Abstract
One hundred twenty CTX-M-15-producing Escherichia coli strains isolated in 10 different hospitals from Paris (France), in the Hospital Charles Nicolle in Tunis (Tunisia), and in the Pasteur Institute in Bangui, Central African Republic (CAR), between 2000 and 2004 were studied. Eighty isolates, recovered from the three countries, were clonally related by repetitive extragenic palindromic PCR and pulsed-field gel electrophoresis. Various resistance profiles were identified among these clonal strains. After conjugation or electroporation of plasmids from E. coli strains representative of each profile and each geographic region, we observed seven resistance profiles in the recipient strains. Incompatibility typing showed that all the plasmids transferred from the clonal strains studied, except one, belonged to the incompatibility group FII. They all shared a multidrug resistance region (MDR) resembling the MDR region located in pC15-1a, a plasmid associated with an outbreak of a CTX-M-15-producing E. coli strain in Canada. They also shared the common backbone of an apparent mosaic plasmid, including several features present in pC15-1a and in pRSB107, a plasmid isolated from a sewage treatment plant. This study suggests that although the plasmid-borne blaCTX-M-15 gene could be transferred horizontally, its dissemination between France, Tunisia, and CAR was due primarily to its residence in an E. coli clone with a strong propensity for dissemination.
While the first extended-spectrum β-lactamases were derived by point mutations from the common TEM- and SHV-type β-lactamases (4), new families of extended-spectrum β-lactamases among which the CTX-M-type β-lactamases (first reported in 1986) have increased dramatically since 1992 have been described more recently (2). These later enzymes are now present in most parts of the world and constitute probably the most widespread enzymes among the Enterobacteriaceae (11). Recent surveys from Canada, Italy, Greece, and the United Kingdom have illustrated an alarming trend in the association between these CTX-M enzymes and resistance to other classes of antimicrobial agents (12). This is explained by the finding that the blaCTX-M genes are commonly found on plasmids often carrying genes that confer resistance to multiple antibiotics, including aminoglycosides, chloramphenicol, sulfonamide, trimethoprim, and tetracycline (2, 10). One of the CTX-M-type β-lactamases, CTX-M-15, first described in 2001 (8) and differing from CTX-M-3 only by an Asp-240-Gly substitution which increases its catalytic efficiency against ceftazidime (2), is now found worldwide. Found mainly in strains of Escherichia coli isolated from both hospital and community settings, it was involved in several outbreaks in France, the United Kingdom, and Canada (3, 9, 10, 17). The complete sequence of the plasmid pC15-1a that was found associated with the Canadian outbreak has recently been reported (3). This plasmid harbored a multidrug resistance (MDR) region which consists of many transposable elements and all drug resistance genes found in pC15-1a, including blaCTX-M-15 (3). In a previous study, the DNA sequence of the surrounding region of blaCTX-M-15 of a representative isolate from our hospital was 100% identical to the corresponding region of pC15-1a (7).
In the present study, we first determined the genetic relatedness of a collection of multiresistant CTX-M-15-producing E. coli strains isolated in different hospitals in France, Tunisia, and the Central African Republic (CAR); then we analyzed the backbone and the particular structures of the MDR regions of the CTX-M-15-encoding plasmids present in representative strains.
MATERIALS AND METHODS
Bacterial strains.
One hundred twenty isolates of CTX-M-15-producing E. coli recovered, between 2000 and 2004, in the Charles Nicolle Hospital in Tunis (Tunisia), in the Pasteur Institute in Bangui (CAR), and in 10 different hospitals in Paris (France) were studied (Table 1). More than 80% of these clinical isolates were recovered from urinary specimens. CTX-M-15 in these strains was characterized as described previously (6).
TABLE 1.
CTX-M-15-producing E. coli clinical isolates
| Country | Hospital | Yr of isolation | No. of isolates | No. of clonally related isolatesa |
|---|---|---|---|---|
| France | Tenon | 2000-2004 | 35 | 20 |
| Lagny | 2003 | 10 | 9 | |
| Paul Brousse | 2001-2003 | 26 | 26 | |
| Emile Roux | 2004 | 7 | 7 | |
| Other hospitals (n = 6) | 2000-2004 | 6 | 0 | |
| Tunisia | Charles Nicolle de Tunis | 2000-2003 | 26 | 16 |
| CAR | Institut Pasteur de Bangui | 2003-2004 | 10 | 2 |
| All | 120 | 80 |
By REP-PCR and PFGE.
Molecular typing.
Repetitive extragenic palindromic (REP) PCR using a GeneAmp PCR system 9700 (PE Biosystems, Courtaboeuf, France) was performed with genomic DNA as described previously (6). Pulsed-field gel electrophoresis (PFGE) was performed using a GenePath system (Bio-Rad, Marnes-la-Coquette, France) with genomic DNA (5) digested once with XbaI and once with NotI (Ozyme, Saint Quentin en Yvelines, France). Isolates were considered to be genetically related if their DNA macrorestriction patterns differed by fewer than seven bands (16).
Antimicrobial susceptibility.
Susceptibility to antimicrobial agents was determined using an antibiotic disk (Bio-Rad) diffusion method on Mueller-Hinton agar (Bio-Rad). The results were interpreted according to the standards of the French Antibiogram Committee (http://www.sfm.asso.fr).
Conjugation and transformation experiments.
Mating experiments were performed as previously described (6) using E. coli J53-2 (met pro; Rifr) as a recipient strain. Transconjugants were then selected on Trypticase soy (Bio-Rad) agar plates containing rifampin (250 μg/ml) and cefotaxime (2.5 μg/ml).
For transformation, plasmid DNA, isolated with a QIAGEN plasmid DNA midi kit (QIAGEN, Courtaboeuf, France), was transferred by electroporation into E. coli DH10B cells (Invitrogen SARL, Cergy-Pontoise, France). Transformants were incubated for 1.5 h at 37°C and plated on Trypticase soy agar supplemented with cefotaxime (2.5 μg/ml).
PCR experiments.
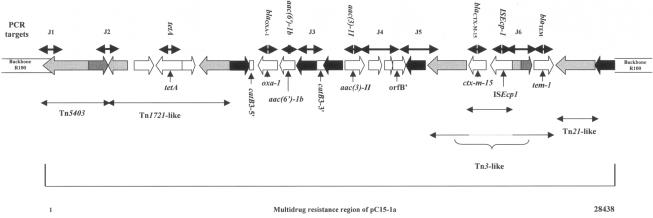
Plasmid DNA extracted from transconjugants or transformants of different E. coli strains was used as a template for PCR. Incompatibility typing was based on PCR amplification of conserved regions within the plasmid replicon (13). The sequence of the MDR region of pC15-1a (accession number AY458016) (3) was used for the design of PCR primers used in this study (Table 2) and the corresponding amplified regions shown in Fig. 1.
TABLE 2.
Primers used in this study
| PCR target | Primer name | Primer sequence | Source or reference |
|---|---|---|---|
| Junction 1 | PemK up | 5′-CACGGGGCGGAAAACGACT-3′ | This study |
| TnpATn5403 low | 5′-ACAGAGGGGCAGCACTACC-3′ | ||
| Junction 2 | TnpR Tn 5403 up | 5′-GACCAAAATCAGACCGACTT-3′ | This study |
| TnpA Tn 1721 low | 5′-AGCCGCCACTTCACCCACTT-3′ | ||
| tetA | tetA up | 5′-GTTTCGGGTTCGGGATGGTC-3′ | This study |
| tetA low | 5′-GCAGGCAGAGCAAGTAGAGG-3′ | ||
| blaOXA-1 | OXA-1 up | 5′-TATCAACTTCGCTATTTTTTTA-3′ | This study |
| OXA-1 low | 5′-TTTAGTGTGTTTAGAATGGTGA-3′ | ||
| aac(6′)-1b | AAC6′-1b up | 5′-ATGACTGAGCATGACCTT-3′ | 9 |
| AAC6′-1b low | 5′-GAAGGGTTAGGCATCACT-3′ | ||
| Junction 3 | Cat B3 up | 5′-CATCATCTTTCCCGTTCTTTT-3′ | This study |
| AAC 3-II low | 5′-ACCGTCTCCGCTCCTCCTTC-3′ | ||
| aac(3)-II | AAC3-II up | 5′-CAATAACGGAGGCAATTCG-3′ | 9 |
| AAC3-II low | 5′-GATTATCATTGTCGACGG-3′ | ||
| Junction 4 | AAC 3- II up | 5′-CGTGGGCTTTGCTCAGTGCT-3′ | This study |
| ORF B′ low | 5′-GTGTTCGGGCGGCGGTAGAT-3′ | ||
| Junction 5 | ORF B′ up | 5′-GGGTGCGTGGCGAGACAATG-3′ | This study |
| Tnp A Tn 3 low | 5′-ACTGGACAAAAGCGAACTAT-3′ | ||
| blaCTX-M | M13 upper | 5′-GGTTAAAAAATCACTGCGTC-3′ | 6 |
| M13 lower | 5′-TTGGTGACGATTTTAGCCGC-3′ | ||
| ISEcp1 | ISEcp1 début | 5′-TTCAAAAAGCATAATCAAAGCC-3′ | This study |
| PCR inv ISEcp1 272 | 5′-TTCAATAAAATCAAAAATCCCA-3′ | ||
| Junction 6 | PCR inv ISEcp1 272 | 5′-TTCAATAAAATCAAAAATCCCA-3′ | This study |
| Tem 1 low | 5′-ATACCGCACCACATAGCAGA-3′ | ||
| blaTEM | OT3 | 5′-ATGAGTATTCAACATTTCCG-3′ | 6 |
| OT4 | 5′-CCAATGCTTAATCAGTGAGG-3′ |
FIG. 1.
Schematic drawing showing the multidrug resistance region of pC15-1a (accession number AY458016) (3) and the position of the PCR targets with the primers listed in Table 2. Black arrow, IS26; light-gray arrow, tnpA; dark-gray arrow, tnpR.
Hybridization of plasmid DNA.
HpaI-digested plasmid DNA was transferred onto a nylon membrane, Hybond N+ (Amersham Biosciences, Saclay, France), using the Southern method (14). Southern hybridizations with blaCTX-M-15, blaOXA-1, blaTEM-1, tetA, aac(6′)-Ib, and aac(3)-II probes were performed using an ECL nonradioactive labeling and detection kit (Amersham Biosciences). The sequences of all PCR products used as probes were verified. Hybridization was also performed with a “backbone” probe consisting of HpaI restriction fragments of less than 9 kb obtained from plasmid EpLA2 (Table 3). Bands were purified with a GFX PCR DNA and gel band purification kit (Amersham Biosciences).
TABLE 3.
Comparative organization of the MDR regions of Inc FII plasmids associated with different profiles of resistance and originating from clonal and nonclonal strains, in relation with the MDR of pC15-1aa
| Profileb | Plasmidc | Inc FII | Junction 1 | Junction 2 | tetA | blaOXA-1 | aac(6′)-Ib | Junction 3 | aac(3)-II | Junction 4 | Junction 5 | blaCTX-M | ISEcp1 | Junction 6 | blaTEM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | EpTN03 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 1 | EpPB01 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 1 | TcTN17 | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| 1 | TcTN15 | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| 1 | TcTN20 | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| 2 | TcTN49 | + | + | + | + | + | + | + | + | + | − | + | + | − | + |
| 2 | EpCAF | + | + | + | + | + | + | + | + | + | − | + | + | − | + |
| 3 | TcTN08 | + | + | + | + | + | + | − | − | − | − | + | + | − | + |
| 4 | TcTN36 | + | + | + | + | + | + | − | − | − | − | + | + | − | + |
| 4 | EpLA2 | + | + | + | + | + | + | − | − | − | − | + | + | − | + |
| 5 | TcER15 | + | + | + | − | + | + | − | + | + | + | + | + | + | + |
| 6 | EpTU | + | − | − | − | + | + | + | + | + | − | + | + | − | + |
| 7 | TcTN34 | − | − | − | − | − | − | − | − | − | − | + | + | − | − |
| 8 | TcTN50 | − | − | − | − | − | − | − | − | − | − | + | + | − | − |
The localization of the gene encoding resistance to trimethoprim and sulfamides was not explored. +, presence; −, absence.
Antibiotic resistance profile present in the recipient containing the different plasmids: profile 1, cefotaxime, kanamycin, gentamicin, tobramycin, netilmicin, amikacin, and tetracycline; profile 2, cefotaxime, kanamycin, gentamicin, tobramycin, netilmicin, amikacin, tetracycline, and trimethoprim; profile 3, cefotaxime, kanamycin, tobramycin, netilmicin, amikacin, tetracycline, and trimethoprim; profile 4, cefotaxime, kanamycin, tobramycin, netilmicin, amikacin, and tetracycline; profile 5, cefotaxime, kanamycin, gentamicin, tobramycin, netilmicin, and amikacin; profile 6, cefotaxime, kanamycin, gentamicin, tobramycin, netilmicin, amikacin, trimethoprim, and sulfamides; profile 7, cefotaxime and tetracycline; profile 8, cefotaxime.
Ep, plasmid obtained from electroporant; Tc, plasmid obtained from transconjugant. EpTN03, EpPB01, TcTN49, EpCAF, TcTN08, TcTN36, EpLA2, TcER15, EpTU, and TcTN34 were issued from the clonal E. coli strains. TcTN17, TcTN15, and TcTN20 were issued from the nonclonal E. coli Tenon collection harboring the same MDR as the plasmids issued from the clonal E. coli strains. TcTN50 harboring CTX-M-15 was used as a control.
Partial plasmid sequencing.
HpaI fragments of less than 9 kb (backbone) of plasmid EpPB01 were cloned into the ScaI (Ozyme) site of phagemid pBK-CMV (Stratagene, La Jolla, Calif.) and introduced into E. coli DH10B by electroporation. The molecular sizes of 115 clones were estimated after electrophoresis in 1% agarose gels, using a supercoiled DNA ladder (Invitrogen SARL) as a size standard. Forty-five clones containing fragments of between 1 and 8 kb were selected and totally or partially sequenced (up to 1,600 bp per clone) using universal M13 and M13 reverse primers.
RESULTS
Molecular typing.
One hundred twenty CTX-M-15-producing E. coli isolates were studied by REP-PCR. Among these, 40 isolates were not clonally related while 80 isolates, recovered in six different centers in the three countries, were related to the E. coli strain TN03, originally identified in the hospital Tenon (6) and later associated with outbreaks in two other hospitals of the Paris area (Lagny and Paul Brousse) (Table 1) (9). This observation was confirmed by PFGE (data not shown).
Antimicrobial susceptibility of clonally related strains and their transconjugants and transformants.
All clonally related strains were resistant to ciprofloxacin in addition to extended-spectrum cephalosporins. Taking into account the susceptibilities to aminoglycosides, tetracycline, trimethoprim, and sulfamides, different profiles were identified. One E. coli strain representing each profile and each geographic region was studied further: TN03, TN08, TN34, TN36, TN49 (from Tenon Hospital), ER15 (from Emile Roux Hospital), LA2 (from Lagny Hospital), PB01 (from Paul Brousse Hospital), TU (from Tunisia), and CAF (from CAR). In addition, the clonally unrelated TN50 strain, susceptible to all antibiotics except cefotaxime, was used as a negative control. Cefotaxime resistance was transferred by conjugation into E. coli J53-2 from six of these E. coli strains, including TN50, and by electroporation of plasmid DNA from the five other strains into E. coli DH10B. Eight resistance patterns were observed in the transconjugants and transformants (including the control strain) and were compared to those observed in the transconjugants obtained from nonclonally related E. coli isolates, TN17, TN15, and TN20 (Table 3). The quinolone resistance was not transferred in any case.
Plasmid DNA fingerprinting and hybridization in clonally related strains.
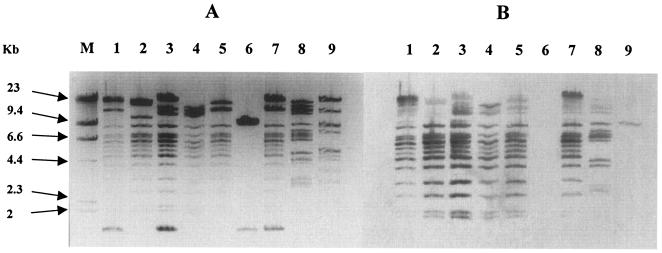
After HpaI restriction, the sizes of the plasmids extracted from each of the transconjugants and transformants were estimated to be 95 kb to 130 kb, except for plasmids pTcTN34 and pTcTN50 (control), the sizes of which were 10 kb and 80 kb, respectively (Fig. 2A). While identical fingerprints were found for plasmids pEpTN03 and pEpPB01 (Fig. 2A), similar patterns of HpaI-digested fragments of less than 9 kb were observed for the remaining plasmids, except pTcTN34 and pTcTN50. These results suggested that a common backbone was present in most of these plasmids. This was confirmed by hybridization analysis using the backbone probe generated from plasmid EpLA2 (Fig. 2B).
FIG. 2.
Fingerprint of Inc FII plasmids harboring CTX-M-15 β-lactamase. (A) HpaI-digested plasmid profiles of transconjugants or electroporants producing CTX-M-type β-lactamases. (B) Southern blot analysis results of the same HpaI-digested plasmids, using the backbone probe. Lane M, DNA molecular weight marker II (Roche Diagnostics); lane 1, EpTN03; lane 2, TcTN36; lane 3, TcTN49; lane 4, TcER15; lane 5, EpLA2; lane 6, TcTN34; lane 7, EpPB01; lane 8, EpTU; lane 9, TcTN50.
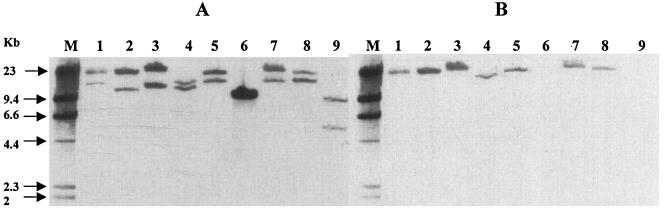
Southern hybridization analysis with the blaCTX-M-15, blaOXA-1, blaTEM, tetA, aac(6′)-Ib, and aac(3)-II probes showed, for each plasmid, that all these resistance genes were located on one or two high-molecular-weight, HpaI-generated fragments of over 9 kb (Fig. 3 and data not shown). The blaCTX-M-15 probe always hybridized with both fragments, due to the presence of an HpaI restriction site in blaCTX-M-15 (Fig. 3) indicating that these were two neighboring fragments on the plasmid HpaI restriction map. tetA, blaOXA-1, aac(6′)-1b, and aac(3)-II, when present, were located on the larger fragment, while blaTEM was present on the smaller fragment (Fig. 3 and data not shown). Plasmid pTcTN34 (profile 7) from one of the clonal E. coli strains harbored, after electroporation, only blaCTX-M-15 and a tet resistance gene different from tetA (Table 3, Fig. 3, and data not shown). The control plasmid pTcTN50 (profile 8) harbored only blaCTX-M-15 (Fig. 3 and data not shown).
FIG. 3.
Hybridization of HpaI-digested plasmids, using blaCTX-M-15 (A) and blaOXA-1 (B) probes associated to a DNA molecular weight marker II (Roche Diagnostics) probe. Lane M, DNA molecular weight marker II probe; lane 1, EpTN03; lane 2, TcTN36; lane 3, TcTN49; lane 4, TcER15; lane 5, EpLA2; lane 6, TcTN34; lane 7, EpPB01; lane 8, EpTU; lane 9, TcTN50.
Detection of common sequences with the pC15-1a MDR region.
Incompatibility typing based on PCR amplification of conserved regions within the plasmid replicons showed that all the plasmids transferred from the clonal strains studied, except TcTN34, belonged to the incompatibility (Inc) group FII (data not shown). This, in addition to the presence on the same DNA fragment of the particular set of resistance genes described above, including blaCTX-M-15, suggested the presence of an MDR region similar to that described for plasmid pC15-1a by Boyd et al. (3). The latter was isolated from a multiresistant E. coli strain during a major outbreak in Canada. Thus, oligonucleotides were designed (Table 2) to detect PCR products which would allow the identification of junction regions within this MDR region (Fig. 1 and Table 2).
Since these plasmids could have disseminated horizontally, we sought to determine whether a similar MDR region existed on plasmids of some of the nonclonal E. coli strains harboring CTX-M-15. Among 15 nonclonal E. coli isolates from Tenon Hospital, three (TN15, TN17, and TN20) were shown to harbor an Inc FII group plasmid. After conjugation and selection on cefotaxime, the Inc FII plasmids present in E. coli J53-2 were restricted with HpaI and shown to have the same backbone as the other plasmids derived from the clonal strains described above (data not shown). These three transconjugants had the susceptibility profile 1 (Table 3).
Interestingly, all the Inc FII group plasmids isolated from the clonal strains and those from the Tenon nonclonal strains showed an organization compatible with that of the MDR region present in pC15-1a (Table 3). No difference was found between the MDR region organization of pC15-1a (Fig. 1) and those of EpTN03 and EpPB01 (Table 3 and data not shown). Six out of the 12 plasmids showed the absence of both junctions 5 and 6 although blaCTX-M15 and blaTEM-1 were present (Table 3). For TcTN08, TcTN36, and EpLA2, junctions 3 and 4, as well as the aac(3)-II gene that was supposed to be present between these two junctions, were not detected (Table 3).
Partial plasmid characterization.
To better understand the assembly of these plasmids with apparently similar backbones (see above), we cloned fragments of less than 9 kb of one representative plasmid, EpPB01. Forty-five clones containing fragments of between 1 and 8 kb were selected, and their extremities (ca. 800 bp) were sequenced using M13 universal primers. Nonredundant sequences allowed partial identification of 16 open reading frames (ORFs). Four ORFs showed >95% identity with psiA, psiB, traT, and yhfA present in pC15-1a and R100 (3). Three ORFs showed >97% identity with ECs1338, ECs1339, and yehA present in pC15-1a and pRSB107 (15) but absent in R100 (3, 15). One ORF (ydaA) was found to be present in these three plasmids. Eight ORFs showed >98% identity with orf101, orf102, orf105, orf106, orf132, orf133, orf134, and orf135 present only in pRSB107 (15).
DISCUSSION
The prevalence of CTX-M-type β-lactamases has increased dramatically since 1992 (2), and these are now probably the most widespread enzymes in Enterobacteriaceae (11). This phenomenon could be explained by horizontal transfer of plasmids carrying blaCTX-M among nonrelated Enterobacteriaceae as well as clonal dissemination of CTX-M-producing microorganisms. Among the CTX-M-type β-lactamases, the CTX-M-15 variant which occurs mainly in E. coli seems to be epidemic in most countries (3, 9, 10, 17). Unexpectedly, in this study, 80 out of 120 CTX-M-15-producing E. coli strains isolated in different centers in the Paris area, in Tunis and, to a lesser extent, in Bangui (CAR) were found to be clonally related.
With one exception, all the CTX-M-15-encoding plasmids present in the transformants or transconjugants, obtained from selected clonal strains isolated at different locations and specifying different antibiotic susceptibility profiles, belonged to the incompatibilty group FII. This was reminiscent of the pC15-1a plasmid encoding CTX-M-15 in Canada, as described by Boyd et al. (3). An MDR region resembling that of pC15-1a containing different resistance genes, including blaCTX-M15, was present in these plasmids. While two plasmids showed the exact same organization as that seen in the MDR region of pC15-1a, an MDR region close to that of pC15-1a was present in all other plasmids, suggesting different rearrangements in their MDR regions. In pC15-1a, the MDR region consists mostly of transposons or partial transposons and five copies of the insertion sequence IS26. In particular, the aac(3)-II gene encoding gentamicin resistance and surrounded by two IS26 elements was absent in 3 out of the 12 plasmids studied. A similar variability in the presence of the the aac(3)-II gene has previously been observed in clonally related CTX-M-15-producing E. coli isolates in a Parisian geriatric hospital (10).
Fingerprinting with HpaI of the plasmids studied here revealed a backbone of common fragments of less than 9 kb, but their profiles differed from that described for pC15-1a (3). Sequence analysis of different HpaI-generated backbone fragments of the representative plasmid EpPB01 (Table 3) showed that it was a mosaic plasmid which shares common ORFs with R100, pC15-1a, and pRSB107, a multiresistance IncF plasmid isolated from activated sludge bacteria of a wastewater treatment plant (15). If the MDR region present in the strains studied here resembles that of pC15-1a, it is very different (in terms of resistance genes) from those harbored by pRSB107 and R100. Thus, it is likely that EpPB01 and the related plasmids were gradually assembled by integration of different horizontally acquired DNA segments via transposition or homologous recombination (15). It is noteworthy that CTX-M-15-encoding plasmids from nonclonal Inc FII strains had an MDR similar to that of the CTX-M-15-encoding plasmids present in the clonal strains, suggesting horizontal plasmid transfer between these strains.
In conclusion, we demonstrated that the current dissemination of CTX-M-15-producing strains of E. coli in the Paris area is due mainly to the diffusion of an epidemic clone, reminiscent of the dissemination of SHV-4-producing strains of Klebsiella pneumoniae previously attributed to a French epidemic clone (1). Surprisingly, clonally related E. coli strains harboring very close Inc FII, CTX-15-encoding plasmids were also found in Tunisia and the Central African Republic. This suggests the existence of a particular CTX-M-15-producing E. coli clone that may have additional characteristics which would explain its extraordinary propensity for colonization. On the other hand, dissemination of the CTX-M-15-encoding mosaic plasmids by horizontal transfer was also very likely since they were similarly found in clonally unrelated strains that were not implicated in outbreaks.
Acknowledgments
This work was supported by grants from Faculté de Médecine Pierre et Marie Curie, Université Paris VI, and from the European Community (6th PCRD contract; LSHM-CT 2003-503335).
We thank Laurent Gutmann and Ekkehard Collatz for critical reading of the manuscript.
REFERENCES
- 1.Arlet, G., M. Rouveau, I. Casin, P. J. Bouvet, P. H. Lagrange, and A. Philippon. 1994. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decre, D., B. Gachot, J. C. Lucet, G. Arlet, E. Bergogne-Berezin, and B. Regnier. 1998. Clinical and bacteriologic epidemiology of extended-spectrum beta-lactamase-producing strains of Klebsiella pneumoniae in a medical intensive care unit. Clin. Infect. Dis. 27:834-844. [DOI] [PubMed] [Google Scholar]
- 6.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 8.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 9.Kassis-Chikhani, N., S. Vimont, K. Asselat, C. Trivalle, B. Minassian, C. Sengelin, V. Gautier, D. Mathieu, E. Dussaix, and G. Arlet. 2004. CTX-M beta-lactamase-producing Escherichia coli in long-term care facilities, France. Emerg. Infect. Dis. 10:1697-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leflon-Guibout, V., C. Jurand, S. Bonacorsi, F. Espinasse, M. C. Guelfi, F. Duportail, B. Heym, E. Bingen, and M. H. Nicolas-Chanoine. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitout, J. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 13.Sherley, M., D. M. Gordon, and P. J. Collignon. 2003. Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid 49:79-85. [DOI] [PubMed] [Google Scholar]
- 14.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 15.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Puhler, and A. Schluter. 2005. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]