-fold lower than in planktonic cells grown to 6 h (see Table S1 in the supplemental material). Next, we performed Western blotting analyses and found that Adh1p levels were lower in biofilm than in planktonic cells at all time points (Fig. 2A). Additionally, Northern blot analysis also revealed that as biofilm formation progressed through developmental phases, the level of ADH1 expression decreased in C. albicans biofilms compared to planktonically grown cells (Fig. 2B). These results confirmed the identity of Adh1p, as well as its reduced expression in C. albicans biofilm.
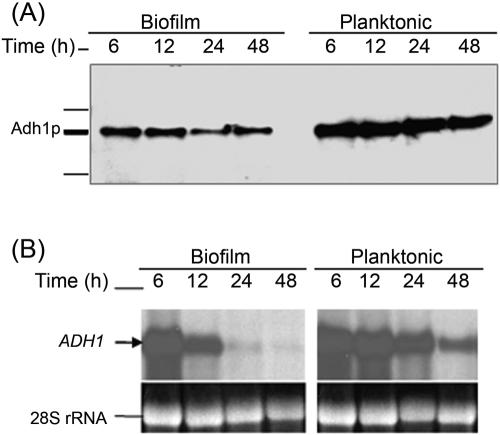
FIG. 2.
Correlation between protein and mRNA level expressions of Adh1p and its corresponding gene in C. albicans biofilms grown to different developmental phases. Temporal expression of ADH1 in C. albicans biofilm during development was determined by (A) Western and (B) Northern blot analyses. As can be seen in panel A, Adh1p protein levels were lower in biofilms than in planktonic cells. (B) Levels of ADH1 mRNA decrease in C. albicans biofilms with time, with no signal detected in mature biofilm.
Molecular and biochemical evidence shows that ADH1 restricts the ability of Candida to form biofilm. (i) Targeted disruption of the ADH1 gene enhances the ability of C. albicans to form biofilm in vitro.
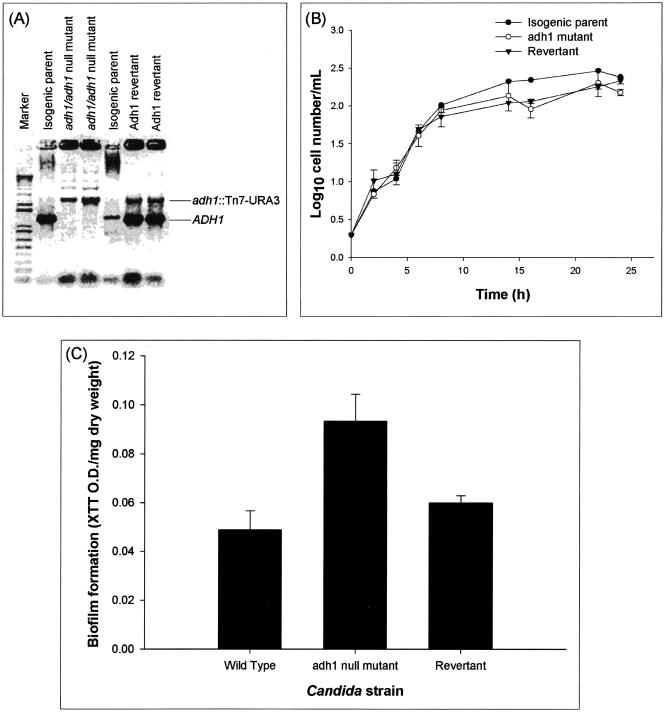
Since Adh1p protein levels were lower in biofilms than planktonic forms of C. albicans, we hypothesized that disruption of the ADH1 gene, which encodes this protein, would result in increased biofilm formation. To test this hypothesis and to unequivocally prove the role of Adh1p in C. albicans biofilm formation, we disrupted the C. albicans ADH1 gene using transposon insertion (16). Disruption and reintroduction of the ADH1 gene were confirmed by PCR amplification, which revealed the presence of the expected 1,297-bp product in the wild-type parent strain and the ADH1 revertant strain, but not in the adh1 mutant (Fig. 3A). Phenotypic characterization of the isogenic adh1 mutant, revertant, and wild-type parent strains revealed that both growth rate (Fig. 3B) and germ tube formation were similar to those of the corresponding isogenic parental strains (Table 2), demonstrating that disruption of the ADH1 gene affected neither the growth of C. albicans nor its ability to form hyphae. Evaluation of metabolic activity and dry-weight biomass (XTT optical density [OD]/mg dry weight) showed that the adh1 mutant formed significantly more biofilm than the isogenic parental C. albicans strain (Fig. 3C) (P = 0.0046). Moreover, confocal microscopic examination revealed that at both early and mature phases, the adh1 mutant formed thicker biofilm than the wild-type strain (78 μm and 210 μm thick for the adh1 mutant and 48 μm and 140 μm thick for the parent strain at early [6 h] and mature [48 h] phases, respectively). The biofilm formed by the revertant strain was similar to that formed by the isogenic parental strain (data not shown). These results provided unequivocal genetic evidence that Adh1p restricts the ability of C. albicans to form biofilm.
FIG. 3.
Disruption of the C. albicans ADH1 gene increases the ability of C. albicans to form biofilm in vitro. (A) Disruption of ADH1 gene. (B) Effect of disruption of the ADH1 gene on the growth curve of planktonic C. albicans. (C) Biofilm formation by the wild-type, adh1 mutant, and revertant strains of C. albicans. The adh1/adh1 mutant strain was constructed from strain BWP17 through use of a Tn7-UAU1 transposon insertion in the middle of the ADH1/orf19.3997 open reading frame as described by Davis et al. (16). The complemented strain was created by cloning the ADH1 ORF, along with 1,000 bp of 5′ flanking sequence and 500 bp of 3′ flanking sequence, in the HIS1 vector pDDB78. The resulting plasmid was transformed into the adh1/adh1 mutant with selection for His+ complementation. Disruption and reintroduction of the ADH1 gene were confirmed by PCR amplification, which revealed the presence of the expected 1,297-bp product in the wild-type parent strain and the ADH1 revertant strain, but not in the adh1 mutant. Biofilm formation by the isogenic strains was quantified as XTT OD/mg (dry weight) (biomass) of biofilm formed. Growth curve studies were performed twice, and a representative set of graphs is shown in panel B. The error bars represent standard deviations.
TABLE 2.
Effects of disruption of the ADH1 gene on the germination ability of C. albicans
| Strain | Mean germinationa (%) | SD | P valueb |
|---|---|---|---|
| Wild-type parent | 93.25 | 2.21 | 0.572 |
| adh1 mutant | 92.25 | 1.71 | |
| Revertant | 91.50 | 0.57 | 0.319 |
Percent germination was determined by counting the number of germinating Candida cells for each isogenic strain incubated in 10% fetal bovine serum for 180 min.
P values were determined by Student's t test, compared to the adh1 mutant strain. A P value of <0.05 was considered statistically significant.
(ii) Downregulation of Adh1p enhances the ability of C. albicans to form biofilm on EHOM and mediates invasion and damage of tissues.
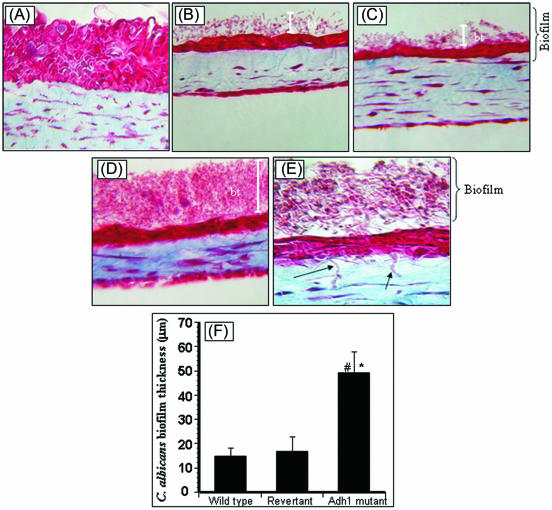
To move our studies closer to the in vivo setting, we determined the effect of Adh1p on the ability of C. albicans to form biofilm in the in vivo-like EHOM model, which mimics the physiologic conditions and complex structure of the oral mucosa in terms of three-dimensional structure, the relationship between the different cell types, and secretion of soluble factors (58). Our results revealed that while all the isogenic strains tested formed biofilms on the EHOM layers and led to disorganization of the tissue layers, the adh1 mutant caused maximum disruption of EHOM layers (Fig. 4). Importantly, hyphae of the adh1 mutant were observed infiltrating into the lamina propria of EHOM, indicating invasion of the tissue layers by the mutant strain (Fig. 4D and E), but not by the isogenic parental and revertant strains. Quantitative analysis revealed that biofilms formed by the adh1 mutant strain on EHOM layers were significantly thicker (P < 0.05) than those formed by the parent or revertant strain (Fig. 4F). These studies demonstrated that disruption of the ADH1 gene resulted in increased biofilm-forming ability in C. albicans and enhanced its ability to invade and damage host mucosal tissues.
FIG. 4.
Effect of disruption of the ADH1 gene on the ability of C. albicans to form biofilms on EHOM tissue layers. Histological features of EHOM following exposure to isogenic C. albicans strains are shown. EHOM tissue was infected with (A) no Candida cells (uninfected control), (B) the parental C. albicans strain, (C) the revertant strain, or (D) the adh1 mutant strain. (E) Hyphae of the adh1 mutant infiltrating into the lamina propria of EHOM (arrow). (F) Quantitative assessment of the thickness of biofilm formed on EHOM tissues. Representative photographs of three different experiments are shown (two EHOMs per experiment). Magnification, ×250 for panel A, ×200 for panels B to D, and ×500 for panel E. * and #, P < 0.0001 compared to the wild-type or revertant C. albicans strain, respectively. The error bars represent standard deviations.
(iii) Scanning electron microscopy revealed that the C. albicans adh1 mutant strain forms a confluent biofilm with dense extracellular matrix on catheters inserted in rats.
The EHOM model provides information about biofilm formation and the penetration and damage induced by C. albicans biofilm. However, the model lacks some in vivo modulatory factors, such as salivary histatins and immunoglobulin A, mucosal dendritic cells, defensins, and lysozyme, and does not completely mimic an in vivo situation. To determine whether Adh1p affects biofilm formation in an in vivo environment, we compared the biofilm-forming abilities of the isogenic set of the adh1 mutant, wild-type, and revertant strains using a recently established rat model of catheter-associated biofilm formation (1). SEM examinations revealed that the wild-type and ADH1 revertant C. albicans strains produced mature biofilm by 24 h with adherent yeast and filamentous populations and matrix (data not shown). Although these strains formed biofilms in vivo, several areas of the catheter surface were visible, indicating nonconfluent and patchy biofilm formation. In contrast, biofilm formed by the adh1 C. albicans mutant was more extensive and completely covered the catheter surface, with dense extracellular matrix (data not shown). To confirm that biofilms formed by the adh1 mutant were thicker than those of the parent or revertant strain on catheters inserted in rats, we used CSLM to visualize the penetration of ConA through the biofilms formed. We detected ConA staining on biofilms formed by the adh1 mutant on internal surfaces of catheters obtained from the in vivo rat model (data not shown). Moreover, the ECM of these biofilms was very dense, which precluded visualization of individual yeast and hyphal cells and prevented the stain from reaching the basal surface of the biofilm adjacent to the catheter surface (data not shown). In contrast, ConA penetrated biofilms formed by C. albicans parent and revertant strains, which exhibited both yeast and filamentous forms, suggesting that the dye penetrated to the basal biofilm layer on the catheter surface (data not shown) and indicating that biofilms formed by the parent and revertant strains were not as thick as those formed by the adh1 mutant. These results correlate well with our in vitro quantitative and microscopy analyses, as well as EHOM results, which revealed that the adh1 mutant forms thicker biofilms than the wild-type and revertant C. albicans strains. Taken together, our studies demonstrated that disruption of the ADH1 gene increased the ability of C. albicans to form biofilm in an in vivo-like EHOM in vitro, and in vivo models, thereby unequivocally proving that Adh1p restricts the ability of C. albicans to form biofilm.
(iv) Biochemical inhibition of ADH enhances its ability to form biofilm.
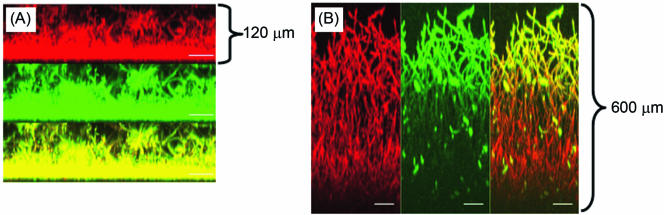
Having shown that disruption of the ADH1 gene enhances the ability of C. albicans to form biofilms, we examined whether inhibiting the enzymatic activity of Adh1p with a known ADH inhibitor, disulfiram (a zinc-chelating agent) (40, 41), modulates the biofilm-forming ability of this organism. Although disulfiram is known to inhibit ADH, it also inhibits aldehyde dehydrogenase (29). Therefore, we included 4-methylpyrazole, which inhibits ADH specifically by binding to the active-site zinc of the enzyme and to the enzyme-cofactor complex (20, 21, 63), in our studies to test the effect of specific ADH inhibition on biofilm formation. Our data showed that upon incubation, C. albicans formed significantly more biofilm in the presence of disulfiram than in its absence (XTT OD/mg dry biomass, 183.4 ± 6.09 and 79.895 ± 5.8, respectively; P < 0.05). A similar pattern of biofilm formation was observed for biofilm formed in the presence of 4-methylpyrazole (XTT OD/mg dry biomass, 107.692 ± 13.3988) compared to that formed in its absence (XTT OD/mg dry biomass, 79.895 ± 5.8; P = 0.034). Next, we used CSLM to examine whether ADH inhibitors affect the morphology and architecture of C. albicans biofilm. CSLM analysis showed that disulfiram treatment resulted in biofilm that was thicker (∼600 μm) than untreated control biofilm (120 μm) (Fig. 5A and B). Moreover, biofilms formed in the presence of disulfiram had a complex architecture with an elaborate network of C. albicans hyphae enmeshed in a carbohydrate-rich ECM. Taken together, these studies provide biochemical confirmation that Adh1p restricts the ability of C. albicans to form biofilm.
FIG. 5.
Effect of ADH inhibitor on C. albicans biofilms. CSLM analysis of biofilm formed in the (A) absence or (B) presence of disulfiram. Side view reconstructions of CSLM images revealed that incubation of biofilm with the inhibitor increased the thickness of mature-phase biofilm from (A) 120 μm to (B) 600 μm. Metabolically active fungal biofilm is indicated in red (due to FUN-1 conversion), while polysaccharide-rich cell wall and matrix are shown in green (due to ConA binding). The yellow regions in both panels represent merged red and green channels of the CSLM-derived images. The bars represent 50 μm in panels C and D (magnification, ×19.5).
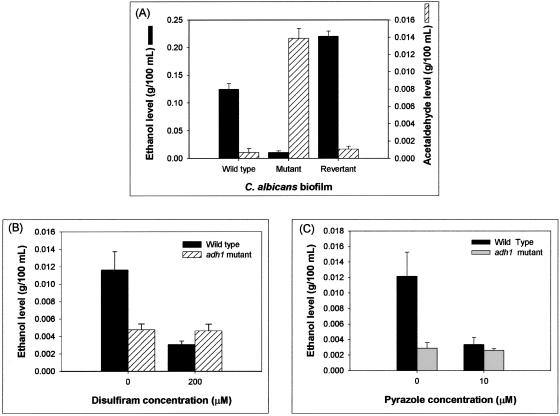
ADH1 downregulates Candida biofilm formation through an ethanol-dependent mechanism. (i) Deletion of the ADH1 gene and inhibition of the ADH enzyme decrease ethanol production and increase the acetaldehyde levels in C. albicans biofilm.
Since disulfiram and 4-methylpyrazole target ADH enzyme activity, we hypothesized that the role of Adh1p in C. albicans biofilm formation is mediated by its enzymatic activity. Adh1p is known to catalyze the reversible conversion of acetaldehyde to ethanol (4). To determine whether this reaction is also catalyzed by Adh1p in C. albicans biofilm, we compared the levels of ethanol and acetaldehyde in biofilms formed by the adh1 mutant, wild-type, and revertant strains. Biofilms formed by the mutant strain produced significantly smaller amounts of ethanol, with concomitantly larger amounts of acetaldehyde (18- to 20-fold higher), than biofilms formed by the parent and revertant strains (P < 0.0001 for both comparisons) (Fig. 6A). These results suggested that the effect of Adh1p on biofilm formation is mediated by its enzymatic activity and not by a general change in cellular metabolism. To further confirm this observation, we determined the effect of inhibition of ADH on ethanol levels in wild-type and adh1 mutant strains of C. albicans. Inhibiting the enzyme activities of biofilms formed by the parent strain (using the specific inhibitors) resulted in significant reduction in ethanol levels (P < 0.001, Fig. 6B and C). However, such reduction was not detected in biofilms formed by the adh1 mutant strain (P > 0.05) (Fig. 6B and C), which was expected, since the mutant strain lacks a functional ADH enzyme. These studies demonstrated that in C. albicans biofilm, Adh1p catalyzes the production of ethanol, suggesting that the mechanism by which Adh1p modulates C. albicans biofilm formation is mediated by its enzymatic activity.
FIG. 6.
Adh1p catalyzes ethanol production in C. albicans biofilm. Ethanol levels in biofilms incubated with inhibitors were calculated as percentages of ethanol levels in biofilm formed in the absence of ethanol, which were considered as 100%. Supernatants (4 ml each) were collected from biofilms grown to mature phase and then used as samples to determine the ethanol and acetaldehyde levels using specific assay kits according to the manufacturer's instructions. The levels of ethanol and acetaldehyde were expressed as g/100 ml of sample. Experiments were repeated in triplicate (n = 3). The error bars represent standard deviations.
(ii) Ethanol inhibits the ability of C. albicans to form biofilm in vitro and in vivo.
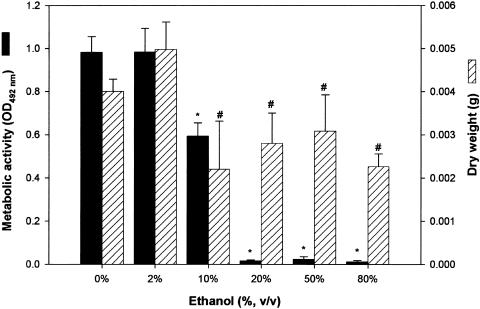
Since disruption of the ADH1 gene and inhibition of the Adh1p enzyme resulted in significantly increased biofilm formation with a concomitant decrease in ethanol levels, we hypothesized that ethanol inhibits C. albicans biofilm formation. To test this hypothesis, using the XTT metabolic-activity assay, we evaluated the effects of different concentrations of ethanol (10% to 80%) on the ability of C. albicans to form biofilm in vitro. Fluorescence microscopy analysis of biofilms formed in the presence of ethanol (and subsequently stained with the fluorescent dye Calcofluor White) revealed distinct changes in biofilm morphologies in the presence of ethanol at concentrations of >10% (data not shown). Only remnants of ECM were detected in biofilm incubated with 20% ethanol, while biofilm structures appeared shriveled and thin in the presence of 50% ethanol. In the presence of 80% ethanol, the biofilm was completely dehydrated, and no ECM was detected (data not shown). As shown in Fig. 7, quantitative analyses of metabolic activity and dry biomass revealed that while 2% ethanol had no effect on biofilm formation, there was a significant decrease in both the XTT activity and dry weight of Candida biofilm grown in the presence of 10% ethanol compared to control biofilms (grown in the absence of ethanol), and interestingly, even in the presence of 80% ethanol, which may be due to residual cells and ECM remnants that contain no viable cells. The metabolic activity of biofilms reached a significantly reduced basal level in the presence of 20% ethanol and remained at this basal level in the presence of higher concentrations of ethanol (50% and 80%). While 20% to 80% ethanol also induced a significant decrease in the dry weights of biofilms, the biomasses of these biofilms were not significantly different from that of biofilm grown in the presence of 10% ethanol (Fig. 7). These studies indicated that 10% ethanol can significantly decrease the biofilm-forming ability of C. albicans.
FIG. 7.
Effects of ethanol on the ability of C. albicans to form biofilm in vitro and in vivo. In vitro biofilm formation in the presence of ethanol was quantitated as metabolic activity (XTT activity) and dry biomass (g) (mean plus standard deviation). Medium containing ethanol at the indicated concentration was added to adhered C. albicans cells, and a biofilm was allowed to form. Significant reduction in Candida biofilm formation was observed in vitro in the presence of 10% ethanol, while biofilm formation was completely inhibited at ethanol concentrations of ≥15%. *, P < 0.001; #, P < 0.05 compared to biofilm formed in the absence of ethanol (0% ethanol control).
To determine whether our in vitro findings were relevant to biofilm inhibition in vivo, we used a rabbit model of C. albicans catheter biofilm (62) to determine whether ethanol inhibits biofilms formed in vivo by C. albicans. We used Staphylococcus epidermidis and S. aureus as controls. Staphyloccus species were selected as controls for two reasons: (i) unlike its effect on Candida biofilms, ethanol is known to enhance the production of Staphylococcus biofilm (10-12, 32-34), and (ii) in contrast to Candida biofilm, ADH1 expression is known to be upregulated in Staphylococcus biofilms (3, 22). The rabbit model was used in these studies because the model has been used to screen the antibiofilm activities of antifungal agents (24, 61) and has been shown to be clinically relevant (5). Our data showed that lock therapy with 50% ethanol inhibited biofilms formed by C. albicans on in vivo catheters but had no effect on the biofilm-forming ability of S. epidermidis or S. aureus (Table 3). In contrast, in vitro studies showed that planktonically grown C. albicans and Staphylococcus were completely inhibited when grown in the presence of 50% ethanol (data not shown). Taken together, these studies demonstrated that (i) ethanol inhibits biofilm formation by C. albicans in vitro and in vivo, (ii) planktonic forms of both C. albicans and S. aureus are susceptible to the inhibitory effect of ethanol, and (iii) the effect of ethanol on in vivo biofilms is specific to C. albicans, and it does not affect Staphylococcus biofilms.
TABLE 3.
Effects of ethanol on biofilm formation in vivo in the rabbit catheter model of Candida biofilmsa
| Organism | Quantitative catheter culture (log10CFU/catheter segment) (mean ± SD)
|
|
|---|---|---|
| Untreated | 50% Ethanol | |
| C. albicans | 2.191 ± 1.008 | 0.000 ± 0.000 |
| S. epidermidis | 3.005 ± 1.553 | 2.761 ± 2.380 |
| S. aureus | 3.429 ± 2.468 | 3.101 ± 1.898 |
Lock therapy with 50% ethanol resulted in complete clearance of the C. albicans biofilm formed on catheters inserted in rabbits but not of the biofilm formed by Staphylococcus spp.
DISCUSSION
In this study, we used a proteomic approach and showed that 24 proteins are differentially expressed in early-phase C. albicans biofilms. Our data are similar to those reported for bacterial biofilms, where a number of proteins were shown to be differentially expressed. Oosthuizen et al. (51) showed that four proteins were uniquely present in biofilm formed by Bacillus cereus, while seven proteins were absent. In a subsequent study using a different B. cereus strain, the same group (52) reported that 15 unique proteins were detected in early-phase biofilm, while seven proteins were overexpressed in the mature biofilm. In a separate study, Sauer and Camper (60) reported that 15 proteins were upregulated following bacterial adhesion and 30 proteins were downregulated in biofilm formed by the plant saprophyte Pseudomonas putida on a silicone surface.
One of the proteins found to be downregulated in our analysis was ADH. We focused on ADH because it is differentially expressed in bacterial biofilms (3, 22), has been shown to have immunogenic properties (45, 55), and may have a role in fungal-host cell interactions mediated by its ability to bind host structural proteins (e.g., plasminogen and integrins) (13, 31). In contrast to our finding with Candida biofilm, where we showed ADH is downregulated, Becker et al. (3) reported that ADH1 was upregulated in S. aureus biofilms. Such differential roles of the same enzyme in different microbial biofilms may have implications for the interactions between C. albicans and bacteria in a mixed-species milieu. For example, it is likely that bacteria, by overexpressing ADH, will enhance their ability to form biofilm while at the same time inhibiting Candida biofilm formation. In this regard, preliminary studies by our group showed that addition of exogenous bacterial ADH to Candida reduced its ability to form biofilm on a catheter surface (M. A. Ghannoum and P. K. Mukherjee, unpublished data).
Molecular and biochemical evidence presented in this study showed that Adh1p restricts the ability of Candida to form biofilm, as revealed by enhanced biofilm formation in response to disruption of the ADH1 gene or biochemical inhibition of the enzyme. In contrast to our finding with Candida biofilm, Finelli et al. (22) showed that disruption of a putative P. aeruginosa alcohol dehydrogenase had no effect on planktonic growth but caused defects in biofilm formation in static and flowing systems. In the same study, competition experiments showed that the corresponding mutant demonstrated reduced fitness compared with the parent strain. The differences in the contributions of ADH to biofilm formation by P. aeruginosa and C. albicans make it tempting to speculate that the enzyme may be involved in the bacterial-fungal interactions that exist in the environment. In this regard, Hogan and Kolter (28) suggested a link between biofilm formation and the activities of some eukaryotic specific virulence factors toward fungal cells and that antagonism between bacteria and fungi may contribute to the evolution and maintenance of many pathogenesis-related genes.
Our studies extended investigation of the role of Adh1p in Candida biofilm formation from the in vitro to the in vivo setting, where we used an in vivo-like EHOM model, as well as a catheter-associated biofilm rat model, and confirmed that Adh1p restricts the ability of Candida to form biofilms. This investigation is the first to demonstrate a correlation between the activity of a protein and biofilm formed in vitro and in vivo and brings us closer to the clinical setting. The observation that an adh1 mutant of C. albicans formed increased biofilm and could invade the lamina propria of EHOM suggests that biofilm plays a role in the ability of Candida to invade host tissues. It is likely that having the ability to form biofilm in the gastrointestinal tract allows Candida to transmigrate across the gastrointestinal submucosa and cause systemic infection. In the nonbiofilm (planktonic) form of C. albicans, Adh1p has been suggested to have multiple roles, including interactions with the host immune system (55) and binding host proteins (31). Crowe et al. (13) used a proteomics-based approach to identify Adh1p as one of eight C. albicans cell wall proteins that were able to bind plasminogen and that activated plasmin release. Since plasmin is known to have proteolytic activity, these authors suggested that Adh1p may also contribute to fungal invasion of host tissues. Since our studies showed that the adh1 mutant had greater ability to penetrate EHOM layers, one mechanism of action of Adh1p in biofilm formation may be modulating host tissue invasion by Candida biofilm. This aspect of the mechanism of action of Adh1p in Candida biofilm formation remains to be investigated.
The mechanism by which Adh1p regulates biofilm formation is unknown. Our studies showed that Adh1p catalyzes the production of ethanol in Candida biofilm and that deletion of the C. albicans ADH1 gene resulted in a decrease in ethanol and an increase in acetaldehyde levels. Similarly, inhibition of Adh1p with specific inhibitors also increased the ability of C. albicans to form biofilms. These results suggested that the effect of Adh1p on biofilm formation is mediated by the activity of the ADH enzyme itself and is not due to changes in metabolism, but is specific. Ethanol is a general disinfectant (30) and has been shown to have additional roles, e.g., mediation of microbial synergy and signaling (9, 18, 64). Our studies showed that ethanol inhibited Candida biofilm formation both in vitro and in vivo, indicating that ADH downregulates Candida biofilm formation through an ethanol-dependent mechanism. One possible explanation for the role of ADH in C. albicans biofilm formation could be based on the concept that ethanol is an inhibitory end product of metabolism in this pathogenic yeast. Under conditions of waste product inhibition, ADH may be downregulated as a way of avoiding ethanol toxicity.
Our observation that ethanol lock therapy did not affect bacterial biofilms while inhibiting Candida biofilm, but had an inhibitory effect on the growth of planktonic cells (Candida as well as bacteria), suggests that the inhibitory effect of ethanol on biofilm formation is specific to C. albicans and is not related to its antiseptic activity per se. Our finding that ethanol has no effect on Staphylococcus biofilms in vivo is consistent with previous findings showing that ethanol enhances biofilm formation by these bacterial species (10-12, 26, 32-34, 44). Our results are likely to have important clinical implications, especially since some previous studies have suggested the use of ethanol lock therapy to treat central venous line infections in the adult and pediatric populations (15, 54, 59). Although the focus of our study was not to test the efficacy of ethanol as a therapeutic alternative in the treatment of catheter-associated infections, the fact that ethanol does not affect bacterial biofilm formed on catheters in vivo suggests caution in the random use of ethanol lock therapy. Such use of ethanol can result in enhanced bacterial biofilm formation on intravenous catheters, similar to that resulting from the use of ethanol-based skin disinfectants, which can induce increased biofilm formation by S. epidermidis (33).
In conclusion, our studies revealed that Adh1p contributes to the ability of C. albicans to form biofilms in vitro and in vivo and that the protein restricts the ability of Candida to form biofilms through an ethanol-dependent mechanism. Our findings provide insight not only into Candida biofilms, but also into the interactions between fungi and bacteria in mixed-species environments. These results are clinically relevant and have important implications for understanding the formation of Candida biofilms on medical devices and the mechanism of C. albicans-mediated host tissue damage and may point to novel treatment strategies for these infections.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance provided by Guangyin Zhou, Jian Li for assistance in biofilm formation, and Jeniel Nett for technical help in collection of in vivo biofilm images.
This work was supported by funds from the NIH (1R01 DE13932-01A1), a Bristol Myers Squibb Freedom to Discover Award to M.A.G., and American Heart Association (SDG 0335313N) and NIH/SDRC Pilot and Feasibility Award (P30-AR-39750) funds to P.K.M. The assistance of the Confocal Scanning Laser Microscopy core facility (NCI grant P30CA43703-12) at Case Western Reserve University is gratefully acknowledged.
Editor: A. Casadevall
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andes, D., J. Nett, P. Oschel, R. Albrecht, K. Marchillo, and A. Pitula. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 72:6023-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P., W. Hufnagle, G. Peters, and M. Herrmann. 2001. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 67:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram, G., R. K. Swoboda, G. W. Gooday, N. A. Gow, and A. J. Brown. 1996. Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast 12:115-127. [DOI] [PubMed] [Google Scholar]
- 5.Castagnola, E., M. G. Marazzi, A. Tacchella, and R. Giacchino. 2005. Broviac catheter-related candidemia. Pediatr. Infect. Dis. J. 24:747. [DOI] [PubMed] [Google Scholar]
- 6.Chakir, J., M. Laviolette, M. Boutet, R. Laliberte, J. Dube, and L. P. Boulet. 1996. Lower airways remodeling in nonasthmatic subjects with allergic rhinitis. Lab. Investig. 75:735-744. [PubMed] [Google Scholar]
- 7.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans—development, architecture and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra, J., P. K. Mukherjee, S. D. Leidich, F. F. Faddoul, L. L. Hoyer, L. J. Douglas, and M. A. Ghannoum. 2001. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903-908. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., D. L. Clemens, A. I. Cederbaum, and B. Gao. 2001. Ethanol inhibits the JAK-STAT signaling pathway in freshly isolated rat hepatocytes but not in cultured hepatocytes or HepG2 cells: evidence for a lack of involvement of ethanol metabolism. Clin. Biochem. 34:203-209. [DOI] [PubMed] [Google Scholar]
- 10.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:171-177. [DOI] [PubMed] [Google Scholar]
- 13.Crowe, J. D., I. K. Sievwright, G. C. Auld, N. R. Moore, N. A. Gow, and N. A. Booth. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 47:1637-1651. [DOI] [PubMed] [Google Scholar]
- 14.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 15.Dannenberg, C., U. Bierbach, A. Rothe, J. Beer, and D. Korholz. 2003. Ethanol-lock technique in the treatment of bloodstream infections in pediatric oncology patients with broviac catheter. J. Pediatr. Hematol. Oncol. 25:616-621. [DOI] [PubMed] [Google Scholar]
- 16.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Groot, P. W. J., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domenicotti, C., D. Paola, A. Vitali, M. Nitti, D. Cottalasso, G. Poli, M. A. Pronzato, and U. M. Marinari. 1999. Primary role of alcohol dehydrogenase pathway in acute ethanol-induced impairment of protein kinase C-dependent signaling system. Adv. Exp. Med. Biol. 463:321-330. [DOI] [PubMed] [Google Scholar]
- 19.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 20.Eklund, H., B. V. Plapp, J. P. Samama, and C. I. Branden. 1982. Binding of substrate in a ternary complex of horse liver alcohol dehydrogenase. J. Biol. Chem. 257:14349-14358. [PubMed] [Google Scholar]
- 21.Eklund, H., J. P. Samama, and L. Wallen. 1982. Pyrazole binding in crystalline binary and ternary complexes with liver alcohol dehydrogenase. Biochemistry 21:4858-4866. [DOI] [PubMed] [Google Scholar]
- 22.Finelli, A., C. V. Gallant, K. Jarvi, and L. L. Burrows. 2003. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:2700-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman, D. B., S. Hill, J. W. Keller, N. B. Merchant, S. E. Levy, R. J. Coffey, and R. M. Caprioli. 2004. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 4:793-811. [DOI] [PubMed] [Google Scholar]
- 24.Ghannoum, M., L. A. Long, H. G. Kim, R. Munyon, V. Rotondo, J. Chandra, and P. K. Mukherjee. 2005. Amphotericin B lipid complex (ABLC) antifungal lock therapy (AFLT) is effective in the treatment of Candida albicans catheter-associated biofilm infections. Presented at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy.
- 25.Ghannoum, M. A., S. G. Filler, A. S. Ibrahim, Y. Fu, and J. E. Edwards, Jr. 1992. Modulation of interactions of Candida albicans and endothelial cells by fluconazole and amphotericin B. Antimicrob. Agents Chemother. 36:2239-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch, J. P., C. Dietzel, and J. Kurjan. 1991. The carboxyl terminus of Scg1, the G alpha subunit involved in yeast mating, is implicated in interactions with the pheromone receptors. Genes Dev. 5:467-474. [DOI] [PubMed] [Google Scholar]
- 28.Hogan, D. A., and R. Kolter. 2002. Pseudomonas- Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 29.Jornvall, H., J. Hempel, and B. Vallee. 1987. Structures of human alcohol and aldehyde dehydrogenases. Enzyme 37:5-18. [DOI] [PubMed] [Google Scholar]
- 30.Kampf, G., and A. Kramer. 2004. Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin. Microbiol. Rev. 17:863-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klotz, S. A., M. L. Pendrak, and R. C. Hein. 2001. Antibodies to α5β1 and αvb3 integrins react with Candida albicans alcohol dehydrogenase. Microbiology 147:3159-3164. [DOI] [PubMed] [Google Scholar]
- 32.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knobloch, J. K., M. A. Horstkotte, H. Rohde, P. M. Kaulfers, and D. Mack. 2002. Alcoholic ingredients in skin disinfectants increase biofilm expression of Staphylococcus epidermidis. J. Antimicrob. Chemother. 49:683-687. [DOI] [PubMed] [Google Scholar]
- 34.Knobloch, J. K., S. Jager, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kojic, E. M., and R. O. Darouiche. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn, D. M., J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn, D. M., T. George, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46:1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn, D. M., and M. A. Ghannoum. 2004. Candida biofilms: antifungal resistance and emerging therapeutic options. Curr. Opin. Investig. Drugs 5:186-197. [PubMed] [Google Scholar]
- 39.Kumamoto, C. A. 2002. Candida biofilms. Curr. Opin. Microbiol. 5:608-611. [DOI] [PubMed] [Google Scholar]
- 40.Langeland, B. T., and J. S. Kinley-McKee. 1996. The effects of disulfiram on equine hepatic alcohol dehydrogenase and its efficiency against alcoholism: vinegar effect. Alcohol 31:75-80. [DOI] [PubMed] [Google Scholar]
- 41.Langeland, B. T., and J. S. McKinley-McKee. 1997. The effects of disulfiram and related compounds on equine hepatic alcohol dehydrogenase. Comp. Biochem. Physiol. C 117:55-61. [DOI] [PubMed] [Google Scholar]
- 42.Leidich, S. D., A. S. Ibrahim, Y. Fu, A. Koul, C. Jessup, J. Vitullo, W. Fonzi, F. Mirbod, S. Nakashima, Y. Nozawa, and M. A. Ghannoum. 1998. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273:26078-26086. [DOI] [PubMed] [Google Scholar]
- 43.Lilley, K. S., and D. B. Friedman. 2004. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Exp. Rev. Proteomics 1:401-409. [DOI] [PubMed] [Google Scholar]
- 44.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez, J. P., M. L. Gil, J. L. Lopez-Ribot, and W. L. Chaffin. 1998. Serologic response to cell wall mannoproteins and proteins of Candida albicans. Clin. Microbiol. Rev. 11:121-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee, P. K., J. Chandra, D. M. Kuhn, and M. A. Ghannoum. 2003. Mechanism of azole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 71:4333-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee, P. K., S. Mohamed, J. Chandra, M. K. Schinabeck, and M. A. Ghannoum. 2003. Proteomic identification of phase-specific cell wall proteins expressed during Candida albicans biofilm formation, abstr. M-381. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, D.C.
- 48.Mukherjee, P. K., and R. Prasad. 1998. Purified arginine permease of Candida albicans is functionally active in a reconstituted system. Yeast 14:335-345. [DOI] [PubMed] [Google Scholar]
- 49.Nucci, M., and E. Anaissie. 2002. Should vascular catheters be removed from all patients with candidemia? An evidence-based review. Clin. Infect. Dis. 34:591-599. [DOI] [PubMed] [Google Scholar]
- 50.Ojha, A., M. Anand, A. Bhatt, L. Kremer, W. R. Jacobs, Jr., and G. F. Hatfull. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861-873. [DOI] [PubMed] [Google Scholar]
- 51.Oosthuizen, M. C., B. Steyn, D. Lindsay, V. S. Brozel, and A. von Holy. 2001. Novel method for the proteomic investigation of a dairy-associated Bacillus cereus biofilm. FEMS Microbiol. Lett. 194:47-51. [DOI] [PubMed] [Google Scholar]
- 52.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. von Holy, and V. S. Brozel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pappas, P. G., J. H. Rex, J. D. Sobel, S. G. Filler, W. E. Dismukes, T. J. Walsh, and J. E. Edwards. 2004. Guidelines for treatment of candidiasis. Clin. Infect. Dis. 38:161-189. [DOI] [PubMed] [Google Scholar]
- 54.Pennington, C. R., and A. D. Pithie. 1987. Ethanol lock in the management of catheter occlusion. JPEN J. Parenter. Enteral. Nutr. 11:507-508. [DOI] [PubMed] [Google Scholar]
- 55.Pitarch, A., J. Abian, M. Carrascal, M. Sanchez, C. Nombela, and C. Gil. 2004. Proteomics-based identification of novel Candida albicans antigens for diagnosis of systemic candidiasis in patients with underlying hematological malignancies. Proteomics 4:3084-3106. [DOI] [PubMed] [Google Scholar]
- 56.Pitarch, A., M. Sanchez, C. Nombela, and C. Gil. 2003. Analysis of the Candida albicans proteome. I. Strategies and applications. J. Chromatogr. B 787:101-128. [DOI] [PubMed] [Google Scholar]
- 57.Pitarch, A., M. Sanchez, C. Nombela, and C. Gil. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteom. 1:967-982. [DOI] [PubMed] [Google Scholar]
- 58.Rouabhia, M., and N. Deslauriers. 2002. Production and characterization of an in vitro engineered human oral mucosa. Biochem. Cell Biol. 80:189-195. [DOI] [PubMed] [Google Scholar]
- 59.Salzman, M. B., H. D. Isenberg, and L. G. Rubin. 1993. Use of disinfectants to reduce microbial contamination of hubs of vascular catheters. J. Clin. Microbiol. 31:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schinabeck, M. K., M. D'Angelo, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2004. Anidulafungin inhibits Candida albicans biofilms in vitro. Presented at the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy.
- 62.Schinabeck, M. K., L. A. Long, M. A. Hossain, J. Chandra, P. K. Mukherjee, S. Mohamed, and M. A. Ghannoum. 2004. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob. Agents Chemother. 48:1727-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sigman, D. S. 1967. Interactions of substrates, inhibitors, and coenzymes at the active site of horse liver alcohol dehydrogenase. J. Biol. Chem. 242:3815-3824. [PubMed] [Google Scholar]
- 64.Smith, M. G., S. G. Des Etages, and M. Snyder. 2004. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 24:3874-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spreghini, E., D. A. Davis, R. Subaran, M. Kim, and A. P. Mitchell. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell. 2:746-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vilain, S., P. Cosette, M. Hubert, C. Lange, G. A. Junter, and T. Jouenne. 2004. Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal. Biochem. 329:120-130. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.