Abstract
Glycopeptide resistance, in a set of in vitro step-selected teicoplanin-resistant mutants derived from susceptible Staphylococcus aureus SA113, was associated with slower growth, thickening of the bacterial cell wall, increased N-acetylglucosamine incorporation, and decreased hemolysis. Differential transcriptome analysis showed that as resistance increased, some virulence-associated genes became downregulated. In a mouse tissue cage infection model, an inoculum of 104 CFU of strain SA113 rapidly produced a high-bacterial-load infection, which triggered MIP-2 release, leukocyte infiltration, and reduced leukocyte viability. In contrast, with the same inoculum of the isogenic glycopeptide-resistant derivative NM67, CFU initially decreased, resulting in the elimination of the mutant in three out of seven cages. In the four cages in which NM67 survived, it partially regained wild-type characteristics, including thinning of the cell wall, reduced N-acetylglucosamine uptake, and increased hemolysis; however, the survivors also became teicoplanin hypersusceptible. The elimination of the teicoplanin-resistant mutants and selection of teicoplanin-hypersusceptible survivors in the tissue cages indicated that glycopeptide resistance imposes a fitness burden on S. aureus and is selected against in vivo, with restoration of fitness incurring the price of resistance loss.
Over 15 years ago, the first clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates with decreased susceptibility to the glycopeptide antibiotic teicoplanin were described (27). In the interceding years, glycopeptide intermediate-resistant S. aureus (GISA) isolates have been recovered from most parts of the world (reviewed in references 24 and 58). GISA is the general term used to describe strains with intermediate glycopeptide MICs ranging from 4 to 16 μg/ml that are usually isolated from patients after prolonged glycopeptide exposure (54). Resistance arises intrinsically upon glycopeptide exposure, as the result of multiple mutations and/or alterations in gene expression (47, 49, 51). Several of the clinical and laboratory GISA strains described share phenotypic similarities, most commonly a modification of the cell wall, reducing the amount of glycopeptide able to reach its target at the cell membrane (13, 14, 23, 24, 50). Common GISA features include cell wall thickening, decreased peptidoglycan cross-linking, decreased growth rate and hemolysis, alterations in rates of autolysis, and changes in the structure and/or abundance of cell wall teichoic acids (7-9, 13, 41, 42, 49, 50). However, there is still little known about the genetic basis of this phenotype, and there is no universal genetic marker typical for all GISA isolates.
Certain genetic observations have been frequently documented in both clinical and laboratory-derived GISA, such as increased penicillin binding protein (PBP) 2 and decreased PBP4 expression (10, 23, 38, 43, 48), which are associated with cell wall modifications leading to increased cell wall synthesis and decreased peptidoglycan cross-linking. However, this observation is not true for all GISA strains, and these changes in PBP abundance are not sufficient in themselves to create a GISA phenotype (23). Several genetic alterations have been shown to only contribute to increased resistance in a single or restricted number of strains, including (i) overexpression of the global regulator SigB (2, 51) and the two-component sensor transducer VraSR (32); (ii) defective agr function (36, 46); (iii) alterations in the expression of genes encoding autolysins or affecting autolytic function (7, 8, 28, 41, 49) and genes involved in carbohydrate metabolism and cell wall synthesis (29); (iv) inactivation of tcaA, a membrane protein of unknown function (34); and (v) inactivation of mprF (fmtC), a membrane protein involved in the biosynthesis of the positively charged cell membrane lipid lysylphosphatidylglycerol, that was found to increase resistance levels in glycopeptide susceptible strains but to decrease resistance in GISA (39, 45).
The limited number of global transcriptome analyses performed have indicated that multiple mutational and regulatory events are causing numerous metabolic changes in GISA (12, 33, 37). The full genetic basis of GISA formation has not been elucidated in any strain, but it appears that there are likely to be several different, as-yet-undiscovered, loci and pathways involved in resistance formation.
Increased production of peptidoglycan, required to facilitate the construction of a thicker cell wall, is an energetically unfavorable phenotype, as evidenced by the slower growth rate of GISA strains, and is probably the main reason that GISA strains are not isolated more frequently (13, 35, 49). This also contributes to the reported instability of the resistance phenotype in the absence of selection pressure (6, 13). Reversion from a GISA to a GSSA phenotype could be driven by a combination of the reversal of regulatory events and forward mutations, which lead to reclaimed fitness at the cost of decreased resistance. It is speculated that in the absence of drug pressure mutants with greater fitness emerge spontaneously and dominate the population (13).
The fitness of S. aureus cells inside a host is governed by intrinsic properties such as the rate at which they reproduce and the rate at which they are cleared by host defense mechanisms. Since the fitness of glycopeptide-resistant strains in infected hosts is unknown, we chose to investigate the in vivo behavior of a teicoplanin-resistant strain in a defined murine infection model. The tissue cage model was first described and extensively characterized in the guinea pig (62) and then adapted to the mouse (31). Low inocula, 103 CFU, of S. aureus cause a persistent local infection, which never becomes systemic. The absence of vascularization limits serum factors and the presence of leukocytes, which are attracted by the polymer implant before infection and show weak functional capacity (61), contribute to pathogenesis in this model. Persistence is facilitated by progressive leukocyte apoptosis and necrosis (30). This model accurately mimics orthopedic implant infections. Because bacteria are inoculated directly into the cage, with no adherence and invasion step through epithelia, the minimal infective dose of staphylococci which is required for a persistent infection reflects virulence. Accordingly, virulence is dependent on the resistance of S. aureus to extracellular phagocyte-dependent killing in the immunocompetent host. Therefore, this model differentiates S. aureus strains that have altered susceptibilities to bactericidal mechanisms. The host response is mediated exclusively by phagocytes and comprises defensins, reactive oxygen species, cytokines, chemokines, leukocyte infiltration, and apoptosis.
S. aureus SA113 was passaged several times on teicoplanin to obtain an isogenic mutant with intrinsically acquired glycopeptide resistance. The stability of the resistance phenotype and accompanying cell wall characteristics were monitored in vitro, prior to and during infection in a mouse tissue cage infection model, to assess the impact of the GISA phenotype on in vivo survival. A transcriptome comparison of the mutant and wild type was performed, and the transcription of selected differentially regulated genes was profiled in vitro and from tissue cage isolates.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains were routinely cultured at 37°C on sheep blood agar or in brain heart infusion (BHI) broth (BBL Becton Dickinson, Maryland) and stored as frozen stocks in skim milk at −80°C. A series of isogenic strains with increasing glycopeptide resistance levels was generated from susceptible S. aureus strain SA113 (26) by plating dilutions of an overnight culture on BHI agar containing increasing concentrations of teicoplanin. Ten single colonies growing at the highest concentration were selected and subcultured twice on nonselective agar to ensure the stability of the phenotype. Their resistance levels were compared to the susceptible parent on teicoplanin gradient plates. The strain with the highest increase in teicoplanin resistance, first-step mutant NM18, was then used to repeat the selection and to obtain the most resistant second-step mutant NM30, which was used to obtain a third-step mutant NM67. To ensure that clonality was maintained, all strains were tested by pulsed-field gel electrophoresis of SmaI-digested chromosomal DNA according to the protocol of Wada et al. (57). Growth rate experiments were performed as previously described (18).
For measurement of N-acetylglucosamine uptake and release, high-pressure liquid chromatography, autolysis, and transmission electron microscopy on cells recovered from tissue cages, 50 μl of tissue cage fluid (TCF) was centrifuged at 4,000 rpm for 5 min. The pellet was resuspended in BHI medium and grown overnight at 37°C. The overnight culture was pelleted and transferred into cryovials containing beads in cryopreservative for storage (Pro Lab Microbank Bacterial Preservation System Green, Basel, Switzerland). After vials were inverted four to five times, cryopreservative was aspirated, and the beads were stored at −70°C.
Stability of resistant mutants in vitro.
The stability of the teicoplanin resistance phenotype was monitored under in vitro culture conditions. BHI cultures (20 ml) of SA113 and NM67 were either subjected to prolonged stationary culture conditions at 37°C for 8 days or subcultured daily in 20 ml of fresh broth, using a dilution factor of 104, over the 8-day period.
Resistance tests.
MICs of glycopeptides were determined by E-test (AB-Biodisk, Solna, Sweden) on BHI (BBL) plates with an inoculum of 2 McFarland standards and incubation at 37°C for 48 h. Lysostaphin MICs were determined by broth microdilution in BHI as recommended by the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards [25]). For population analysis profiles, dilutions of an overnight culture were spread on BHI agar plates containing increasing concentrations of teicoplanin. For population analyses of strains ex vivo, 50 μl of TCF was grown overnight and then adjusted to McFarland 0.5 before plating. The CFU were determined after 48 h of incubation at 37°C.
Hemolysis assays.
Hemolytic activities were compared on sheep blood agar plates. Wells were stamped out of the agar and filled with 100 μl of filtered supernatant from cultures grown in BHI for 24 h. Plates were incubated overnight at 37°C.
Tissue cage model.
Male C57BL/6 mice (12 to 16 weeks old) were anesthetized, and sterile cylindrical Teflon tissue cages were implanted subcutaneously in their backs, as described previously (31). Two weeks after surgery, the sterility of tissue cages was verified, and 200 μl of a suspension from stationary overnight cultures containing 104 CFU was injected percutaneously. Mice never developed bacteremia and showed no weight change during 8 days of infection.
Mice were anesthetized after 1, 2, 5, and 8 days of infection, and TCF samples of 150 μl were collected by percutaneous aspiration and transferred into tubes containing 15 μl of 1.5% EDTA. The load of planktonic bacteria in TCF was determined by serial dilutions of the samples on BHI agar plates. Leukocytes from TCF were quantified with a Coulter counter. The percentage of viable leukocytes was assessed by trypan blue exclusion. The MIP-2 content was determined by sandwich enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, MN). Mice were kept under specific-pathogen-free conditions in the Animal House of the Department of Research, University Hospital Basel, and animal experimentation guidelines were followed according to the regulations of Swiss veterinary law.
Statistics.
Median and lowest quartiles were calculated from each group of strains on all days of investigation. Analysis of variance for repeated measures, with Scheffé posthoc tests, was used to compare SA113 and NM67 during infection.
Sampling, RNA isolation, and transcriptional profiling.
Differential microarray analysis of the transcriptomes of SA113 and NM67 was carried out on exponentially growing cultures in Luria-Bertani (LB) medium, harvested at an optical density at 600 nm (OD600) of 2, as previously described (3).
RNA extraction and Northern hybridization.
RNA isolation from bacteria grown in shaking LB cultures and Northern blotting were performed as described earlier (34). A total of 10 μg of total RNA from each sample were separated through a 1.5% agarose-20 mM guanidine thiocyanate gel in 1× Tris-borate-EDTA running buffer (21). Open reading frames (ORFs) selected for probe amplification and the primers used are shown in Table 1. All Northern blot analyses were performed at least twice on independently isolated RNA samples.
TABLE 1.
Primers used in this study
| Gene and/or ORFa | Primer | Sequence (5′ to 3′) |
|---|---|---|
| spa (SA0107) | spa F | TGTAGGTATTGCATCTGTAA |
| spa R | AAGTTAGGCATATTCAAGAT | |
| SA0112 | SA0112 F | GTTAGAGTATATGAATACCT |
| SA0112 R | TACTCATACCCATAATGCTA | |
| SA0184 | SA0184 F | CAAGTGACTTACCTCATAGA |
| SA0184 R | AACACTTCAGATACACCAGT | |
| geh (SA0309) | geh F | TCATGCTGAACGTAATGGAT |
| geh R | CACTTACACTTGCTTGATGT | |
| SA1007 | SA1007 F | TAATGAATCCTGTCGCTAAT |
| SA1007 R | TTCAGTGTATGACCAATCGA | |
| nuc (SA1160) | nuc F | GTAGGTGTATTAGCATTTCA |
| nuc R | GTACATACGATCTTTACTTA | |
| SA1898 | SA1898 F | ATGGCGCAATCAAATGATCA |
| SA1898 R | GATGAGCATTCACATTTAGA | |
| SA2097 | SA2097 F | TAGTAGGTCAAGCACATCAT |
| SA2097 R | CCTTCAGAAGATTGTAGGAT | |
| sbi (SA2206) | sbi F | AGAACGTGCACAAGAAGTAT |
| sbi R | CTACTAATGCGTCTAATTGT | |
| opp-1A (SA2255) | opp-1A F | CTTGTACGTAACACGAAAGA |
| opp-1A R | TTAACTTGATAGTCACCTCT | |
| gbsA (SA2406) | gbsA F | CGTTAGAAGAATCATATGCA |
| gbsA R | AAGTTCCAAGGCAATATTCG | |
| icaA (SA2459) | icaA F | TGGATGAATTAGAAGGCATT |
| icaA R | CGGTTCATACTTAATACGAT |
ORF numbers correspond to the genome annotation of S. aureus N315.
Incorporation and release of N-acetylglucosamine.
The incorporation of 14C-labeled N-acetylglucosamine (Amersham) into cells in resting medium and the release of 14C-labeled N-acetylglucosamine into the culture medium were measured essentially as described by Hanaki et al. (23), except that the cells were pregrown in BHI.
Cell wall composition.
Cell walls were purified from bacteria grown to exponential or stationary phase in BHI broth. Teichoic acids were removed from the cell walls by gently mixing the cell wall suspension in hydrofluoric acid (49%) for 48 h at 4°C. The peptidoglycan was recovered, digested with muramidase (Sigma), and reduced, and the muropeptides were separated by high-pressure liquid chromatography as described by de Jonge et al. (17). Eluted material was detected by its absorbance at 210 nm, and principal peaks were identified by using muropeptide peak libraries from S. aureus COL (5). The degree of muropeptide cross-linking was calculated by quantification of the relative amounts of monomers, dimers, trimers, and oligomers in the muropeptide digest according to the method of Snowden and Perkins (52).
Spontaneous autolysis.
Cells grown to an OD600 of 0.7 in BHI were pelleted, washed once with saline, and resuspended in 0.01 M sodium phosphate buffer (pH 7.0) to an OD600 of 0.8. The cell suspension was incubated at 37°C with continuous gentle shaking. The decrease in the OD600 was monitored every hour for 5 h.
Zymography.
Exponentially growing cells, cultured in BHI, were harvested at an OD600 of 1.0. Autolysins were extracted and analyzed according to the method described by Hanaki et al. (23), using cell wall extracts of S. aureus SA113 as the substrate. Band intensities were scanned and quantified.
Transmission electron microscopy.
Cells grown overnight were processed for thin-sectioning electron microscopy as described by Pante et al. (40). Electron micrographs were recorded with a Zeiss LEO 910 transmission electron microscope.
RESULTS
Phenotype and in vitro stability of the teicoplanin-resistant mutants.
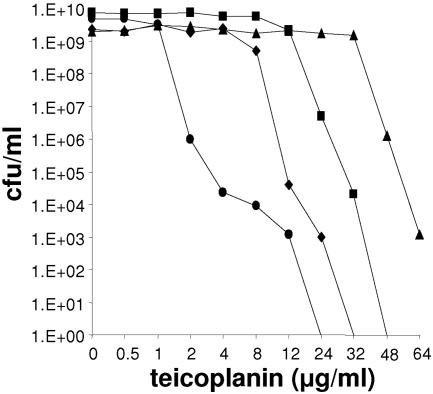
A series of three mutants—NM18, NM30, and NM67—with sequentially increasing teicoplanin MICs of 16, 48, and 64 μg/ml, respectively, were generated from the susceptible parent strain SA113 (teicoplanin MIC of 3 μg/ml). The mutants showed heterogeneous resistance profiles on teicoplanin, segregating subpopulations resistant to higher concentrations of the drug (Fig. 1). Parallel to the increasing teicoplanin resistance, the vancomycin MIC rose from 4 μg/ml in SA113 to 24 μg/ml in NM67; the minimum doubling time increased from 25.7 ± 1.7 min in SA113 to 37.6 ± 1.4 min in NM67; and hemolysin production, which was clearly visible in SA113, was undetectable in NM67 (refer to Fig. 6B). Initial studies demonstrated that NM67 incorporated, within 120 min, approximately 2.7 times (n = 3, P < 0.05) more 14C-labeled N-acetylglucosamine than SA113, a feature of many clinical GISA isolates that is associated with increased cell wall thickness (20, 23). There were no significant differences between SA113 and NM67 in their release rates of 14C-labeled N-acetylglucosamine into the culture supernatant (data not shown). The cells of the teicoplanin-resistant strain NM67 were significantly smaller, but their cell walls were significantly thicker (cytoplasm diameter of 797.4 ± 21.7 nm; cell wall thickness of 54.9 ± 2.3 nm) compared to the parent strain SA113 (961.1 ± 38.3 nm, P < 0.001; 47.3 ± 2.2 nm, P < 0.05). Muropeptide analysis showed that the degree of peptidoglycan cross-linking in SA113 (71.3%) and NM67 (70.9%) did not differ significantly.
FIG. 1.
Resistance profiles of teicoplanin-selected mutants. Population analysis profiles of susceptible strain SA113 (•), first-step mutant NM18 (⧫), second-step mutant NM30 (▪), and third-step mutant NM67 (▴).
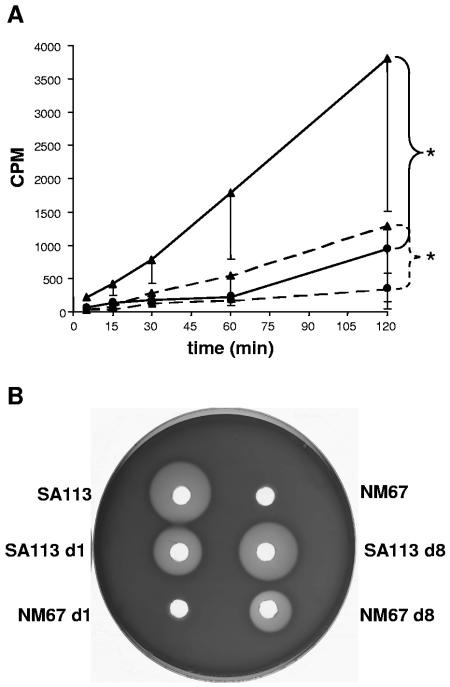
FIG. 6.
Changes in 14C-labeled N-acetylglucosamine incorporation and hemolysis. (A) 14C-labeled N-acetylglucosamine incorporation. Key: filled circle with solid line, SA113 on day 1; filled triangle with solid line, NM67 on day 1; filled circle with dashed line, SA113 on day 8; filled triangle with dashed line NM67 on day 8 after infection. Median values from four cages were determined, and the lowest quartiles are indicated for each group. According to analysis of variance for repeated measures, P < 0.05 (,) for NM67 versus SA113 on days 1 and 8, respectively. (B) Hemolysis zones on sheep blood agar. One representative TCF isolate is shown for each strain from days 1 and 8.
Glycopeptide resistance phenotypes were stable upon storage of the strains at −80°C and through several subcultures on drug-free sheep blood agar plates. However, upon prolonged incubation or repeated daily subculture, over 8 days in liquid culture, decreases in teicoplanin resistance occurred. Loss of resistance was very stochastic with repeated experiments giving widely varying results. After 8 days, resistance had always decreased, with MICs ranging from 6 to 24 μg/ml (data not shown).
Downregulation of virulence-associated genes in NM67.
Transcriptional differences were evaluated between the teicoplanin-resistant mutant NM67 and its susceptible parent SA113, in the mid-exponential-growth phase. In the mutant, there were no genes upregulated by threefold or more over the parent, despite the relevant increase in cell wall thickness and N-acetylglucosamine uptake. In contrast, 26 ORFs were downregulated more than threefold in NM67 (Table 2). Downregulated ORFs comprised 12 loci, consisting of seven single gene loci and five multigene clusters, most of which are likely to play a role in virulence. These included, lipase (geh), alpha-hemolysin precursor (SA1007), nuclease (nuc), and the secretory antigen (SA2097), which is similar to the secretory antigen precursor ssaA (1); protein A (spa) and immunoglobulin G-binding surface sprotein (sbi), which are involved in the evasion of host defenses (60); betA and gbsA playing a role in osmoprotection (4); the icaA operon, which is linked to biofilm formation and increased virulence in mouse tissue cage infections (19); and the large operon SA0112-SA120, which is involved in siderophore biosynthesis (15).
TABLE 2.
Genes downregulated in the step-selected teicoplanin-resistant mutant
| ORF no.a | N315 gene | Description and/or putative functionb | Fold downregulation |
|---|---|---|---|
| SA0107 | spa | Protein A | 4.5 |
| SA0112 | Siderophore biosynthesis | 8.2 | |
| SA0113 | Siderophore biosynthesis | 7.6 | |
| SA0114 | Siderophore biosynthesis | 6.1 | |
| SA0115 | Siderophore biosynthesis | 4.4 | |
| SA0116 | Siderophore biosynthesis | 7.7 | |
| SA0117 | Siderophore biosynthesis | 5.7 | |
| SA0118 | Siderophore biosynthesis | 6.4 | |
| SA0119 | Siderophore biosynthesis | 5.7 | |
| SA0120 | Siderophore biosynthesis | 4.6 | |
| SA0184 | Conserved hypothetical protein of unknown function | 4.0 | |
| SA0185 | Glucokinase regulator-related protein (carbohydrate transport and metabolism) | 3.7 | |
| SA0186 | Homologue of PTS system IIBC components (carbohydrate transport and metabolism) | 4.2 | |
| SA0309 | geh | Glycerol ester hydrolase-lipase | 3.8 |
| SA1007 | Alpha-hemolysin precursor | 5.1 | |
| SA1160 | nuc | Thermostable staphylococcal nuclease | 6.2 |
| SA1898 | Hypothetical, similar to SceD precursor | 7.6 | |
| SA2097 | Secretory antigen precursor SsaA homologue | 4.3 | |
| SA2206 | sbi | IgG-binding protein SBI | 6.3 |
| SA2255 | opp-1A | Oligopeptide transporter substrate-binding domain | 4.4 |
| SA2252 | opp-1D | Oligopeptide transporter ATPase domain | 3.8 |
| SA2405 | betA | Choline dehydrogenase (osmoprotection) | 3.7 |
| SA2406 | gbsA | Glycine betaine aldehyde dehydrogenase (osmoprotection) | 3.7 |
| SA2459 | icaA | Intercellular adhesion protein A (biofilm production) | 5.2 |
| SA2460 | icaD | Intercellular adhesion protein D (biofilm production) | 5.4 |
| SA2461 | icaB | Intercellular adhesion protein B (biofilm production) | 5.3 |
ORF numbers correspond to the S. aureus N315 genome sequence.
Functions and/or descriptions are derived from the N315 and COL genome annotations. IgG, immunoglobulin G.
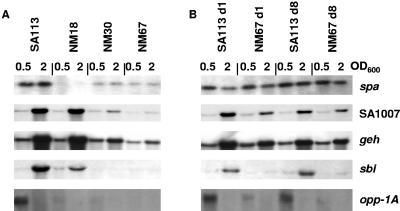
Northern blots of RNA harvested in early-exponential- and mid-exponential-growth phase substantiated the microarray results and in some cases showed at which step during the three-step selection process transcription had decreased. Transcription of spa and opp-1A was already downregulated after the first selection step in NM18, the transcript abundance of sbi was slightly lower in the first-step mutant but decreased more significantly after the second selection step in NM30, whereas transcription of SA1007 and geh was only downregulated after the second selection step (Fig. 2A). The transcription levels of these five genes correlated with the microarray results, confirming the overall validity of the microarrays. Four of the probes, SA0112, icaA, gbsA, and SA0184 produced unreadable Northern blots (data not shown), most likely due the large size of polycistronic transcripts but possibly also due to poor transfer, degradation, or low expression levels. The remaining genes either showed no alteration in transcription, such as SA2097, or gave results that did not agree with the microarray, with the transcript abundance of SA1898 and nuc appearing to have slightly increased in the teicoplanin-resistant mutants (data not shown). This discrepancy could indicate periodic fluctuations in gene expression that were not captured in the intervals measured or could simply reflect subtle differences between the growth conditions or extraction methods used to prepare the RNA for the microarrays and the Northern blots.
FIG. 2.
Changes in gene expression upon acquisition and loss of glycopeptide resistance. (A) Transcription of differentially expressed genes in SA113 and the three step-selected teicoplanin-resistant mutants NM18, NM30, and NM67 at early-exponential (OD600 = 0.5) and mid-exponential (OD600 = 2) phases. (B) Transcription profiles of the same genes from SA113 and NM67 cells recovered from mouse TCF on days 1 and 8 of infection.
Survival in the mouse cage infection model.
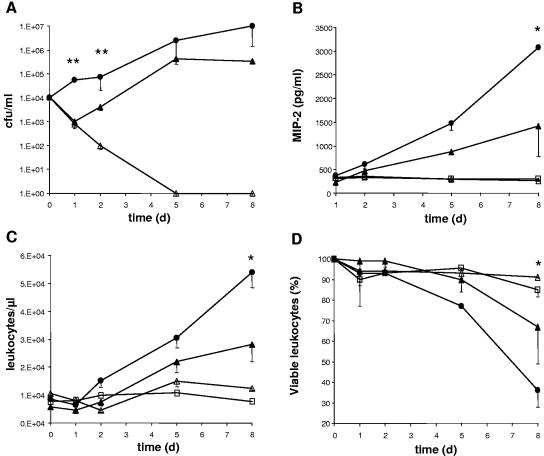
To assess tissue cage survival, equal numbers of SA113 and NM67 cells were inoculated into mouse tissue cages, and infection was monitored over 8 days. The bacterial load of SA113 steadily increased over the 8 days in the TCF. However, three of seven mice infected with an identical dose of NM67 completely cleared the bacteria within 5 days. In the remaining four mice, NM67 decreased in numbers after the first day postinfection, as in mice clearing the bacteria (P < 0.01 for day 1, P < 0.05 for day 2, Fig. 3A) and then resumed growth. The recovering bacteria reached the same final cell density as the wild type after 8 days (Fig. 3A). This suggested that NM67 was less fit than the wild-type parent and that it was actively cleared in the tissue cages, giving rise to the outgrowth of a surviving population.
FIG. 3.
Bacterial load and host response after infection of tissue cages with strains SA113 and NM67. In vivo bacterial growth and host response over 8 days in tissue cages infected with 104 CFU of S. aureus. Symbols: •, SA113 (n = 5); ▴, NM67 growing in tissue cages (n = 4); ▵, NM67 cleared from tissue cages during infection (n = 3); □, values in uninfected cages. Time zero refers to values before infection. (A) Bacterial load in TCF. (B) MIP-2 concentrations in TCF. (C) Leukocyte numbers in TCF. (D) Viable leukocytes in TCF. Median values are plotted for each group, and the lowest quartiles are indicated.  , P < 0.05;
, P < 0.05; 
 , P < 0.01.
, P < 0.01.
Host response.
During infection, inflammation in the TCF is marked by granulocyte influx, apoptosis, and chemokine release (30, 31). Baseline leukocyte counts of 1 × 104 ± 3.1 × 103 cells/μl, consisting of a larger polymorphonuclear neutrophil (91% ± 3%) and a smaller monocyte (9% ± 3%) fraction were found in uninfected tissue cages, in agreement with previous reports (16, 62). The macrophage inflammatory protein (MIP-2, CXCL2) is a murine neutrophil chemotactic factor with a function similar to interleukin-8 in humans (55). After infection with SA113, the MIP-2 concentration in TCF steadily increased from day 1 through day 8. NM67 survivors induced twofold less MIP-2 than SA113 after the 8 days (P < 0.05, Fig. 3B).
MIP-2 is the major leukocyte attractant; accordingly, leukocyte infiltration into tissue cages inoculated with SA113 or NM67 showed a time-dependent increase during infection and correlated with the bacterial load of the TCF (Fig. 3C). Eight days after infection, the concentration of leukocytes was twofold higher in cages containing SA113 than in those with the surviving NM67 (P < 0.05). Mice that were mock infected or that were able to clear the infection showed no increased leukocyte infiltration (Fig. 3C). Since the MIP-2 levels and consequent leukocyte infiltration were proportional to the bacterial load in both strains, it seemed likely that the surviving NM67 cells may have had a similar capacity to induce MIP-2 and a chemoattractant potential similar to that of SA113.
The leukocyte viability within the TCF during infection was evaluated because staphylococci are known to kill eukaryotic cells (22). In cages infected with SA113 a significant decline of the proportion of viable leukocytes (from 100 to 36%) was observed on day 8 postinfection (P < 0.05). Leukocytes in mice that cleared NM67 did not lose viability. In mice successfully infected with NM67, however, the decrease in leukocyte viability started only 2 days postinfection (Fig. 3D), coinciding in time with the emergence of the surviving NM67 subpopulation.
Host-driven selection of NM67 “survivors.”
The clearing or delayed growth of NM67 in the tissue cages points to a decreased virulence of the teicoplanin-resistant mutant. The surviving population that emerged from NM67 may consequently represent the selection and outgrowth of a variant or mutant subpopulation from NM67. Cells isolated on days 1 and 8 postinfection were therefore analyzed for host-induced changes in their phenotype and gene expression.
Teicoplanin resistance.
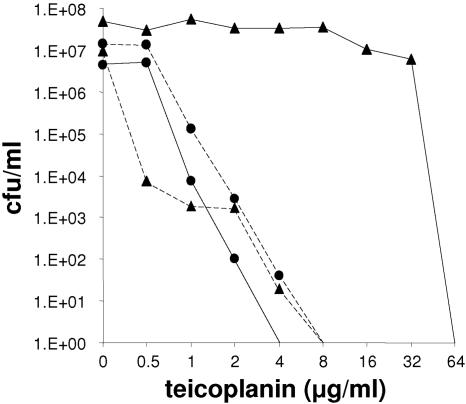
Teicoplanin MICs for NM67 isolates dropped to a median of 48 μg/ml after 1 day in the tissue cages and then decreased further to median values of 0.36 and 0.5 μg/ml on days 5 and 8, respectively. This was well below the MICs for SA113 isolates, which remained at a constant 2 μg/ml throughout the 8 days. The survivors of NM67 collected from tissue cages after 8 days showed a heterogeneous resistance profile, but the majority of the cells had become hypersusceptible compared to SA113 (Fig. 4).
FIG. 4.
Teicoplanin resistance profiles after passage in tissue cages. Median values of population analysis profiles on increasing concentrations of teicoplanin for strains SA113 and NM67 isolated from tissue cages on days 1 and 8. Key: filled triangles with solid line, strain NM67 on day 1 (n = 5); filled triangles with dashed line, strain NM67 on day 8 (n = 4); filled circles with solid line, strain SA113 on day 1 (n = 4); filled circles with dashed line, SA113 on day 8 (n = 4).
Cell size.
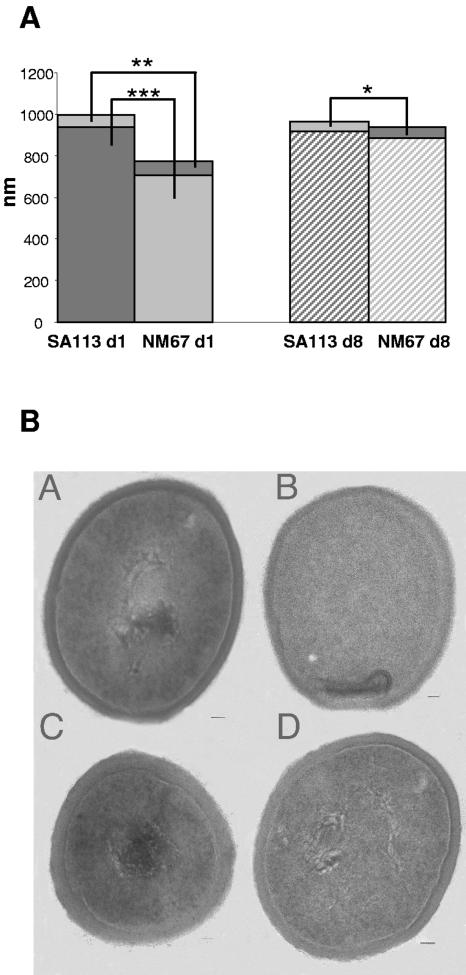
On day 1 of infection, cell size and cell wall thickness measurements of SA113 and NM67 tissue cage isolates were similar to the original in vitro values; NM67 cells were still significantly smaller, with thicker cell walls than SA113 (Fig. 5). After 8 days of infection, the cell walls of NM67 survivors became thinner, and their cell diameters increased, whereas both overall cell wall thickness and cytoplasm diameter did not change in wild-type SA113 (Fig. 5). On day 8, the cell wall of NM67 was still slightly thicker (P < 0.05) than that of SA113; however, the cytoplasm diameter was now similar in both strains.
FIG. 5.
Transmission electron microscopy of SA113 and NM67. (A) Diameter of cytoplasm and cell wall thickness of SA113 and NM67 ex vivo on days 1 and 8 of infection. Bars show the average cytoplasm diameter and cell wall thickness in nanometers. Mean values from at least 20 electron microscopy pictures, made from bacteria that were harvested from three mice infected with either NM67 or SA113, are shown. 
 , P < 0.01 NM67 versus SA113 cell wall thickness on day 1;
, P < 0.01 NM67 versus SA113 cell wall thickness on day 1; 

 , P < 0.001 NM67 versus SA113 cytoplasm diameter on day 1; , P < 0.05 NM67 versus SA113 cell wall thickness on day 8. (B) Representative TEM pictures of SA113 (A and B) and NM67 (C and D) on days 1 (left) and 8 (right). Scale bars in the bottom right hand corners of the pictures correspond to 50 nm.
, P < 0.001 NM67 versus SA113 cytoplasm diameter on day 1; , P < 0.05 NM67 versus SA113 cell wall thickness on day 8. (B) Representative TEM pictures of SA113 (A and B) and NM67 (C and D) on days 1 (left) and 8 (right). Scale bars in the bottom right hand corners of the pictures correspond to 50 nm.
N-Acetylglucosamine incorporation and turnover.
The incorporation of 14C-labeled N-acetylglucosamine in cells isolated from TCF was significantly (4.5-fold, P < 0.05) higher in NM67 than in SA113 on day 1. Incorporation decreased with time during infection in both strains. However, on day 8 the surviving NM67 cells still showed a higher incorporation rate (2.5-fold, P < 0.05) than SA113 (Fig. 6A). Surprisingly, cell wall turnover remained unchanged, with no significant differences between SA113 and NM67 in the release rates of 14C-labeled N-acetylglucosamine into the culture supernatant after passage in the tissue cages (data not shown).
Growth rate and hemolysin production.
The minimum doubling time of NM67 (37.6 ± 1.4 min) decreased in the tissue cage survivors to mean values of 36.2 ± 1.6 min after day 1 and 32.5 ± 3.6 min after day 8. Meanwhile, the minimum doubling time of SA113 (25.7 ± 1.7 min) remained relatively stable, with recovered isolates exhibiting mean minimal doubling times of 26.0 ± 0.6 min and 25.7 ± 1.0 min on days 1 and 8 of infection, respectively. Hemolysin production, which was undetectable in NM67 in vitro, was partially restored in the NM67 survivors isolated on day 8 of infection, whereas the hemolytic activities of SA113 supernatants remained relatively stable during the course of infection (Fig. 6B).
Cell wall properties.
Spontaneous autolysis rates for NM67 and SA113 isolates remained similar over the 8-day infection period (data not shown). Zymography, performed on SA113 and NM67 isolates collected during infection, showed no alterations in abundances of the 62-kDa amidase, the 50-kDa N-acetylglucosaminidase, and an undefined 36-kDa enzyme band in the SA113 and NM67 isolates (data not shown). SA113 and NM67 were both similarly lysostaphin susceptible, as were all isolates recovered from the tissue cages (data not shown). The proportions of peptidoglycan monomers and multimers were similar, with peptidoglycan cross-linking degrees of 65.9% on day 1 and 66.1% on day 8 of infection for NM67 and 68.5% and 71.2% on days 1 and 8 of infection, respectively, for SA113. Therefore, the physical qualities of the cell wall, such as resistance to osmotic stress, peptidoglycan cross-linking, susceptibility to lysostaphin, and the autolysin banding pattern, were very similar in the wild type and mutant and did not change remarkably during infection.
Northern analysis of ex vivo isolates.
In ex vivo SA113 isolates, the transcription levels of all ORFs analyzed remained the same as the parent. Transcription levels in the ex vivo NM67 survivors were more variable. For spa, SA1007, and geh, transcription levels had increased again after passage in vivo, with the signals approaching wild-type levels again by day 1 of infection. However, transcription of sbi and opp-1A remained low in the NM67 survivors (Fig. 2B).
DISCUSSION
The fitness cost of glycopeptide intermediate resistance in S. aureus has been evidenced in several studies by the decreasing growth rates of resistant strains and correlated in several cases with increases in cell wall thickness (42). This fitness burden, imposed by the accommodation of gross alterations in cell wall morphology, is thought to account for the widely observed instability of the resistance phenotype. However, it has also been previously reported that the conditions in a tissue cage infection model promoted the emergence of teicoplanin-intermediate-resistant mutants in the absence of teicoplanin pressure (56), suggesting that certain in vivo conditions favored the development, and thereby potentially the maintenance, of a teicoplanin resistance phenotype.
Here we assessed the fitness of the in vitro-selected teicoplanin-resistant strain NM67 in a murine tissue cage model. Although the MIC for NM67 was above the teicoplanin breakpoint for intermediate level resistance (25), we define NM67 as being glycopeptide intermediate resistant because of the genetic basis of its resistance mechanism, which is distinct from the van gene-based resistance of clinical vancomycin-resistant S. aureus (11). GISA phenotypes and accompanying genetic alterations are known to vary from strain to strain; therefore, not all GISA isolates share identical characteristics. NM67 did, however, show several typical GISA phenotypes such as decreased growth rate, increased cell wall thickness, and enhanced N-acetylglucosamine uptake compared to its isogenic, susceptible parent. The tissue cage is a closed in vivo system, where neither bacteria nor attracted leukocytes can escape. The balance is in favor of bacterial survival, since phagocytes in tissue cage fluid show a decreased phagocytic and bactericidal activity compared to blood granulocytes (61). This particular situation reflects infection of an orthopedic implant, which explains why such low numbers of wild-type S. aureus can cause a chronic persistent infection in the mice (31).
In contrast to the parent strain, which replicated over the 8-day period and elicited bacterial load-dependent MIP-2 release, leukocyte infiltration, and reduction of leukocyte viability, the teicoplanin-resistant mutant was either immediately cleared from the tissue cages or began to lose viability and then recovered and started to grow after 2 days. The clearing or delayed growth of NM67 strongly suggested that the original teicoplanin resistant NM67 had lost its ability to cause a persistent infection and was actively cleared in the tissue cages, whereas the survivors appeared to have regained wild-type virulence. The survivors had, however, also become hypersusceptible to teicoplanin, with MICs approximately fourfold less than the susceptible wild-type SA113.
Characteristics of the reemerging NM67 survivors, such as their higher growth rate, the conversion to teicoplanin hypersusceptibility, their partial loss of the thickened cell wall paired with a reduction in the high N-acetylglucosamine incorporation rate, and the restitution of transcription of some of the downregulated virulence determinants, suggested that the survivors were phenotypic revertants of NM67, which arose by forward mutation, since not all wild-type characteristics had been restored. Similar forward mutations producing susceptible variants with slightly altered phenotypes have been observed earlier (13, 43).
Despite maintaining a slightly thicker cell wall and higher N-acetylglucosamine uptake, the teicoplanin MICs for NM67 survivors isolated on day 8 were lower than for SA113 isolates. Interestingly, resistance profiles of the wild-type SA113 remained relatively stable over the 8-day period; spontaneously resistant mutants did not arise as had been described earlier in a similar rat tissue cage infection model (56). Several factors may explain the absence of such spontaneous highly resistant mutants in our model. Teicoplanin-resistant mutants were observed in the rat model as late as 3 weeks after infection and were stable only when cultured continuously on teicoplanin-containing agar (56), whereas our study covered just the 8 days needed to reach the maximal numbers of CFU in the murine tissue cage model. Also, we used a susceptible S. aureus strain, whereas the previous study used a multiresistant, clinical methicillin-resistant S. aureus isolate. The NCTC8325 strain lineage used here has no functional IS elements (53), whereas clinical multiresistant MRSA isolates may carry active IS elements and thus have additional means to form glycopeptide-resistant mutants by rearrangements and movement of the IS elements. IS256 transposition was shown to facilitate mutations contributing to resistance levels in a clinical GISA isolate (34).
Altered autolytic properties are sometimes observed in glycopeptide-resistant strains (28, 59). However, neither cell wall cross linking and turnover nor the accessibility of the lysostaphin target were affected by teicoplanin resistance in our in vitro-selected strain. Here, glycopeptide resistance seemed to be mainly characterized by the thickened cell wall, although microarray analysis did not detect any significant upregulation of ORFs or any transcriptional changes that might account for these phenotypes, such as transcriptional alterations that have been described for other GISA strains (12, 33, 37). The microarray performed here probably does not reflect all transcriptional changes that occurred during the resistance development of NM67 since it was performed on cultures harvested at a single time point during in vitro growth and should therefore be regarded as a snapshot of some of the transcriptional changes that occurred during resistance formation. More detailed transcriptional analyses would be required to obtain a global picture of the differences between these strains.
Altered virulence factor expression has been previously described in other GISA strains; spa transcription was decreased in Mu50 (33), and fibronectin-binding protein production decreased in strain 14-4 (44). The decreased in vivo survival of NM67 may have been predicted by the gradual turning down of gene expression of virulence-associated genes as increased resistance was selected. It is likely that the downregulation of genes involved in biofilm formation, lipase, hemolysin, and secretory antigen production and evasion of host immune responses, contributed to the poor survival of NM67 in vivo. The downregulation of virulence genes was probably a consequence of, but unlikely to be the cause of increased teicoplanin resistance.
Transcription of some of the virulence-associated genes was partially restored upon loss of the resistance phenotype in the in vivo-selected survivors. This possible link between resistance profile and the transcription of virulence genes may be one explanation for why the resistance phenotype was lost so rapidly in the tissue cages that did not clear the teicoplanin-resistant bacteria. In these cases the mutation(s) causing reversion of resistance and restoration of virulence gene expression must have occurred before the bacteria were cleared.
In conclusion, intrinsic glycopeptide resistance acquisition can impose a fitness burden on S. aureus, creating selection pressure for the loss of the resistance phenotype in the absence of glycopeptides. In the present study, the glycopeptide resistance level of NM67 gradually decreased upon in vitro culture in glycopeptide-free media; however, in vivo, in the presence of the host defense system, resistance was lost extremely rapidly due to the elimination of the resistant strain and subsequent selection for fitter, susceptible variants. The link between resistance acquisition and the decreased expression of virulence-associated genes is likely to contribute to the strong selection for the clearance of the resistant strain in vivo. It is also possible that the decreased growth rate, which accompanied the high-level resistance, had also contributed to the poor in vivo survival of NM67. Compensatory mutations leading to decreased cell wall thickness indicate that metabolism reverts to normal, with the concomitant increase in virulence factor production indicating that the easiest way to restore fitness and virulence is at the expense of resistance.
Acknowledgments
We thank Ursula Sauder, Zentrum Mikroskopie Biocenter, Basel, Switzerland, for the electron microscopy pictures. We are also grateful to S. Projan, P. Dunman, and the WYETH Antimicrobial Research Department for providing us with the necessary materials for the microarray experiments.
This study was supported by Swiss National Science Foundation grants NRP 49-63201 to B.B.-B., NRP 49106295/1 to R.L., and 3200B0-103793 to P.M. and by the Forschungskredit der Universität Zürich grant 560030.
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, M., and B. Berger-Bachi. 2001. Teicoplanin stress-selected mutations increasing sigma(B) activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boncompagni, E., M. Osteras, M. C. Poggi, and D. le Rudulier. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 65:2072-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boneca, I. G., N. Xu, D. A. Gage, B. L. de Jonge, and A. Tomasz. 1997. Structural characterization of an abnormally cross-linked muropeptide dimer that is accumulated in the peptidoglycan of methicillin- and cefotaxime-resistant mutants of Staphylococcus aureus. J. Biol. Chem. 272:29053-29059. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., R. B. Carey, and R. S. Daum. 2001. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J. Antimicrob. Chemother. 48:617-625. [DOI] [PubMed] [Google Scholar]
- 8.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle-Vavra, S., S. H. Yin, M. Challapalli, and R. S. Daum. 2003. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob. Agents Chemother. 47:1776-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, et al. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 12.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, L., X. X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, L. Z., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale, S. E., M. T. Sebulsky, and D. E. Heinrichs. 2004. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 186:8356-8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson, J., C. Rordorf-Adam, T. Geiger, H. Towbin, S. Kunz, H. Nguyen, O. Zingel, D. Chaplin, and K. Vosbeck. 1993. Interleukin-1 (IL-1) production in a mouse tissue chamber model of inflammation. I. Development and initial characterisation of the model. Agents Actions 38:247-254. [DOI] [PubMed] [Google Scholar]
- 17.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain: the role of penicillin binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 18.Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bachi. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fluckiger, U., M. Ulrich, A. Steinhuber, G. Doring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisel, R., F. J. Schmitz, A. C. Fluit, and H. Labischinski. 2001. Emergence, mechanism, and clinical implications of reduced glycopeptide susceptibility in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:685-697. [DOI] [PubMed] [Google Scholar]
- 21.Goda, S. K., and N. P. Minton. 1995. A simple procedure for gel electrophoresis and northern blotting of RNA. Nucleic Acids Res. 16:3357-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gresham, H. D., J. H. Lowrance, T. E. Caver, B. S. Wilson, A. L. Cheung, and F. P. Lindberg. 2000. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J. Immunol. 164:3713-3722. [DOI] [PubMed] [Google Scholar]
- 23.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 24.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: 15th informational supplement. CLSI/NCCLS document M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 26.Iordanescu, I., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 27.Kaatz, G. W., S. M. Seo, N. J. Dorman, and S. A. Lerner. 1990. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J. Infect. Dis. 162:103-108. [DOI] [PubMed] [Google Scholar]
- 28.Koehl, J. L., A. Muthaiyan, R. K. Jayaswal, K. Ehlert, H. Labischinski, and B. J. Wilkinson. 2004. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3749-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsuzawa, H., T. Fujiwara, H. Nishi, S. Yamada, M. Ohara, N. McCallum, B. Berger-Bachi, and M. Sugai. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221-1231. [DOI] [PubMed] [Google Scholar]
- 30.Kristian, S. A., T. Golda, F. Ferracin, S. E. Cramton, B. Neumeister, A. Peschel, F. Gotz, and R. Landmann. 2004. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb. Pathog. 36:237-245. [DOI] [PubMed] [Google Scholar]
- 31.Kristian, S. A., X. Lauth, V. Nizet, F. Goetz, B. Neumeister, A. Peschel, and R. Landmann. 2003. Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J. Infect. Dis. 188:414-423. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 33.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 34.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bachi. 2004. TcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midolo, P. D., T. M. Korman, D. Kotsanas, P. Russo, and T. G. Kerr. 2003. Laboratory detection and investigation of reduced susceptibility to vancomycin in oxacillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 22:199-201. [DOI] [PubMed] [Google Scholar]
- 36.Moise-Broder, P. A., G. Sakoulas, G. M. Eliopoulos, J. J. Schentag, A. Forrest, and R. C. J. Moellering. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700-1705. [DOI] [PubMed] [Google Scholar]
- 37.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreira, B., S. BoyleVavra, B. L. M. deJonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishi, H., H. Komatsuzawa, T. Fujiwara, N. McCallum, and M. Sugai. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4800-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peschel, A., C. Vuong, M. Otto, and F. Gotz. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reipert, A., K. Ehlert, T. Kast, and G. Bierbaum. 2003. Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin. Antimicrob. Agents Chemother. 47:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renzoni, A., P. Francois, D. M. Li, W. L. Kelley, D. P. Lew, P. Vaudaux, and J. Schrenzel. 2004. Modulation of fibronectin adhesins and other virulence factors in a teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruzin, A., A. Severin, S. L. Moghazeh, J. Etienne, P. A. Bradford, S. J. Projan, and D. M. Shlaes. 2003. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. BBA-Gen. Subjects 1621:117-121. [DOI] [PubMed] [Google Scholar]
- 46.Sakoulas, G., G. M. Eliopoulos, V. G. J. Fowler, R. C. J. Moellering, R. P. Novick, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 49.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, V. K., J. L. Schmidt, R. K. Jayaswal, and B. J. Wilkinson. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int. J. Antimicrob. Agents 21:256-261. [DOI] [PubMed] [Google Scholar]
- 52.Snowden, M. A., and H. R. Perkins. 1990. Peptidoglycan cross-linking in Staphylococcus aureus: an apparent random polymerisation process. Eur. J. Biochem. 191:373-377. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenover, F. C., J. W. Biddle, and M. V. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsujimoto, H., S. Ono, H. Mochizuki, S. Aosasa, T. Majima, C. Ueno, and A. Matsumoto. 2002. Role of macrophage inflammatory protein 2 in acute lung injury in murine peritonitis. J. Surg. Res. 103:61-67. [DOI] [PubMed] [Google Scholar]
- 56.Vaudaux, P., P. Francois, B. Berger-Bachi, and D. P. Lew. 2001. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 47:163-170. [DOI] [PubMed] [Google Scholar]
- 57.Wada, A., Y. Katayama, K. Hiramatsu, and T. Yokota. 1991. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 176:1319-1325. [DOI] [PubMed] [Google Scholar]
- 58.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 59.Wootton, M., P. M. Bennett, A. P. Macgowan, and T. R. Walsh. 2005. Reduced expression of the atl autolysin gene and susceptibility to autolysis in clinical heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA) and GISA strains. J. Antimicrob. Chemother. 56:944-947. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, L., A. Rosander, K. Jacobsson, M. Lindberg, and L. Frykberg. 2000. Expression of staphylococcal protein Sbi is induced by human IgG. FEMS Immunol. Med. Microbiol. 28:211-218. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerli, W., P. D. Lew, and F. A. Waldvogel. 1984. Pathogenesis of foreign body infection: evidence for a local granulocyte defect. J. Clin. Investig. 73:1191-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerli, W., F. A. Waldvogel, P. Vaudaux, and U. E. Nydegger. 1982. Pathogenesis of foreign body infection: description and characteristics of an animal model. J. Infect. Dis. 146:487-497. [DOI] [PubMed] [Google Scholar]