Abstract
The molecular mechanisms of drug resistance in 19 strains of Vibrio fluvialis isolated from 1998 to 2002 in Kolkata, India, were investigated. Class 1 integrons were detected in eight strains, and four strains were found to carry SXT integrases. In the presence of carbonyl cyanide m-chlorophenylhydrazone or reserpine, all nalidixic acid- and ciprofloxacin-resistant strains became sensitive, suggesting that drug efflux plays a major role in quinolone resistance in V. fluvialis. It was further seen that strains which had MICs of >25 μg/ml for nalidixic acid had a sense mutation (Ser to Ile) at position 83 of the quinolone resistance-determining region of gyrA. All except one of the integron- and SXT integrase-bearing strains belonged to the same ribotype.
Vibrio fluvialis, a halophilic Vibrio species, has been associated with sporadic outbreaks of diarrhea worldwide (10, 11, 13), which is clinically very similar to cholera. It is being isolated with an increased frequency from hospitalized patients in Kolkata, India, with cholera-like illnesses (our unpublished observations). The V. fluvialis strains isolated in the past were found to be resistant to several antibiotics (13). However, there is a paucity of information on the genetic basis of drug resistance in these strains. Antibiotic resistance arises through multiple means and can be mediated by plasmids, integrons, and transposons, besides mutations in target genes and the overexpression of efflux systems (4, 5, 6, 8). Only two reports (1, 2) have addressed this issue. In those studies, the presence of the drug cassettes aac(3)-Id and aadA7, which confer resistance to gentamicin, streptomycin, and spectinomycin, were identified in V. fluvialis, as was an SXT constin (6). In this study, we performed antibiotic susceptibility tests with 19 strains of V. fluvialis isolated from hospitalized patients in Kolkata, India, recovered from 1998 to 2002 and also analyzed their drug resistance profiles.
MATERIALS AND METHODS
V. fluvialis strains were isolated from patients with acute cholera-like diarrhea admitted to the Infectious Diseases Hospital, Kolkata, India, between 1998 and 2002.
Bacteriology.
V. fluvialis isolates were identified by using the API 20E system (bioMerieux, Marcy l'Etoile, France) and also by 16S rRNA gene sequencing.
Antimicrobial susceptibility testing and MIC determination.
The isolates were examined for antibiotic resistance as described previously (17). The MICs of ciprofloxacin and nalidixic acid were determined as described in the CLSI standards (7).
Bacterial genomic DNA and plasmid isolation.
Genomic and plasmid DNA was extracted from the isolates by a method described earlier (17), except that for plasmid isolation, 0.75 ml of culture and twice the suggested volumes of all three solutions were used.
Bacterial transformation and plating on selective media.
Escherichia coli JM109 electrocompetent cells, prepared according to the manufacturer's recommendations (Bio-Rad Laboratories, Richmond, Calif.), were transformed with 30 ng of plasmid preparations and selected as described before (17).
Ribotyping.
The rRNA gene restriction patterns (ribotypes) of the test strains were determined exactly as described before (16).
PCR amplifications, sequence analyses, and GenBank accession numbers.
PCRs for determination of the presence of class 1 integrons, SXT integrases, and mutations in the quinolone resistance-determining region (QRDRs) of the gyrA, gyrB, parC, and parE genes of quinolone-resistant strains were carried out with the appropriate primers (Table 1). Elution of the amplicons was done as described previously. Sequencing was carried out by following the manufacturer's instructions (BigDye Terminator kit; Applied Biosystems) with an ABI Prism 310 instrument. The identities of the sequences were established through a database search by using the BLAST program (3), and mutations were identified by comparison of the sequences with the V. cholerae genome sequence (9). Sequences with the following accession numbers were obtained from GenBank: AY605692, strain BD146 (dfrA15); AY605688, strain BD73 (dfrA1); AY605684, strain BD123 (aadA7); AY605683, strain BD51 [aac(3)-Id and aadA7]; and AY103456, strain PL78/6 (dfrA1).
TABLE 1.
Primers used in this study
| Primer | Sequence | GenBank accession no. | Position | Size of amplicon (kb) | Reference |
|---|---|---|---|---|---|
| gyrA-F | AATGTGCTGGGCAACGACTGG | AE003852 | 157-177 | 0.25 | 5 |
| gyrA-R | GTGCGCGATTTTCGACATACG | AE003852 | 376-396 | 0.25 | 5 |
| gyrB-F | GGAAATGACTCGCCGTAAAGG | AE003852 | 1170-1190 | 0.3 | 5 |
| gyrB-R | GTTGTGATAACGCAGTTTATCTGGG | AE003852 | 1455-1479 | 0.3 | 5 |
| parC-F | GTCTGAGTTGGGTCTCTCGGC | AE003852 | 162-188 | 0.3 | 5 |
| parC-R | AGAATCTCGGCAAACTTTGACAG | AE003852 | 388-411 | 0.3 | 5 |
| parE-F | ATGCGTGCCAGCAAGAAAGTG | AE003852 | 1138-1161 | 0.3 | 5 |
| parE-R | TTATCGCTGTCAGGGTCAATCC | AE003852 | 1408-1428 | 0.3 | 5 |
| in-F | GGC ATC CAA GCA GCA AGC | U12338 | 1416-1433 | Variable | 8 |
| in-B | AAG CAG ACT TGA CCT GAT | U12338 | 4831-4814 | Variable | 8 |
| qacEΔ1-F | ATCGCAATAGTTGGCGAAGT | X15370 | 211-230 | 0.8 | 8 |
| Sul1-B | GCAAGGCGGAAACCCGCC | X12869 | 1360-1341 | 0.8 | 8 |
| L2 | GACGATGCGTGGAGACC | M73819 | 910-926 | 0.3 | 14 |
| L3 | CTTGCTGCTTGGAtGCC | M73819 | 1206-1190 | 0.3 | 14 |
| SXT-F | TTATCGTTTCGATGGC | AF099172 | 129-144 | 0.8 | 17 |
| SXT-B | GCTCTTCTTGTCCGTTC | AF099172 | 915-931 | 0.8 | 17 |
Accumulation studies with quinolones.
The accumulation of quinolones was examined by the method of Mortimer and Piddock (15), with minor modifications: phosphate-buffered saline (0.136 M NaCl, 0.0026 M KCl, 0.01 M Na2HPO4, 0.00176 M KH2PO4; pH 7.0) was used instead of sodium phosphate buffer, and cell lysis was carried out for 6 h. Carbonyl cyanide m-chlorophenylhydrazone (CCCP) or reserpine was added to a final concentration of 20 μg/ml. The fluorescence of the supernatant was measured with a spectrofluorimeter (Tegimenta AG type SFM 25).
RESULTS
Drug resistance in V. fluvialis.
The V. fluvialis strains were found to be resistant to one or more antibiotics, with 75% of the strains being resistant to ampicillin, furazolidone, nalidixic acid, and streptomycin (Table 2).
TABLE 2.
Phenotypic and genotypic characterization of V. fluvialis strains
| Strain name | Antibiograma | Yr of isolation | Presence of class I integrons | Size of amplicon (kb) | Gene cassette | Presence of SXT | Plasmid size (kb) | gyrAb mutation | MIC (μg/ml)
|
Efflux pump
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | PMF for NAL | ABC for NAL | PMF for CIP | ABC for CIP | |||||||||
| PL78/6 | AMP, CHL, GEN, NAL, NEO, SM, STR, SZ, TET, TMP | 1998 | + | 1.25 | dfrA1 orfC | + | 240 | Ser-Iso | 100 | <0.5 | + | + | − | − |
| PL45 | AMP, FUR, SM, STR, SX, SZ, TMP | 1998 | − | − | 2-8 | <0.5 | <0.5 | − | − | − | − | |||
| PL78/7b | AMP, GEN, NAL, NEO, SM, STR, SX, SZ, TMP | 1998 | + | Null | + | 2-8 | Ser-Iso | 100 | <0.5 | + | + | − | − | |
| PG39 | AMP, FUR, NEO | 1998 | − | − | <0.5 | <0.5 | − | − | − | − | ||||
| PG41 | AMP | 1998 | − | − | <0.5 | <0.5 | − | − | − | − | ||||
| PG152 | AMP, FUR, NEO | 1998 | − | − | <0.5 | <0.5 | − | − | − | − | ||||
| PL169b | AMP, FUR, NEO | 1998 | − | − | <0.5 | <0.5 | − | − | − | − | ||||
| PL171b | AMP, CHL, CIP, FUR, NAL, NEO, SM, STR, SX, SZ, TET | 1998 | + | Null | + | Ser-Iso | 100 | 10 | + | + | + | + | ||
| CRC159 | AMP, FUR, NAL, NEO | 2000 | − | − | 10 | <0.5 | + | + | − | − | ||||
| CRC233 | AMP, FUR, GEN, NAL, NEO, STR, SX, SZ, TET | 2000 | + | Null | − | Ser-Iso | 25 | <0.5 | + | + | − | − | ||
| CRC111 | AMP, FUR, NAL, SM, SZ | 2000 | − | − | 6 | 10 | <0.5 | + | + | − | − | |||
| BD51 | AMP, CHL, FUR, GEN, NAL, STR | 2002 | + | 1.5 | aac(3)-Id aadA7 | + | 2-8 | Ser-Iso | 100 | <0.5 | + | + | − | − |
| BD73 | AMP, CHL, FUR, NAL, STR | 2002 | + | 1.25 | dfrA1 orfC | − | 15 | <0.5 | + | + | − | − | ||
| BD123 | AMP, CHL, FUR, GEN, NAL, STR | 2002 | + | 1.0 | AadA7 | − | 2-8 | Ser-Iso | 100 | <0.5 | + | + | − | − |
| BD146 | AMP, CHL, CIP, GEN, NAL, STR | 2002 | + | 0.7 | dfrA15 | − | 2-8 | Ser-Iso | 100 | 10 | + | + | + | + |
| BD36 | AMP, FUR, GEN, NAL, STR | 2002 | − | − | 2-8 | 10 | <0.5 | + | + | − | − | |||
| BD56 | AMP, FUR, NAL, STR | 2002 | − | − | 2-8 | 10 | <0.5 | + | + | − | − | |||
| BD135 | CHL, NAL, STR | 2002 | − | − | 2-8 | Ser-Iso | 100 | <0.5 | + | + | − | − | ||
| BD159 | AMP, FUR, GEN, NAL, STR | 2002 | − | − | 2-8 | Ser-Iso | 100 | <0.5 | + | + | − | − | ||
Abbreviations: AMP, ampicillin (10 μg); CHL, chloramphenicol (30 μg); FUR, furazolidone (50 μg); GEN, gentamicin (10 μg); STR, streptomycin (10 μg); TET, tetracycline (30 μg); NEO, neomycin (30 μg); SM, sulfamethizole (300 μg); SZ, sulfamethoxazole (50 μg); SX, sulfadiazine (100 μg); and TMP, trimethoprim (25 μg); NAL, nalidixic acid; CIP, ciprofloxacin. Interpretation of the results was done by using the criteria recommended by the CLSI (7), except that no distinction was made between intermediate and full resistance. Escherichia coli ATCC 25922 was used for quality control.
gyrA mutations were identified by comparing the sequences obtained with the corresponding sequences of V. cholerae (9).
Class 1 integrons and mapping of resistance gene cassettes.
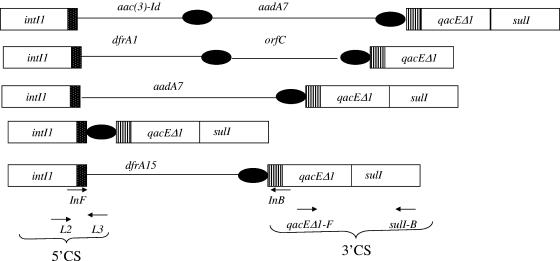
Eight of the 19 strains produced a 0.8-kb amplicon with the primers qacEΔ1-F and Sul1B (Table 1; Fig. 1). A 0.3-kb amplicon was obtained with the L2 and L3 primer pair (Table 1; Fig. 1). Sequencing of the 0.8-kb and 0.3-kb amplicons followed by BLAST analysis revealed 100% sequence identity of these amplicons with the 3′ conserved sequence (3′ CS) and 5′ CS, respectively, of class 1 integrons (8, 12). PCR carried out with primers in-F (5′ CS) and in-B (3′ CS) (Table 1; Fig. 1) produced amplicons of four distinct size classes: 0.7, 1.0, 1.25, and 1.5 kb. Sequence analysis of the 0.7-kb amplicon revealed the presence of the dfrA15 gene cassette in strain BD146 (Table 2; Fig. 1). The 1.25-kb amplicon, generated by strains PL78/6 and BD73, harbored the dfrA1 gene cassette and the gene cassette of an unknown gene (orfC) (Table 2; Fig. 1). The aadA7 gene cassette was found on the 1.0-kb PCR product obtained with BD123 (Table 2; Fig. 1). The 1.5-kb amplicon from strain BD51 harbored two cassettes, aac(3)-Id and aadA7 (Table 2; Fig. 1). In strains PL78/7b, PL171b, and CRC233, no amplicon could be generated with primer pair in-F and in-B (Table 1; Fig. 1). However, a 1.2-kb amplicon was obtained with primers L2 and Sul1B. Sequencing of this amplicon revealed the presence of a “null” cassette (17) in this class 1 integron.
FIG. 1.
Diagrammatic representation of the different types of gene cassettes identified in the class 1 integrons in V. fluvialis strains. Solid lines, gene cassettes; solid circles, 59-base elements; hatched box, attI recombination site. The locations of the 5′ CSs and the 3′ CSs of class 1 integrons and those of the primer pair qacEΔI-F and Sul1B and primer pair in-F (5′ CS) and in-B (3′ CS) are shown at the bottom.
Occurrence of SXT integrase gene.
To look for SXT, primers SXT-F and SXT-B, whose sequences are internal to that of the SXT integrase gene (SXT int), were used (Table 1) for amplification of the putative SXT int. A 0.8-kb amplicon was obtained with only four strains, namely, strains PL78/6, PL78/7b, PL171b, and BD51, which, upon sequencing, exhibited 100% homology with the relevant portion of the SXT int gene (17), suggesting the presence of SXT constins or parts thereof in these strains.
Resistance to quinolones: analysis for QRDRs in target genes.
Fourteen of the 19 strains were resistant to the nonfluorinated quinolone nalidixic acid and two were additionally resistant to ciprofloxacin. The nalidixic acid-resistant strains could broadly be classified into two groups: one with MICs between 10 and 20 μg/ml and the other with MICs ≥25 μg/ml (Table 2). Quinolone resistance in bacteria is mainly linked to the mutations in the QRDRs of the gyrAB and parCE genes. To check for the mutations in QRDRs, primers gyrA-F and gyrA-R and primers parC-F and parC-R (Table 1), respectively, were used to amplify these regions. Sequencing of the 0.25-kb amplicon obtained with all strains for the gyrA QRDR showed the presence of a mutation at position 83, which led to the replacement of serine with isoleucine in nine strains, which had MICs ≥25 μg/ml for nalidixic acid. No mutation could be detected in any of the amplicons, an amplicon of 0.25 kb for parC and an amplicon of 0.3 kb for both the gyrB and the parE QRDRs, with the appropriate primers (Table 1).
Involvement of efflux pumps.
Although strains BD36, BD56, BD73, CRC111, and CRC159 were resistant to 10 to 15 μg/ml of nalidixic acid, they did not have any mutations in the QRDRs of target genes. This indicated that other mechanisms could be responsible for this level of nalidixic acid resistance observed in these strains. To examine if multidrug resistance pumps are involved in conferring quinolone resistance to V. fluvialis, the level of accumulation of quinolones in the absence and the presence of the efflux pump inhibitors, namely, CCCP (an inhibitor of the proton motive force [PMF]) and reserpine (a plant alkaloid known to inhibit the ATP binding cassette [ABC transporters]), was measured. It was seen that irrespective of the MICs, both CCCP and reserpine could prevent the accumulation of nalidixic acid in all nalidixic acid-resistant strains (Fig. 2). These same two inhibitors could also prevent the accumulation of ciprofloxacin in two strains, strains PL171b and BD146, which were additionally resistant to this drug. The results (Fig. 3) indicated that efflux pumps indeed had a role here also.
FIG. 2.
Accumulation of nalidixic acid (NAL) by V. fluvialis after the addition of CCCP (A) or resperine (B). The fluorescence of the supernatant was measured with a spectrofluorimeter at an excitation wavelength of 330 nm and an emission wavelength of 417 nm for nalidixic acid. The results for three representative strains, namely, strains BD56, CRC233, and PL78/6, are shown here. The arrows indicate the time of addition of CCCP and reserpine. The graphs reflect the differences in fluorescence shown by the bacterial cell in the presence and the absence of the inhibitors. All experiments were carried out at least three times.
FIG. 3.
Studies with ciprofloxacin (CIP) performed as described in the legend to Fig. 2, except that the fluorescence of the supernatant was measured with a spectrofluorimeter at an excitation wavelength of 275 nm and an emission wavelength of 440 nm. The accumulation of ciprofloxacin by V. fluvialis after the addition of CCCP (A) or reserpine (B) is shown. The arrows indicate the time of addition of CCCP and reserpine.
Plasmid carriage and resistance conferred by plasmids.
Eleven of the 19 strains had plasmids, of which 10 had plasmids ranging in size from 2 to 8 kb. Strain PL78/6 had a 240-kb plasmid (Table 2). All plasmids in the 2- to 8-kb range, except the ones from strain CRC111, could transform E. coli JM109 to ampicillin resistance (data not shown). No transformant could be obtained with the 6-kb plasmid from CRC111. The 240-kb plasmid from PL78/6 could transfer resistance to streptomycin, sulfamethoxazole, and ampicillin (data not shown).
DISCUSSION
The study described in this paper was undertaken to unravel the molecular basis of drug resistance in V. fluvialis strains isolated in Kolkata, India. Compared to only 37% of the isolates recovered in 1998, 100% of the isolates recovered in 2002 were resistant to quinolones. Eight strains were found to carry class 1 integrons. Although an integron in a V. fluvialis strain was detected for the first time in 2002 (1), our work revealed that integrons have actually been present in V. fluvialis at least since 1998. Although the “linked” gene cassettes aadA7 and aac(3)-Id were detected in V. fluvialis earlier (1), the aadA7 gene cassette detected in BD123 (Table 2 and Fig. 1), which occurs singly, has not been found in any Vibrio strain so far. Our studies further showed that although all the strains were drug resistant, only five harbored integrons with drug cassettes which accounted for resistance to only a few antibiotics (Table 2). Although almost all of the strains were resistant to trimethoprim, only three strains, strains PL78/6, BD73, and BD146, had dfrA1 and dfrA15 genes, which confer resistance to this drug. Quite surprisingly, with the exception of four strains, SXT integrases could not be detected in any other strain (Table 2). We found that 11 strains harbored plasmids, but except in 2 strains, they carried genes only for ampicillin resistance. It was of interest that except for strain CRC233, all integron- and SXT int-bearing strains belonged to a single ribotype (data not shown). Quinolone resistance in these strains could be traced either to mutations in the QRDRs of gyrA or to the presence of efflux pumps, or both. Efflux pumps alone were responsible for the nalidixic acid resistance phenotype in the strains with MICs between 10 and 15 μg/ml, whereas both mutations in the gyrA QRDR and efflux pumps were responsible for nalidixic acid resistance in strains with MICs ≥25 μg/ml.
Acknowledgments
Financial support was received from the Department of Biotechnology and the Council of Scientific and Industrial Research, Government of India. Vijaya Srinivasan gratefully acknowledges the Council of Scientific and Industrial Research, Government of India, for a senior research fellowship. Rupinder Kaur Virk was supported by a research associateship from the Department of Biotechnology, Government of India.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, A. M., S. Shinoda, and T. Shimamoto. 2005. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242:241-247. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-441. [DOI] [PubMed] [Google Scholar]
- 4.Amita, S. R. Chowdhury, M. Thungapathra, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2003. Class 1 integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9:500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranwal, S., K. Dey, T. Ramamurthy, G. B. Nair, and M. Kundu. 2002. Role of active efflux in association with target gene mutations in fluoroquinolones resistance in clinical isolates of Vibrio cholerae. Antimicrob. Agents Chemother. 46:2676-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical Laboratory Standards Institute. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 2nd ed. CLSI document M31-A2. Clinical Laboratory Standards Institute, Wayne, Pa.
- 8.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 6795:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 5:1176-1183. [DOI] [PubMed] [Google Scholar]
- 11.Huq, M. I., A. K. M. J. Alam, D. J. Brenner, and G. K. Morris. 1980. Isolation of Vibrio-like group EF-6, from patients with diarrhea. J. Clin. Microbiol. 11:621-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazama, H., H. Hamashima, M. Sasatsu, and T. Arai. 1998. Distribution of the antiseptic resistance genes qacE and qacE1 in gram-negative bacteria. FEMS Microbiol. Lett. 59:173-178. [DOI] [PubMed] [Google Scholar]
- 13.Lesmana, M., D. S. Subekti, P. Tjaniadi, C. H. Simanjuntak, N. H. Punjabi, J. R. Campbell, and B. A. Oyofo. 2002. Spectrum of Vibrio species associated with acute diarrhea in North Jakarta, Indonesia. Diagn. Microbiol. Infect. Dis. 2:91-97. [DOI] [PubMed] [Google Scholar]
- 14.Maguire, A. J., D. F. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and non-fermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortimer, P. G., and L. J. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28:639-653. [DOI] [PubMed] [Google Scholar]
- 16.Sharma, C., G. B. Nair, A. K. Mukhopadhyay, S. K. Bhattacharya, R. K. Ghosh, and A. Ghosh. 1997. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J. Infect. Dis. 175:1134-1141. [DOI] [PubMed] [Google Scholar]
- 17.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class 1 integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]