Abstract
Replacement of phenylalanine with leucine at position 391 in squalene epoxidase was identified as being responsible for terbinafine resistance in mutants of Aspergillus nidulans. The equivalent mutation was engineered into the ergA gene of Aspergillus fumigatus, resulting in an F389L substitution that also conferred resistance to this pathogenic mold.
Terbinafine (TRB) is a member of the allylamine group of antimycotics interfering with early-stage ergosterol biosynthesis by inhibiting the enzyme squalene epoxidase (SE) (4, 6, 17). TRB is used routinely to treat superficial mycoses, but it has also been used in the successful treatment of chromoblastomycosis, Fusarium oxysporum infection, pulmonary aspergillosis, sporotrichosis, and leishmaniasis (7, 14, 16, 18). Thus, the therapeutic potential of TRB extends well beyond its current use against superficial mycoses, since it has potential clinical efficacy in the treatment of subcutaneous and certain systemic infections. In this study, we have explored a mechanism for TRB resistance in Aspergillus by first identifying a resistance-associated mutation within the SE of Aspergillus nidulans. The homologous mutation was then introduced into Aspergillus fumigatus, generating a similar resistant phenotype.
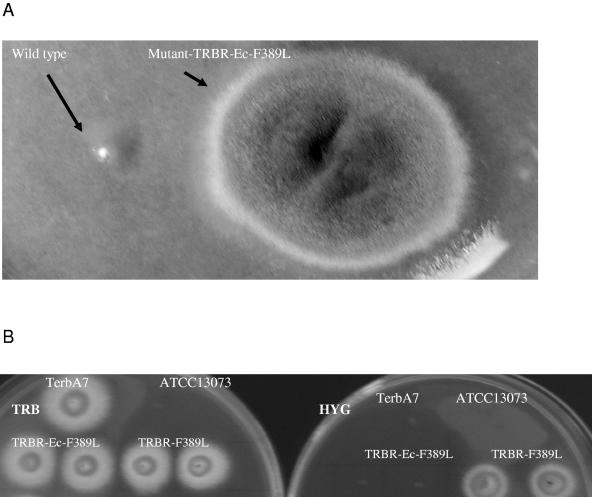
TRB mutants Terb7 and Terb10 (Table 1) were generated from TRB-sensitive A. nidulans strain pabaA1 by UV mutagenesis (15). These mutants exhibited a high level of resistance to TRB (MIC, ≥8.0 μg/ml) in comparison to TRB-sensitive strain pabaA1 (15). Genetic analysis indicated that a single gene (designated tebA), located on chromosome IV, was responsible for resistance to TRB (15). The position of tebA was estimated to be within contig 1.132 because of its proximity to the pyroA marker. Analysis of the sequence adjacent to the pyroA marker identified a putative open reading frame at positions 42692 to 44221 within the contig designated A. nidulans hypothetical protein AN7751.2 (GenBank accession no. EAA61539) (http://www.broad.mit.edu/annotation/fungi/aspergillus). The A. nidulans tebA sequence displayed 40% sequence identity to the Erg1 proteins from Saccharomyces cerevisiae and Candida albicans, respectively (GenBank accession no. CAA97201.1 and BAA13565); thus, the tebA gene was subsequently referred to as the ergA gene (GenBank accession no. DQ391275). Chromosomal DNAs of the Terb7 and Terb10 TRB mutants, which contain the mutations tebA7 and tebA10 (15), were extracted with a FastDNA kit (Qbiogene, Inc., Carlsbad, CA). These served as the template for amplification of the ergA gene of A. nidulans by PCR. The DNA sequences of the PCR products were compared with that of the wild type (pabaA1) and also with that of the FGSC A4 strain obtained from the NCBI (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi). The results showed that each of the ergA alleles of the TRB-resistant mutants contained a single nucleotide exchange (a T-to-C mutation at nucleotide position 1171), which led to the replacement of phenylalanine with leucine at position 391 (F391L) in ErgA from the TRB-resistant mutants (Fig. 1 and Table 1). The ergA nucleotide sequence of A. nidulans was used to BLAST search The Institute for Genomic Research A. fumigatus genome database for homologous sequences (http://www.tigr.org/tdb/fungal/). A nucleotide sequence with 73% sequence similarity to ErgA was obtained from the A. fumigatus genome database (GenBank accession no. EAL91820, AY619002.1, and AY532916) (11). The ergA PCR product of A. fumigatus was cloned into the pCR-TOPO-TA vector (Invitrogen), thus forming plasmid pRG46 (Table 1). A corresponding point mutation was then introduced into the homologous ergA gene at nucleotide position 1664 (T to C), resulting in the replacement of phenylalanine with leucine at position 389 (F389L). Nucleotide alterations were introduced into the 2.5-kb ergA gene fragment with the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), thus forming plasmid pRG47 (Table 1). Plasmid pRG47 also contains a silent SacII site mutation within the ergA gene fragment to facilitate rapid identification of transformants. Transformations of A. fumigatus were carried out by electroporation (3) and protoplasting (5) with the plasmids listed in Table 1. For transformations carried out with plasmid pRG47, TRB-resistant colonies were directly selected upon BD Difco Antibiotic Medium 3 (AM3; Becton Dickinson & Co.) supplemented with 1.0 μg/ml TRB (lot 23925712; stock solution of 25 mg/ml; LKT Laboratories, Inc.) after 3 to 4 days of incubation at 37°C. We isolated several TRB-resistant A. fumigatus strains by transforming plasmid pRG47 into susceptible strain ATCC 13073 (Fig. 2). DNA analysis revealed the ergA gene to be ectopically integrated into the TRBR-Ec (TRB-resistant ectopic) strain, indicating that expression of the ectopic mutant gene was dominant over the wild-type allele. The selection of successful transformants was robust, suggesting potential use of the ergA gene as a selectable marker in A. fumigatus. Strain TRBR-Ec was observed to have a high level of resistance (>500-fold) to TRB in comparison with TRB-sensitive strain ATCC 13073, for which the MIC was approximately 0.3 μg/ml TRB. MICs were determined by the NCCLS M38-P microdilution method (12) in RPMI 1640 medium (Sigma Aldrich, St. Louis, MO) in the presence of 0.03 to 160.0 μg/ml TRB after 24 and 48 h of growth at 37°C. The TRBR-F389L strain did not display cross-resistance to common azole antifungal drugs such as fluconazole, clotrimazole, itraconazole, and ketoconazole (data not shown), suggesting a mechanism-specific resistance to TRB.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| Strains | ||
| pabaA1 | TRB-sensitive A. nidulans strain | 15 |
| TerbA7 | TRB-resistant mutant of pabaA1 isolated after UV mutagenesis | 15 |
| TerbA10 | TRB-resistant mutant of pabaA1 isolated after UV mutagenesis | 15 |
| ATCC 13073 | TRB-sensitive A. fumigatus strain | ATCC (Manassas, VA) |
| TRBR-Ec-F389L | TRB-resistant mutant of ATCC 13073 transformed with pRG47 | This study |
| TRBS | TRB-sensitive strain ATCC 13073 transformed with empty plasmid pRG3-AMA1-HYG | This study |
| TRBR-F389L | TRB-resistant mutant of ATCC 13073 transformed with pRG48 | This study |
| TRBS | Strain derived from TRBR-F389L after plasmid eviction | This study |
| TRBS | TRB-sensitive strain ATCC 13073 transformed with pRG49 | This study |
| Plasmids | ||
| pRG3-AMA1 | Contains A. nidulans AMA1 gene | 1, 2 |
| pRG3-AMA1-HYG | pRG3-AMA1 containing 4-kb KpnI fragment of E. coli HYG resistance gene | This study |
| pRG46 | 2.5-kb fragment of ergA in pCR-TOPO-TA | This study |
| pRG47 | pRG46 containing point mutation F389L in ergA | This study |
| pRG48 | pRG3-AMA1-HYG containing 2.5-kb SmaI fragment of ergA from pRG47 | This study |
| pRG49 | pRG3-AMA1-HYG containing 2.5-kb SmaI fragment of ergA from pRG46 | This study |
FIG. 1.
Multiple-sequence alignment of fungal ErgA domains with amino acid substitutions that confer resistance to TRB. Amino acid substitutions within regions of the Erg1 (ErgA) protein which confer resistance to TRB in S. cerevisiae are shown above the sequences (9, 10). The F389L mutation in the ErgA protein that corresponds to the F391L and F402L substitutions observed in the TRB-resistant isolates of A. nidulans and S. cerevisiae (respectively) is indicated. The L393F mutation observed in the TRB-resistant Trichophyton rubrum clinical isolate is also indicated (13).
FIG. 2.
TRB-sensitive and -resistant isolates of A. fumigatus. (A). Growth of strains ATCC 13073 (wild type) and TRBR-Ec-F389L (mutant) upon AM3 with 1.0 μg/ml TRB. (B) Growth of Aspergillus upon AM3 with 1.0 μg/ml TRB (left) and upon MM with 200 μg/ml HYG (right). Top, left to right: control strains TerbA7 (TRB-resistant A. nidulans) and ATCC 13073 (TRB-sensitive A. fumigatus). Bottom, left to right: strains TRBR-Ec-F389L and TRBR-F389L. Strains were grown for 72 h at 37°C.
To determine whether the TRB resistance displayed by strain TRBR-F389L was dependent on the presence of the mutant ergA gene of A. fumigatus, we evicted plasmids pRG48 and pRG49 (Table 1) by subculturing the strain on minimal medium (MM) without drug selection. Loss of the plasmids was associated with concomitant loss of resistance to TRB on AM3 containing TRB (1.0 μg/ml) and hygromycin B (HYG; 200 μg/ml in MM; Roche Applied Science, Inc.), a selectable marker on the plasmid. Colonies that were unable to grow upon either drug-containing medium were considered to have lost the plasmid; all such strains showed fully restored susceptibility to TRB.
Thus, TRB resistance of strain TRBR-F389L was associated with the mutant ergA gene of A. fumigatus. The F389L substitution in ErgA corresponds to the F402L substitution identified in a TRB-resistant isolate of S. cerevisiae, located within a purported drug-binding site (8, 10). It is interesting that this phenylalanine residue is highly conserved among fungi (Fig. 1) and mammals (10). Thus, TRB resistance in strain TRBR-F389L suggests an aberrant TRB-SE interaction. The lack of cross-resistance to other antifungal classes suggested that the mechanism of resistance reflects the specific nature of drug-target interactions at the SE. In this report, we demonstrate that a single mutation in the ergA gene is sufficient to confer TRB resistance in A. fumigatus.
Acknowledgments
This work was supported by NIH grant AI066561 to D.S.P.
E.M.F.R. acknowledges Padmaja Paderu, Juan C. Robles, Rema Suresh, Guillaume Delmas, Sergey V. Balashov, Svetlana Senina, and Steven Cagas for technical support.
REFERENCES
- 1.Aleksenko, A., and A. J. Clutterbuck. 1996. The plasmid replicator AMA1 in Aspergillus nidulans is an inverted duplication of a low-copy-number dispersed genomic repeat. J. Mol. Microbiol. 19:565-574. [DOI] [PubMed] [Google Scholar]
- 2.Aleksenko, A., and A. J. Clutterbuck. 1997. Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet. Biol. 21:373-387. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Favre, B., and N. S. Ryder. 1997. Cloning and expression of squalene epoxidase from the pathogenic yeast Candida albicans. Gene 189:119-126. [DOI] [PubMed] [Google Scholar]
- 5.Fincham, J. R. 1989. Transformation in fungi. Microbiol. Rev. 53:148-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graminha, M. A., E. M. Rocha, R. A. Prade, and N. M. Martinez-Rossi. 2004. Terbinafine resistance mediated by salicylate 1-monooxygenase in Aspergillus nidulans. Antimicrob. Agents Chemother. 48:3530-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay, R. J. 1999. Therapeutic potential of terbinafine in subcutaneous and systemic mycoses. Br. J. Dermatol. 141(Suppl. 56):36-40. [DOI] [PubMed] [Google Scholar]
- 8.Jandrositz, A., F. Turnowsky, and G. Hogenauer. 1991. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene 107:155-160. [DOI] [PubMed] [Google Scholar]
- 9.Klobucnikova, V., P. Kohut, R. Leber, S. Fuchsbichler, N. Schweighofer, F. Turnowsky, and I. Hapala. 2003. Terbinafine resistance in a pleiotropic yeast mutant is caused by a single point mutation in the ERG1 gene. Biochem. Biophys. Res. Commun. 309:666-671. [DOI] [PubMed] [Google Scholar]
- 10.Leber, R., S. Fuchsbichler, V. Klobucnikova, N. Schweighofer, E. Pitters, K. Wohlfarter, M. Lederer, K. Landl, C. Ruckenstuhl, I. Hapala, and F. Turnowsky. 2003. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 47:3890-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, W., G. S. May, M. S. Lionakis, R. E. Lewis, and D. P. Kontoyiannis. 2004. Extra copies of the Aspergillus fumigatus squalene epoxidase gene confer resistance to terbinafine: genetic approach to studying gene dose-dependent resistance to antifungals in A. fumigatus. Antimicrob. Agents Chemother. 48:2490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1998. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi; proposed standard M38-P. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 13.Osborne, C. S., I. Leitner, B. Favre, and N. S. Ryder. 2005. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. Antimicrob. Agents Chemother. 49:2840-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez, A. 1999. Terbinafine: broad new spectrum of indications in several subcutaneous and systemic and parasitic diseases. Mycoses 42(Suppl. 2):111-114. [PubMed] [Google Scholar]
- 15.Rocha, E. M., C. B. Almeida, and N. M. Martinez-Rossi. 2002. Identification of genes involved in terbinafine resistance in Aspergillus nidulans. Lett. Appl. Microbiol. 35:228-232. [DOI] [PubMed] [Google Scholar]
- 16.Rothe, A., M. Seibold, T. Hoppe, H. Seifert, A. Engert, C. Caspar, M. Karthaus, G. Fatkenheuer, U. Bethe, K. Tintelnot, and O. A. Cornely. 2004. Combination therapy of disseminated Fusarium oxysporum infection with terbinafine and amphotericin B. Ann. Hematol. 83:394-397. [DOI] [PubMed] [Google Scholar]
- 17.Ryder, N. S. 1992. Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 126(Suppl. 39):2-7. [DOI] [PubMed] [Google Scholar]
- 18.Ryder, N. S., and I. Leitner. 2001. Synergistic interaction of terbinafine with triazoles or amphotericin B against Aspergillus species. Med. Mycol. 39:91-95. [DOI] [PubMed] [Google Scholar]