Abstract
β-Lactamases are the main cause of bacterial resistance to penicillins and cephalosporins. Class A β-lactamases, the largest group of β-lactamases, have been found in many bacterial strains, including mycobacteria, for which no β-lactamase structure has been previously reported. The crystal structure of the class A β-lactamase from Mycobacterium fortuitum (MFO) has been solved at 2.13-Å resolution. The enzyme is a chromosomally encoded broad-spectrum β-lactamase with low specific activity on cefotaxime. Specific features of the active site of the class A β-lactamase from M. fortuitum are consistent with its specificity profile. Arg278 and Ser237 favor cephalosporinase activity and could explain its broad substrate activity. The MFO active site presents similarities with the CTX-M type extended-spectrum β-lactamases but lacks a specific feature of these enzymes, the VNYN motif (residues 103 to 106), which confers on CTX-M-type extended-spectrum β-lactamases a more efficient cefotaximase activity.
Mycobacteria are important causes of infectious diseases. Although Mycobacterium tuberculosis and Mycobacterium leprae, two slow-growing species, are responsible for the most serious diseases, some fast-growing species, such as Mycobacterium avium, Mycobacterium kansasii, Mycobacterium chelonae, and Mycobacterium fortuitum, may cause opportunistic infections among AIDS patients (24). In addition, M. fortuitum has been reported to be responsible for a wide spectrum of clinical diseases, such as skin or soft tissue infections following surgery, pulmonary infections, accidental penetrating trauma, and wounds that come in contact with soil or water contaminated with these mycobacteria, though nosocomial infections are by far the most common (41, 44).
The cell wall of mycobacteria contains mycolic acid, arabinogalactan, and peptidoglycan, forming a covalent complex. The influx of small hydrophilic agents through the resulting low-permeability envelope is extremely slow and is thought to be one of the major factors involved in the resistance of mycobacteria to β-lactam antibiotics (21, 41). β-Lactamase production, catalyzing β-lactam antibiotic hydrolysis, appears to be the second mechanism by which mycobacteria express β-lactam resistance (9, 21, 35). Chromosomally encoded β-lactamases have been detected in most, but not all, mycobacterial species, including M. fortuitum and M. tuberculosis (43).
The class A β-lactamase of M. fortuitum (MFO) is a chromosomally encoded broad-spectrum β-lactamase hydrolyzing both cephalosporins and penicillins. Its substrate profile includes ureidopenicillins (piperacillin, azlocillin, and mezlocillin), carbenicillin, cephalothin, cefotaxime, and cefuroxime, but not ceftazidime. Cefoxitin, dicloxacillin, imipenem, and aztreonam are poorly recognized by the enzyme, and the protein is inhibited by the penem inhibitor BRL42715 and, to a lesser extent, clavulanic acid. The specificity profile is similar overall to those of CTX-M-type enzymes; some TEM-derived extended-spectrum β-lactamases (ESBLs); the chromosomally encoded ESBLs from Citrobacter sedlakii and Yersinia enterocolitica; and β-lactamases identified in the genera Nocardia, Kluyvera, and Burkholderia (1, 12, 31, 33, 35, 37). At the amino-acid level, the mature protein shares 70% identity with the β-lactamase of Mycobacterium smegmatis and between 40 and 45% with the aforementioned proteins. MFO also shares 41% amino acid identity with the ESBL Toho-1. Many recent studies have been concerned with the structure solution of TEM, SHV, or CTX-M-type ESBLs, but few structural data are available on chromosomally encoded broad-spectrum class A β-lactamases. MFO is the first solved structure of a mycobacterial β-lactamase. Its structure-activity relationship was analyzed on the basis of a comparison between its structure and recently solved ESBL structures (5, 27, 29, 38).
MATERIALS AND METHODS
Production and crystallization.
The gene used for β-lactamase expression was the blaF*-blaF hybrid described by Timm et al. (42). The β-lactamase was produced by M. smegmatis transformed by plasmid piPJ42* in 1 liter of Müller-Hinton broth. After centrifugation, the supernatant was collected and concentrated on an Amicon 8400. The concentrated solution (130 ml) was dialyzed against 10 liters of 10 mM Tris-HCl, pH 8. The crude extract was adsorbed on a High Load Q Sepharose High Performance column (36/10; Pharmacia, Uppsala, Sweden) preequilibrated with the same buffer. The enzyme was eluted by an NaCl linear gradient (0 to 0.5 M over 300 ml). The active fractions were pooled and dialyzed against 25 mM Tris-HCl buffer, pH 6.3. The solution was concentrated to 10 ml and further purified by chromatofocusing over the pH range 7 to 4 on a MonoP HR5/20 column (Pharmacia). The active fractions were concentrated to 2 ml and filtered through a Superdex 75 10/30 column (Pharmacia) to eliminate the polybuffer. Crystals of MFO were grown by the vapor diffusion method under hanging-drop conditions using a protein concentration of 20 mg/ml in 0.1 M Tris HCl buffer, pH 7.3, with polyethylene glycols of different molecular weights as precipitating agents. Typically, drops containing 5 μl of protein and 5 μl of reservoir were allowed to equilibrate at 20°C against a 1-ml reservoir. Long needles of 150 μm by 150 μm by 800 μm, suited for X-ray diffraction analysis, were obtained from 27% polyethylene glycol 10000 with several additives (ethylene glycol, glycerol, methylpentanediol, polyethylene glycol 400 at 5% concentration, and 10 mM spermidine).
Data collection, phase determination, and structure refinement.
X-ray diffraction data were collected on the DW32 synchrotron beamline at the Laboratoire pour l'Utilization du Rayonnement Electromagnétique (DCI, Orsay, France) using a monochromatic radiation of 0.975 Å. The crystal-to-detector (small MarResearch imaging plate) distance was 180 mm. A total of 45 frames (2° oscillation per frame and 300-s exposure time) were collected at room temperature. The data were processed with the MOSFLM 5.55 package (23). The structure was solved by molecular replacement using the program AMoRe (26), and the model was refined using Refmac5 (4) and Coot (8). Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data | Value |
|---|---|
| Crystal data | |
| Space group | P212121 |
| Cell dimensions (Å) | a = 35.66, b = 73.42, c = 91.32 |
| Data set | |
| Highest resolution (Å) | 2.13 |
| Total no. of reflections | 49,318 |
| No. of unique reflections | 11,576 |
| Completeness (%) | |
| All data | 86.89 |
| Highest shell (Å) | 52.98 (2.185-2.129) |
| I/σ (I) overall | 7.86 |
| R sym (I) (%) | 8.0 |
| Refinement | |
| Resolution (Å) | 28.8-2.13 |
| R factor (%) | 16.6 |
| R free (%) (using 5% of reflections) | 22.3 |
| R.m.s. deviations from ideality | |
| Bond length (Å) | 0.02 |
| Bond angles (°) | 1.9 |
| Average B factor (Å2) | 19.0 |
Protein structure accession number.
Atomic coordinates have been deposited in the Brookhaven Protein Data Bank (PDB) with the accession code 2CC1.
RESULTS
Structure determination.
MFO crystallized in space group P212121 (a = 35.66 Å, b = 73.42 Å, c = 91.32 Å) with one molecule per asymmetric unit. The structure was solved by molecular replacement, using as a search model the Streptomyces albus G β-lactamase structure (referred to hereafter as SAG; PDB code 1BSG) (6). After refinement, the final crystallographic R factor was 0.17 at 2.13-Å resolution (Table 1). The unique molecule in the asymmetric unit was well defined by continuous electron density, except for the N-terminal helix. Residues 27 to 29 are poorly defined, and breaks appear in the (2Fo-Fc) map for the side chains of residues 31, 32, 35, 38, and 41. The Ramachandran plot and the secondary-structure analysis with PROCHECK (22) are consistent with a well-defined structure. The only residue in a special conformation is Cys69. This residue lies next to the active-site serine and exhibits this strained conformation in all known three-dimensional structures of class A β-lactamases. The final structure contains 81 water molecules, mainly in the first coordination shell. Among water molecules present in the active site, one lies in the oxyanion hole defined by the backbone nitrogens of Ser70 and Ser237, and a second is firmly held by Ser70, Glu166, and Asn170.
Overall structure.
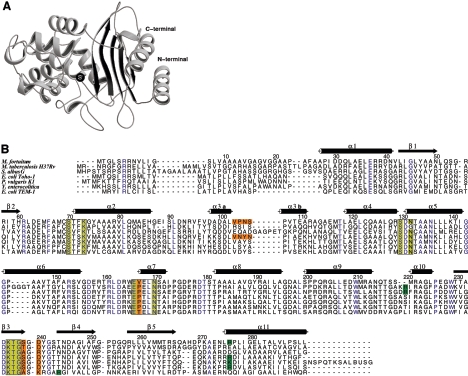
The structure of the MFO β-lactamase is similar to those of other class A β-lactamases (Fig. 1). The active site is at the interface between an α/β domain and an all-α domain. The α/β domain consists of a five-stranded β-sheet covered by three helices on one side and one helix on the other. Besides the conserved motifs defining the active site, the MFO structure shares other well-conserved structural motifs with other class A β-lactamases, suggesting a major role in the stabilization of the folding of this class of proteins. The salt bridge between Glu37 and Arg61 is usually well preserved, like the carboxyl-carboxylate interaction between Asp233 and Asp246. The absence of a side chain for Gly45 creates a dip in the β-sheet surface in which the benzyl group of Phe66 can fit. These two residues are strictly conserved among class A β-lactamases, and Phe66 has been shown to be essential for protein folding and function (32). On the other hand, the deletion of two residues between Ser217 and Ser220 in MFO is a unique feature among class A β-lactamases. This deletion occurs at the N terminus of a short helix in the upper right corner of the active site (with the orientation shown in Fig. 2) and reduces it to a mere single-turn loop. As a consequence of the deletion of residues 218 and 219, MFO residue 220 is in a standard α-helix conformation, whereas this residue is generally constrained in other class A β-lactamases. In view of this deletion, the energetically unfavorable conformation of residue 220 observed in all class A β-lactamases but MFO is surprising. The backbone position of the residues surrounding this deletion is displaced by about 1 Å compared to other class A β-lactamases, resulting in a small backward displacement of strand β3.
FIG. 1.
(Top) Ribbon representation of the class A β-lactamase from Mycobacterium fortuitum. The active-site serine (S) is indicated by a black sphere. (Bottom) Sequence alignment of class A β-lactamases from M. fortuitum (accession no. uniprot Q59517), M. tuberculosis H37Rv (P0A516), S. albus G (P14559), E. coli Toho-1 (Q47066), P. vulgaris K1 (P52664), Y. enterocolitica (Q01166), and E. coli TEM-1 (Q4AE67). The four conserved motifs of class A β-lactamases are boxed. Blue, strictly conserved residues; yellow, active-site conserved residues; green, Arg 220, 244, or 276 (278); orange, other residues discussed in the text.
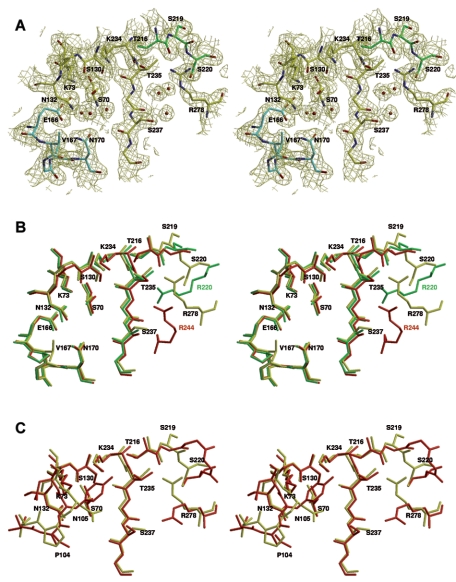
FIG. 2.
Stereo view of the active site of MFO showing the conserved motifs of class A β-lactamases and other residues discussed in the text. (A) Oxygen and nitrogen atoms are red and blue, respectively. Carbon atoms are yellow, green (residues 216 to 220), and cyan (Ω-loop; residues 166 to 170). The (2Fo-Fc) map is contoured at 1.0 σ. (B) Superimposition of the active sites of MFO (PDB code 2CC1) (yellow), S. albus G (1BSG) (green), and TEM-1 (1XPB) (red). MFO residues are labeled in black. SAG Arg220 is labeled in green, and TEM-1 Arg244 is labeled in red. (C) Superimposition of the active sites of MFO (yellow) and Toho-1 (red). Residues 166 to 170 are omitted for clarity.
Another unique feature of MFO is the occurrence of a valine in position 167. The Glu166-Val167 bond is in a cis conformation and is stabilized by a double hydrogen bond between the backbone of Glu166 and the side chain of Asn136. In all class A β-lactamase structures, residue 167 is in a cis conformation and is generally a proline.
Active site.
The active site of MFO, with its dense hydrogen-bonding network, is similar to most class A β-lactamase active sites (Fig. 2). The distances between the essential amino acid side chains are similar to those found in the other class A β-lactamases. Lys73 Nξ is hydrogen bonded to Ser70 Oγ and Asn132 Oδ1. Similarly, Lys234 Nξ hydrogen bonds to Ser130 Oγ and the carbonyl oxygen of Thr235. The distance between Lys73 and Glu166 (3.26 Å) is too long for a regular salt bridge. The relative location of the residues responsible for the catalytic activity (Ser70, Lys73, Ser130, Asn132, Glu166, and Lys234) is similar to that found in the Toho-1 or TEM-1 enzymes (PDB codes 1IYS and 1XPB) (Fig. 2B and C). Following the KTG motif, residue 237 is a serine. The Ser237 hydroxyl group lies near the active site, as does the guanidinium group of Arg278 (Arg278 corresponds to Ambler's numbering position 276 because of the insertion of two residues in positions 274 and 275). These two residues can interact with the substrate carboxylate. The guanidinium group of Arg278, located on the C-terminal helix, is directed toward the active-site cavity and can play a role equivalent to that assigned to Arg220 in SAG or Arg244 in TEM-1. Its exact location in the crystal structure is slightly different from that of Toho-1 or the equivalent groups in SAG and TEM-1, but the flexibility of the Arg278 side chain is sufficient to allow the superimposition of its guanidinium group on those found to be equivalent in Toho-1, SAG, and TEM-1.
The main difference between the MFO and Toho-1 active sites is the loop 103 to 106 at the entrance of the catalytic cleft. The sequence in MFO is VPNS, and the backbone conformation of this segment is similar to that of TEM-1. In contrast, the sequence VNYN in Toho-1 is typical of CTX-M-type ESBLs, and an interaction between Asn106 and the backbone nitrogen of residue 103 induces a different loop conformation.
DISCUSSION
The mechanism by which β-lactamases hydrolyze β-lactam antibiotics is known to follow a three-step pathway involving a Michaelis complex and an acyl-enzyme intermediate. At least five acylation mechanisms, which could be valuable for MFO, have been proposed. They are reviewed and studied by molecular dynamics simulation in reference 28. We will focus on features in or near the active site of MFO that are related to its substrate specificity.
The structural basis for broad-spectrum activity.
As shown in Table 2, MFO is moderately active on most penicillins and cephalosporins. The presence of a serine (or threonine) in position 237 can be related to this broad-spectrum activity. The Ala237Thr mutation in TEM-1 improves the activity toward cephalothin and cephalosporin C but decreases that toward benzylpenicillin and ampicillin (3, 16). Generally, class A β-lactamases devoid of a hydroxyl group in position 237 are more efficient in hydrolyzing penicillins than cephalosporins, while the presence of a hydroxylated residue gives to the enzyme rather broad-spectrum characteristics. Comparison of the specific activities of Klebsiella oxytoca D488 with those of MEN-1 provides a typical example of two enzymes differing mainly by this hydroxyl group. Lacking this group, K. oxytoca D488 is a better penicillinase than cephalosporinase, whereas MEN-1 is rather a broad-spectrum enzyme. Comparable results have been produced by the Ser237Ala mutation in Proteus vulgaris (40) or SME-1 (39). The carboxylate of cephalosporins can more easily hydrogen bond to the Ser237 hydroxyl group than the carboxylate of penicillins. The insertion of transition state models of the acylation step generated at the quantum level (7) into the active site suggests that the hydroxyl group could hinder the movement of one of the methyl groups of the penicillin thiazolidine ring during the course of the acylation process. This can explain the reduction of penicillinase activity resulting from the introduction of the hydroxyl group, as also suggested by Toho-1-benzylpenicillin and Toho-1-cephalothin acyl-enzyme structures (PDB codes 1IYQ and 1IYP) (38).
TABLE 2.
Second-order rate constants of MFO, Toho-1, and TEM-1 against various antibiotics
A second element that can explain the broad substrate profile of MFO is Arg278 (Arg278 corresponds to Ambler's numbering position 276). A specific feature of class A β-lactamases is the guanidinium group of either Arg278, Arg220, or Arg244, which lies in equivalent positions in the three-dimensional structures. The removal of the Arg244 side chain by site-directed mutagenesis has been shown to considerably reduce the enzyme activity against all β-lactam substrates in TEM-1 (45). Similarly, the Arg220Leu mutation in SAG induced a significant decrease in the rates of acylation by most classical substrates. Unexpectedly, the mutation of Arg276 to an uncharged residue in Toho-1 (18) or CTX-M-4 (13) induced only a slight decrease in kcat/Km on third-generation cephalosporins and no change for penicillins and first-generation cephalosporins. The interaction between the substrate carboxylate and the arginine is somewhat neglected in theoretical calculations because the docking of β-lactam substrates in the active site (at the Michaelis complex level) leads to interactions of the carboxylate with Ser130 and Lys234, and the arginine is remote from the carboxylate. The arginine, rather than Lys234, is the electrostatic complement of the carboxylate group. If the interaction between Lys234 and the substrate carboxylate is favorable for recognition and can lead to a decrease in Km, it has to be removed during acylation and will lead to a similar decrease in kcat. The most important thing is not the binding energy resulting from the interaction between the substrate carboxylate and the amine group of the lysine or the guanidinium group of the arginine but the deformation induced in the β-lactam ring by this interaction. One must consider the efficient part of the energy that will help to break the scissile β-lactam bond. The interaction between the guanidinium group and the substrate carboxylate tends to rotate the thiazolidine ring and, together with the attraction of the carbonyl oxygen in the oxyanion hole (17), as well as the interaction between the hydroxyl group of Ser130 and the lone pair of the β-lactam nitrogen, results in a shield stress on the endocyclic amide bond that reduces the activation barrier. From this point of view, the distance between the carboxylate and the guanidinium group, as well as the interaction direction, is an important factor that explains why the enzyme can discriminate between penicillins and cephalosporins, since the orientations of the carboxylate versus the β-lactam amide bonds are different in the two types of antibiotics. An arginine in position 220 or 244 seems more appropriate for an increase in the penicillin acylation rate, while Arg278 seems to only slightly affect third-generation cephalosporin hydrolysis.
Activity against cefotaxime and lack of activity against ceftazidime.
MFO retains a low specific activity on cefotaxime (kcat/Km = 40 mM−1 s−1) intermediate between the activity levels of TEM-1 (kcat/Km = 1.5 mM−1 s−1) (36) and Toho-1 (kcat/Km = 2,100 mM−1 s−1) (18). Arg278 and Ser237, favoring the cephalosporinase activity; the existence in MFO of a larger pocket between the side chains of residues 104, 167, 170, and 240, allowing better accommodation of the cefotaxime side chain; and an aspartic acid in position 240 appear to be the most important factors that can explain the slightly better cefotaxime hydrolysis rate of MFO over TEM-1. The Toho-1-cefotaxime crystal structure shows that Asp240 interacts with the amino group in the aminothiazole ring of the acylamide side chain (38). Nevertheless, unlike CTX-M-type ESBLs, MFO does not seem to be designed for efficient hydrolysis of cefotaxime. The most prevalent difference between MFO and CTX-M-type ESBLs that can explain the higher efficiency of the latter in hydrolyzing cefotaxime is the loop 103-to-106 conformation. In Toho-1, Asn104 interacts with the carbonyl oxygen of the side chain amide group of the substrate, and Toho-1 can fix the bulky side chain of cefotaxime more tightly than MFO.
As for Toho-1 and CTX-M-type ESBLs, ceftazidime is not recognized by MFO due to sterical hindrance between the bulky side chain of ceftazidime and the Ω-loop of the protein. ESBLs have evolved several ways to hydrolyze ceftazidime. TEM-type ESBLs that can hydrolyze ceftazidime harbor mutations of residues located in or near the Ω-loop that destabilize the Ω-loop and help accommodate the bulky side chain of ceftazidime. These mutations are generally detrimental to the hydrolysis of other β-lactams. CTX-M-type ESBLs have evolved differently to gain activity on ceftazidime, privileging substitutions in strand β3. CTX-M-15, CTX-M-16, and CTX-M-27 confer eightfold-higher resistance to ceftazidime than their respective parental enzymes, CTX-M-3, CTX-M-9, and CTX-M-14, from which they differ by the substitution Asp240Gly (2). This substitution also decreases the hydrolytic efficiency against cefotaxime (5). Substitutions in the Ω-loop also arise in CTX-M-type ESBLs in order to extend their hydrolytic activities to ceftazidime. CTX-M-19, which is derived from CTX-M-18 by a Pro167Ser substitution, confers a higher level of resistance to ceftazidime than cefotaxime (34). None of the characteristics encountered in ceftazidime-hydrolyzing ESBLs are found in MFO.
Cefotaxime and ceftazidime have a branched side chain, with one of the branches identical in both substrates. Crystallographic results show that the common branch, the aminothiazole ring, inserts in the active site with sterical hindrance with Asn170, whereas the other branch protrudes from the active site, and the extension specific to ceftazidime makes few contacts with the rest of the protein. Substitutions, whether in the Ω-loop or in the β3-strand, that improve β-lactamase activity on ceftazidime are generally detrimental to the hydrolysis of other β-lactams, because a slight displacement of Glu166 or Asn170 probably results in the reduction of the deacylation rate. In the case of ceftazidime, destabilization of the Ω-loop helps increase the affinity of the enzyme and allows acylation and deacylation rates, though modest, to be sufficient for substrate hydrolysis. The chromosomal class A β-lactamase K1 from Proteus vulgaris, the structure of which is close to that of MFO, can hydrolyze oxyimino β-lactams, such as cefuroxime or ceftazidime (27). MFO harbors a valine in position 167 instead of the proline in K1, and this small difference between the Ω-loops of the two enzymes might result in K1 having a greater activity on ceftazidime than does MFO.
Inhibitors.
Arginine 278 seems important in enzyme inactivation by clavulanic acid. The wild-type TEM enzyme appears to be more sensitive to clavulanic acid than SAG (11) or MFO (12), and the Arg244Ser mutation in TEM-1 strongly decreases its sensitivity to the same inactivator (19). It could be tentatively concluded that an arginine in position 220 or 278 (276) cannot completely compensate for the absence of Arg244 in the catalysis of the clavulanate moiety rearrangement, which results in irreversible enzyme inactivation.
Imipenem and cefoxitin act as poor transient inactivators due to slow hydrolysis and accumulation of the acyl-enzyme. Since the regions surrounding the SDN loop are similar in MFO and TEM-1, the interaction between TEM-1 and imipenem, investigated by crystallography (PDB code 1BTS) (25), is probably equivalent for MFO. Because of sterical hindrance between the Asn132 side chain and the hydroxyethyl group of imipenem, the acyl-enzyme is trapped in a conformation that prevents its deacylation. In the imipenem-hydrolyzing class A β-lactamase NMC-A, the position of the Asn132 side chain enlarges the binding cavity and allows the 6α substituent of imipenem to fit more easily into the active site. This situation also prevails for cefoxitin, as the methoxy side chain on the α face of the β-lactam ring is still bulkier than the hydroxyethyl group of imipenem. In this case, even NMC-A or the SME-1-related β-lactamase is not efficient to hydrolyze this compound. The crystal structure of Bacillus licheniformis BS3 in complex with cefoxitin (PDB code 1I2W) reveals that the sterical hindrance between the 7α-methoxy group of cefoxitin and Asn132 causes a conformational change of the substrate 7β side chain and eliminates the water molecule needed for deacylation (10). The mutation Asn132Ala in SAG has been shown to drastically decrease most β-lactam ratios of hydrolysis (20), but the removal of the Asn132 side chain would leave some free space for the methoxy group and allow a better hydrolysis of cefoxitin. The class A β-lactamase of M. tuberculosis H37Rv, which shares about 61% similar residues with MFO, exhibits a glycine in position 132 (15). This enzyme has a low specific activity on most β-lactam antibiotics, but its ability to hydrolyze cefoxitin is undoubtedly related to the removal of the Asn132 side chain. Similarly, a glycine residue occurs at position 132 of the Streptomyces clavuligerus β-lactamase. Again, the enzyme exhibits a low specific activity but is able to hydrolyze cefoxitin (30). The role of Asn132 is not only to hydrogen bond to the substrate side chain amide group to correctly position the substrate in the Michaelis complex, but rather, it is important to tightly fix the substrate in the enzyme cavity during the course of acylation.
Conclusion.
Compared to TEM-1, MFO shows a broadened and unspecialized spectrum of activity, but MFO did not evolve from intensive use of cefotaxime in antimicrobial therapy, which rather resulted in CTX-M-type ESBL selection. In that sense, “broad spectrum” is more suitable to characterize MFO than “extended spectrum.”
Acknowledgments
This work has been supported by the Belgian Government within the framework of the Pôles d'Attraction Interuniversitaires (PAI no. P4/03) and by EC contracts within the framework of the Biomed (BIO 4-CT96-0126) and Biotech (BIO-CT-0520) programs.
REFERENCES
- 1.Amicosante, G., N. Franceschini, B. Segatore, A. Oratore, L. Fattorini, G. Orefici, J. Van Beeumen, and J. M. Frere. 1990. Characterization of a beta-lactamase produced in Mycobacterium fortuitum D316. Biochem. J. 271:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantu, C., III, W. Huang, and T. Palzkill. 1997. Cephalosporin substrate specificity determinants of TEM-1 beta-lactamase. J. Biol. Chem. 272:29144-29150. [DOI] [PubMed] [Google Scholar]
- 4.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., J. Delmas, J. Sirot, B. Shoichet, and R. Bonnet. 2005. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348:349-362. [DOI] [PubMed] [Google Scholar]
- 6.Dideberg, O., P. Charlier, J. P. Wery, P. Dehottay, J. Dusart, T. Erpicum, J. M. Frere, and J. M. Ghuysen. 1987. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem. J. 245:911-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dive, G., and D. Dehareng. 1999. Serine peptidase catalytic machinery: cooperative one-step mechanism. Int. J. Quant. Chem. 73:161-174. [Google Scholar]
- 8.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 9.Fattorini, L., G. Orefici, S. H. Jin, G. Scardaci, G. Amicosante, N. Franceschini, and I. Chopra. 1992. Resistance to beta-lactams in Mycobacterium fortuitum. Antimicrob. Agents Chemother. 36:1068-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonze, E., M. Vanhove, G. Dive, E. Sauvage, J. M. Frere, and P. Charlier. 2002. Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin. Biochemistry 41:1877-1885. [DOI] [PubMed] [Google Scholar]
- 11.Frere, J. M., C. Dormans, V. M. Lenzini, and C. Duyckaerts. 1982. Interaction of clavulanate with the beta-lactamases of Streptomyces albus G and Actinomadura R39. Biochem. J. 207:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galleni, M., N. Franceschini, B. Quinting, L. Fattorini, G. Orefici, A. Oratore, J. M. Frere, and G. Amicosante. 1994. Use of the chromosomal class A beta-lactamase of Mycobacterium fortuitum D316 to study potentially poor substrates and inhibitory beta-lactam compounds. Antimicrob. Agents Chemother. 38:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazouli, M., N. J. Legakis, and L. S. Tzouvelekis. 1998. Effect of substitution of Asn for Arg-276 in the cefotaxime-hydrolyzing class A beta-lactamase CTX-M-4. FEMS Microbiol. Lett. 169:289-293. [DOI] [PubMed] [Google Scholar]
- 14.Guillaume, G., M. Vanhove, J. Lamotte-Brasseur, P. Ledent, M. Jamin, B. Joris, and J. M. Frere. 1997. Site-directed mutagenesis of glutamate 166 in two beta-lactamases. Kinetic and molecular modeling studies. J. Biol. Chem. 272:5438-5444. [DOI] [PubMed] [Google Scholar]
- 15.Hackbarth, C. J., I. Unsal, and H. F. Chambers. 1997. Cloning and sequence analysis of a class A beta-lactamase from Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 41:1182-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Healey, W. J., M. R. Labgold, and J. H. Richards. 1989. Substrate specificities in class A beta-lactamases: preference for penams vs. cephems. The role of residue 237. Proteins 6:275-283. [DOI] [PubMed] [Google Scholar]
- 17.Hokenson, M. J., G. A. Cope, E. R. Lewis, K. A. Oberg, and A. L. Fink. 2000. Enzyme-induced strain/distortion in the ground-state ES complex in beta-lactamase catalysis revealed by FTIR. Biochemistry 39:6538-6545. [DOI] [PubMed] [Google Scholar]
- 18.Ibuka, A. S., Y. Ishii, M. Galleni, M. Ishiguro, K. Yamaguchi, J. M. Frere, H. Matsuzawa, and H. Sakai. 2003. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry 42:10634-10643. [DOI] [PubMed] [Google Scholar]
- 19.Imtiaz, U., E. K. Manavathu, S. Mobashery, and S. A. Lerner. 1994. Reversal of clavulanate resistance conferred by a Ser-244 mutant of TEM-1 beta-lactamase as a result of a second mutation (Arg to Ser at position 164) that enhances activity against ceftazidime. Antimicrob. Agents Chemother. 38:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob, F., B. Joris, S. Lepage, J. Dusart, and J. M. Frere. 1990. Role of the conserved amino acids of the SDN′ loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem. J. 271:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski, R. A. 2001. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 29:221-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie, A. G. W. 1991. Molecular data processing. Crystallogr. Comput. 5:50-61. [Google Scholar]
- 24.Liu, J., C. E. Barry III, G. S. Besra, and H. Nikaido. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545-29551. [DOI] [PubMed] [Google Scholar]
- 25.Maveyraud, L., I. Saves, O. Burlet-Schiltz, P. Swaren, J. M. Masson, M. Delaire, L. Mourey, J. C. Prome, and J. P. Samama. 1996. Structural basis of extended spectrum TEM beta-lactamases. Crystallographic, kinetic, and mass spectrometric investigations of enzyme mutants. J. Biol. Chem. 271:10482-10489. [DOI] [PubMed] [Google Scholar]
- 26.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57:1367-1372. [DOI] [PubMed] [Google Scholar]
- 27.Nukaga, M., K. Mayama, G. V. Crichlow, and J. R. Knox. 2002. Structure of an extended-spectrum class A beta-lactamase from Proteus vulgaris K1. J. Mol. Biol. 317:109-117. [DOI] [PubMed] [Google Scholar]
- 28.Oliva, M., O. Dideberg, and M. J. Field. 2003. Understanding the acylation mechanisms of active-site serine penicillin-recognizing proteins: a molecular dynamics simulation study. Proteins 53:88-100. [DOI] [PubMed] [Google Scholar]
- 29.Orencia, M. C., J. S. Yoon, J. E. Ness, W. P. Stemmer, and R. C. Stevens. 2001. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 8:238-242. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Llarena, F., J. F. Martin, M. Galleni, J. J. Coque, J. L. Fuente, J. M. Frere, and P. Liras. 1997. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A beta-lactamase of low enzymatic activity. J. Bacteriol. 179:6035-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrella, S., D. Clermont, I. Casin, V. Jarlier, and W. Sougakoff. 2001. Novel class A beta-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of beta-lactamases within the Citrobacter genus. Antimicrob. Agents Chemother. 45:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrosino, J. F., M. Baker, and T. Palzkill. 1999. Susceptibility of beta-lactamase to core amino acid substitutions. Protein Eng. 12:761-769. [DOI] [PubMed] [Google Scholar]
- 33.Poirel, L., F. Laurent, T. Naas, R. Labia, P. Boiron, and P. Nordmann. 2001. Molecular and biochemical analysis of AST-1, a class A beta-lactamase from Nocardia asteroides sensu stricto. Antimicrob. Agents Chemother. 45:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinting, B., J. M. Reyrat, D. Monnaie, G. Amicosante, V. Pelicic, B. Gicquel, J. M. Frere, and M. Galleni. 1997. Contribution of beta-lactamase production to the resistance of mycobacteria to beta-lactam antibiotics. FEBS Lett. 406:275-278. [DOI] [PubMed] [Google Scholar]
- 36.Raquet, X., J. Lamotte-Brasseur, E. Fonze, S. Goussard, P. Courvalin, and J. M. Frere. 1994. TEM beta-lactamase mutants hydrolysing third-generation cephalosporins. A kinetic and molecular modelling analysis. J. Mol. Biol. 244:625-639. [DOI] [PubMed] [Google Scholar]
- 37.Seoane, A., and J. M. Garcia Lobo. 1991. Cloning of chromosomal beta-lactamase genes from Yersinia enterocolitica. J. Gen. Microbiol. 137:141-146. [DOI] [PubMed] [Google Scholar]
- 38.Shimamura, T., A. Ibuka, S. Fushinobu, T. Wakagi, M. Ishiguro, Y. Ishii, and H. Matsuzawa. 2002. Acyl-intermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J. Biol. Chem. 277:46601-46608. [DOI] [PubMed] [Google Scholar]
- 39.Sougakoff, W., T. Naas, P. Nordmann, E. Collatz, and V. Jarlier. 1999. Role of ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A beta-lactamase Sme-1. Biochim. Biophys. Acta 1433:153-158. [DOI] [PubMed] [Google Scholar]
- 40.Tamaki, M., M. Nukaga, and T. Sawai. 1994. Replacement of serine 237 in class A beta-lactamase of Proteus vulgaris modifies its unique substrate specificity. Biochemistry 33:10200-10206. [DOI] [PubMed] [Google Scholar]
- 41.Thami, G. P., S. Kaur, J. Chander, and A. K. Attri. 2002. Post surgical atypical mycobacterial infection due to Mycobacterium fortuitum. J. Infect. 45:210-211. [DOI] [PubMed] [Google Scholar]
- 42.Timm, J., M. G. Perilli, C. Duez, J. Trias, G. Orefici, L. Fattorini, G. Amicosante, A. Oratore, B. Joris, J. M. Frere, et al. 1994. Transcription and expression analysis, using lacZ and phoA gene fusions, of Mycobacterium fortuitum beta-lactamase genes cloned from a natural isolate and a high-level beta-lactamase producer. Mol. Microbiol. 12:491-504. [DOI] [PubMed] [Google Scholar]
- 43.Voladri, R. K., D. L. Lakey, S. H. Hennigan, B. E. Menzies, K. M. Edwards, and D. S. Kernodle. 1998. Recombinant expression and characterization of the major beta-lactamase of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. A. Tschen, and M. S. Stone. 1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5:657-679. [DOI] [PubMed] [Google Scholar]
- 45.Zafaralla, G., E. K. Manavathu, S. A. Lerner, and S. Mobashery. 1992. Elucidation of the role of arginine-244 in the turnover processes of class A beta-lactamases. Biochemistry 31:3847-3852. [DOI] [PubMed] [Google Scholar]